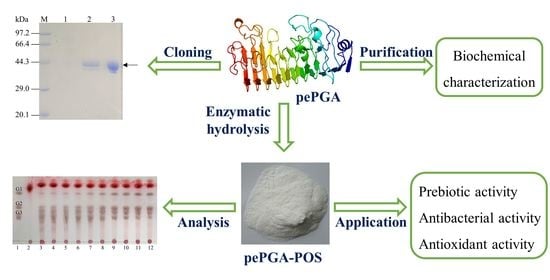
A Novel Endo-Polygalacturonase from Penicillium rolfsii with Prebiotics Production Potential: Cloning, Characterization and Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Vectors, and Chemicals
2.2. Gene Cloning
2.3. Expression and Purification of the Recombinant Enzyme
2.4. SDS-PAGE Analysis and Recombinant Protein Identification
2.5. Enzyme Activity Assay
2.6. Biochemical Characterization of the Purified Recombinant Enzyme
2.7. Analysis of Hydrolysis Products
2.8. In Vitro Antimicrobial, Antioxidant and Prebiotic Activities of pePGA-POS
2.8.1. Prebiotic Activity
2.8.2. Antimicrobial Activity
2.8.3. DPPH Radical Scavenging Activity
3. Results and Discussion
3.1. Expression and Purification of pePGA
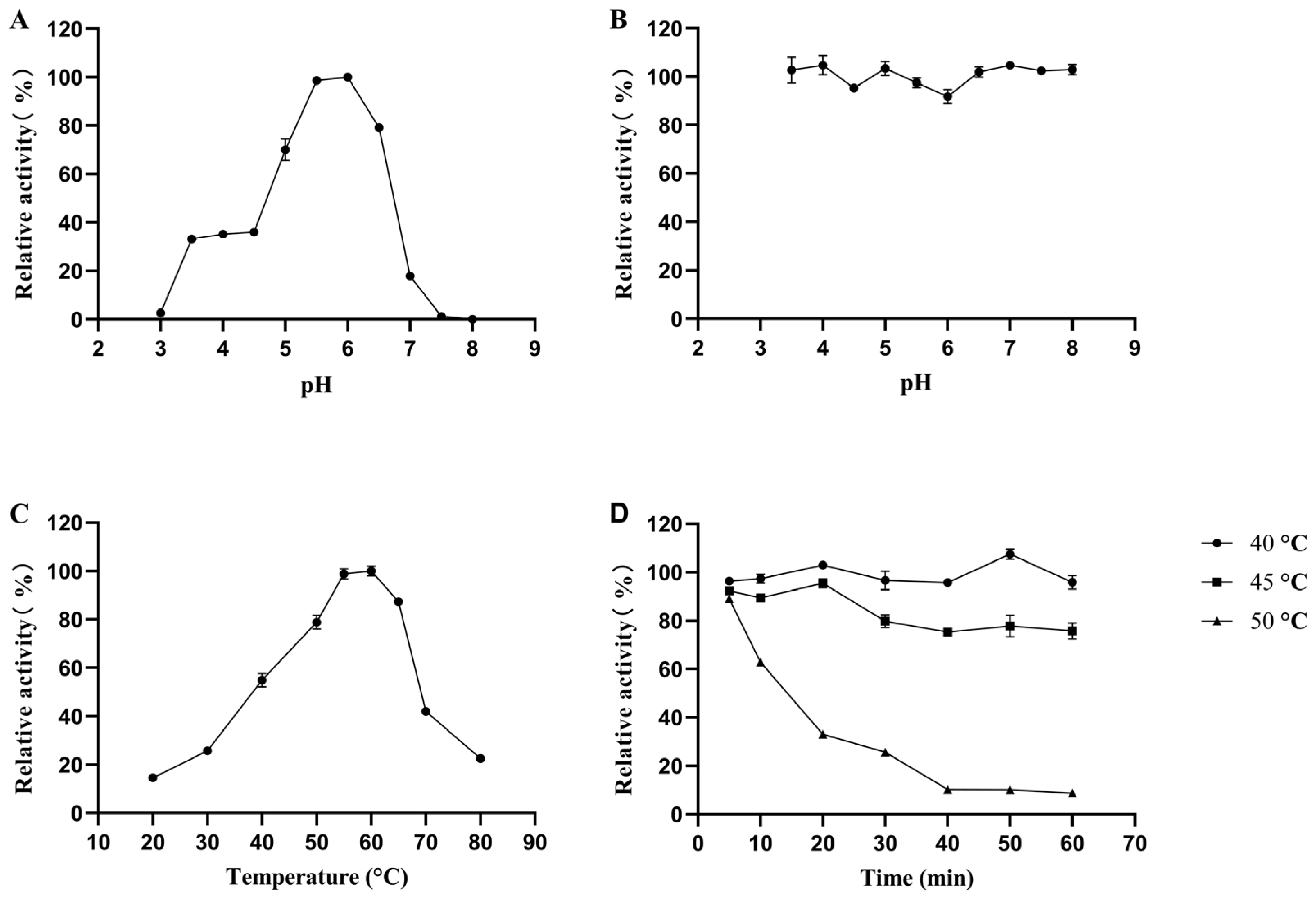
3.2. Biochemical Characterization of the Recombinant pePGA
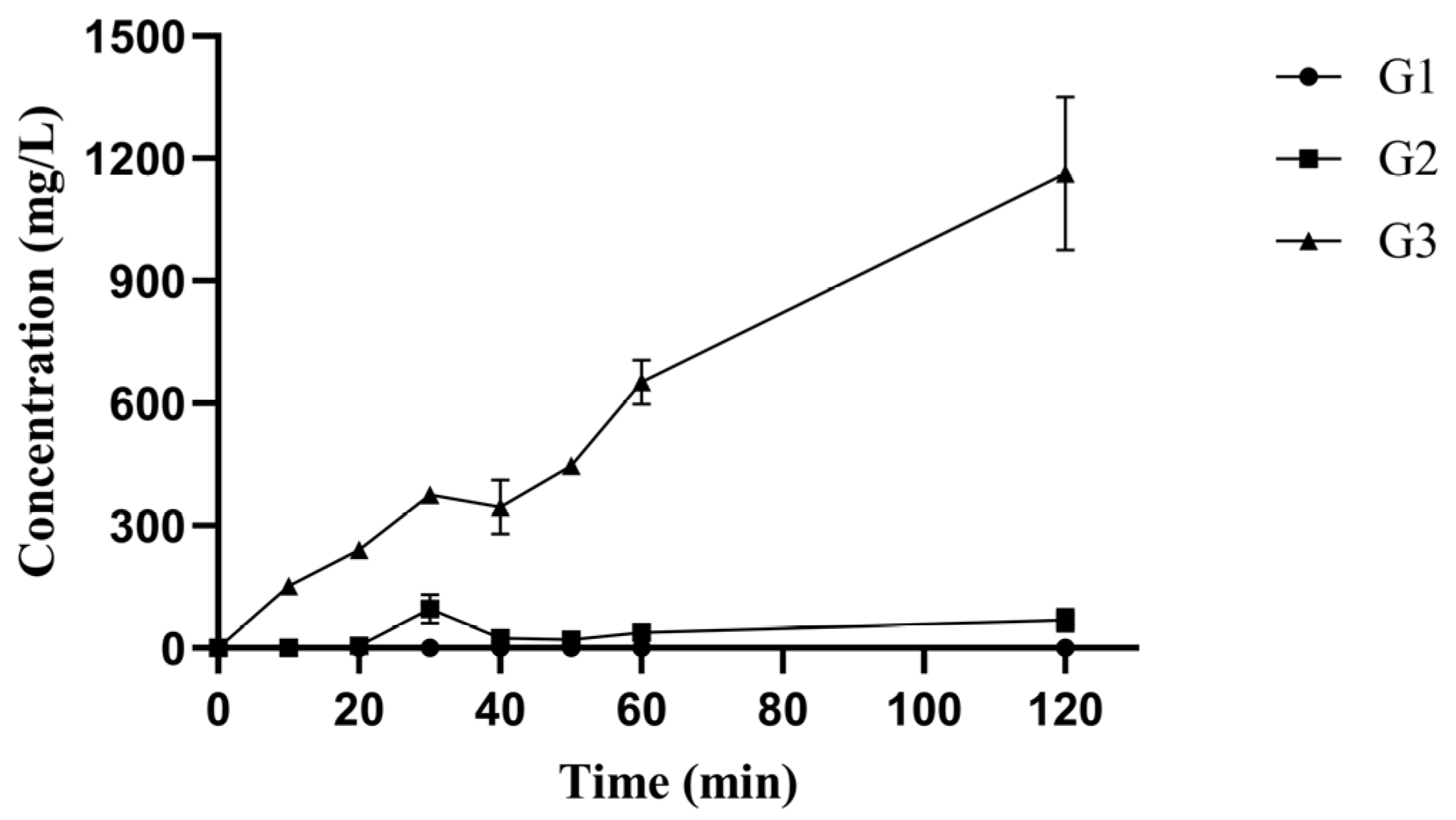
3.3. Analysis of Hydrolysis Products
3.4. Prebiotic Activity of Hydrolysis Products
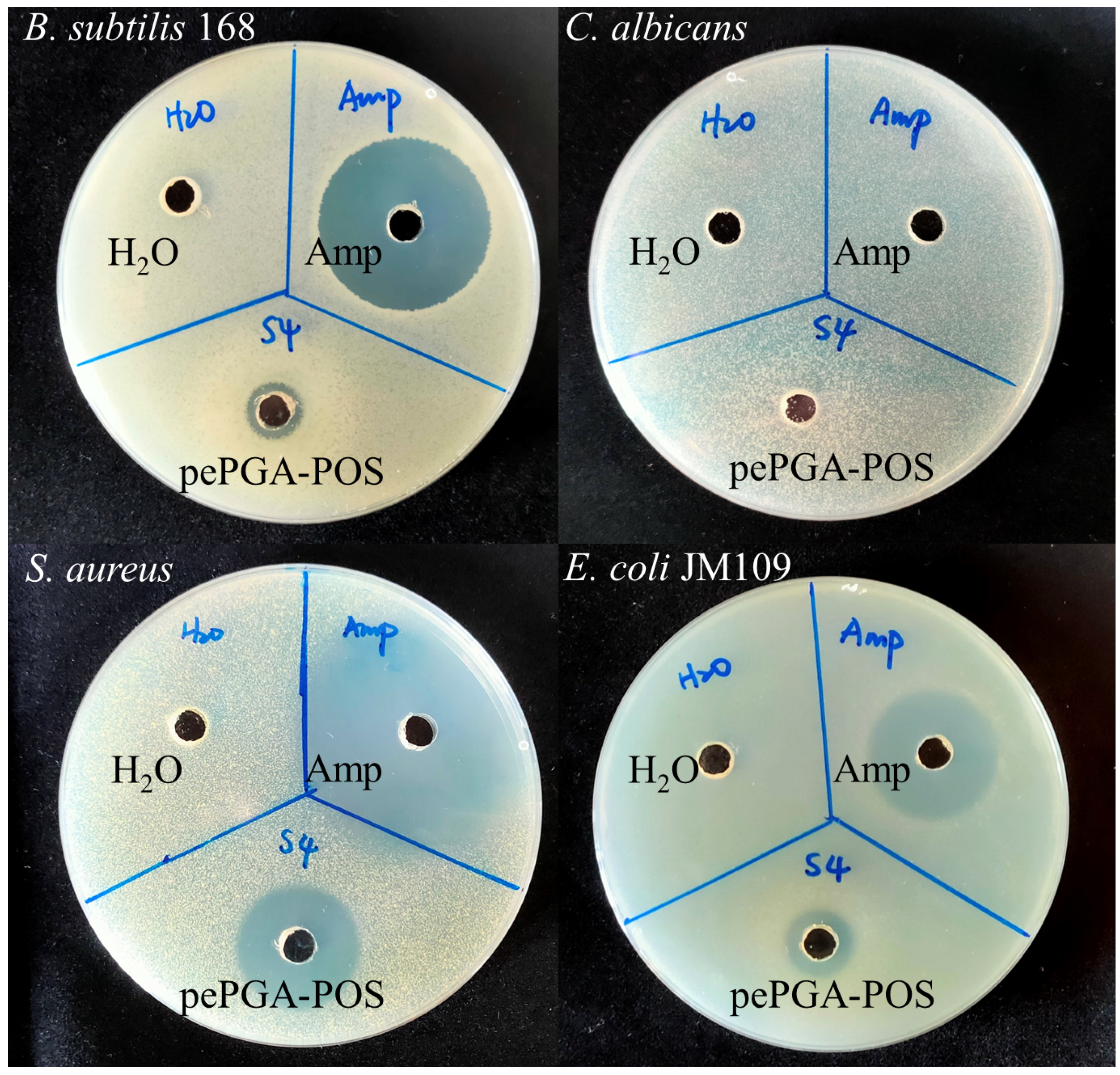
3.5. Antimicrobial Activity of Hydrolysis Products
3.6. DPPH Radical Scavenging Activity of Hydrolysis Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gullón, B.; Gómez, B.; Martínez-Sabajanes, M.; Yáñez, R.; Parajó, J.C.; Alonso, J.L. Pectic oligosaccharides: Manufacture and functional properties. Trends. Food Sci. Technol. 2013, 30, 153–161. [Google Scholar] [CrossRef]
- Kapoor, S.; Dharmesh, S.M. Pectic Oligosaccharide from tomato exhibiting anticancer potential on a gastric cancer cell line: Structure-function relationship. Carbohydr. Polym. 2017, 160, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-J.; Xia, J.-L.; Nie, Z.-Y.; Shan, Y. Pectic oligosaccharides hydrolyzed from orange peel by fungal multi-enzyme complexes and their prebiotic and antibacterial potentials. LWT-Food Sci. Technol. 2016, 69, 203–210. [Google Scholar] [CrossRef]
- Xue, L.; Long, J.; Lu, C.; Li, X.; Xu, X.; Jin, Z. Immobilization of polygalacturonase for the preparation of pectic oligosaccharides from mango peel wastes and assessment of their antibacterial activities. Food Biosci. 2021, 39, 100837. [Google Scholar] [CrossRef]
- Yeung, Y.K.; Kang, Y.-R.; So, B.R.; Jung, S.K.; Chang, Y.H. Structural, antioxidant, prebiotic and anti-inflammatory properties of pectic oligosaccharides hydrolyzed from okra pectin by Fenton reaction. Food Hydrocoll. 2021, 118, 106779. [Google Scholar] [CrossRef]
- Mandalari, G.; Nueno Palop, C.; Tuohy, K.; Gibson, G.R.; Bennett, R.N.; Waldron, K.W.; Bisignano, G.; Narbad, A.; Faulds, C.B. In vitro evaluation of the prebiotic activity of a pectic oligosaccharide-rich extract enzymatically derived from bergamot peel. Appl. Microbiol. Biotechnol. 2007, 73, 1173–1179. [Google Scholar] [CrossRef]
- Gómez, B.; Gullón, B.; Yáñez, R.; Schols, H.; Alonso, J.L. Prebiotic potential of pectins and pectic oligosaccharides derived from lemon peel wastes and sugar beet pulp: A comparative evaluation. J. Funct. Foods 2016, 20, 108–121. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Harholt, J.; Suttangkakul, A.; Vibe Scheller, H. Biosynthesis of pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef]
- Ropartz, D.; Ralet, M.-C. Pectin Structure. In Pectin: Technological and Physiological Properties; Kontogiorgos, V., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 17–36. [Google Scholar] [CrossRef]
- Babbar, N.; Dejonghe, W.; Gatti, M.; Sforza, S.; Elst, K. Pectic oligosaccharides from agricultural by-products: Production, characterization and health benefits. Crit. Rev. Biotechnol. 2016, 36, 594–606. [Google Scholar] [CrossRef]
- Chen, J.; Liang, R.H.; Liu, W.; Li, T.; Liu, C.M.; Wu, S.S.; Wang, Z.J. Pectic-oligosaccharides prepared by dynamic high-pressure microfluidization and their in vitro fermentation properties. Carbohydr. Polym. 2013, 91, 175–182. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, H.; Wang, L.; Liu, F.; Pan, S. Preparation and prebiotic potential of pectin oligosaccharides obtained from citrus peel pectin. Food Chem. 2018, 244, 232–237. [Google Scholar] [CrossRef]
- Wongkaew, M.; Tinpovong, B.; Sringarm, K.; Leksawasdi, N.; Jantanasakulwong, K.; Rachtanapun, P.; Hanmoungjai, P.; Sommano, S.R. Crude Pectic Oligosaccharide Recovery from Thai Chok Anan Mango Peel Using Pectinolytic Enzyme Hydrolysis. Foods 2021, 10, 627. [Google Scholar] [CrossRef]
- Combo, A.M.M.; Aguedo, M.; Goffin, D.; Wathelet, B.; Paquot, M. Enzymatic production of pectic oligosaccharides from polygalacturonic acid with commercial pectinase preparations. Food Bioprod. Process. 2012, 90, 588–596. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, J.; Mahajan, R. Microbial pectinases: An ecofriendly tool of nature for industries. 3 Biotech. 2016, 6, 47. [Google Scholar] [CrossRef]
- Sharma, N.; Rathore, M.; Sharma, M. Microbial pectinase: Sources, characterization and applications. Rev. Environ. Sci. Biotechnol. 2012, 12, 45–60. [Google Scholar] [CrossRef]
- Jayani, R.S.; Saxena, S.; Gupta, R. Microbial pectinolytic enzymes: A review. Process Biochem. 2005, 40, 2931–2944. [Google Scholar] [CrossRef]
- Parenicova, L.; Kester, H.C.M.; Benen, J.A.E.; Visser, J. Characterization of a novel endopolygalacturonase from Aspergillus niger with unique kinetic properties. FEBS Lett. 2000, 467, 333–336. [Google Scholar] [CrossRef]
- Pages, S.; Kester, H.C.M.; Visser, J.; Benen, J.A.E. Changing a Single Amino Acid Residue Switches Processive and Non-processive Behavior of Aspergillus niger Endopolygalacturonase I and II. J. Biol. Chem. 2001, 276, 33652–33656. [Google Scholar] [CrossRef]
- Tounsi, H.; Sassi, A.H.; Ben Romdhane, Z.; Lajnef, M.; Dupuy, J.W.; Lapaillerie, D.; Lomenech, A.M.; Bonneu, M.; Gargouri, A.; Hadj-Taieb, N. Catalytic properties of a highly thermoactive polygalacturonase from the mesophilic fungus Penicillium occitanis and use in juice clarification. J. Mol. Catal. B Enzym. 2016, 127, 56–66. [Google Scholar] [CrossRef]
- Cheng, Z.; Chen, D.; Wang, Q.; Xian, L.; Lu, B.; Wei, Y.; Tang, H.; Lu, Z.; Zhu, Q.; Chen, Y.; et al. Identification of an acidic endo-polygalacturonase from Penicillium oxalicum CZ1028 and its broad use in major tropical and subtropical fruit juices production. J. Biosci. Bioeng. 2017, 123, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Xian, L.; Chen, D.; Lu, J.; Wei, Y.; Du, L.; Wang, Q.; Chen, Y.; Lu, B.; Bi, D.; et al. Development of an Innovative Process for High-Temperature Fruit Juice Extraction Using a Novel Thermophilic Endo-Polygalacturonase From Penicillium oxalicum. Front. Microbiol. 2020, 11, 1200. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Yu, M.; Xia, Y.; Xie, W.; Li, Y.; Yu, X.; Zheng, J.; Zhang, Y. Enzymatic extraction of pectic oligosaccharides from finger citron (Citrus medica L. var. sarcodactylis Swingle) pomace with antioxidant potential. Food Funct. 2021, 12, 9855–9865. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Li, K.; Ma, R.; Shi, P.; Huang, H.; Yang, P.; Meng, K.; Yao, B. Biochemical characterization of three distinct polygalacturonases from Neosartorya fischeri P1. Food Chem. 2015, 188, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Meng, K.; Bai, Y.; Shi, P.; Luo, H.; Wang, Y.; Yang, P.; Zhang, Y.; Zhang, W.; Yao, B. High-yield production of a low-temperature-active polygalacturonase for papaya juice clarification. Food Chem. 2013, 141, 2974–2981. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, P.; Zhang, Y.; Liu, Z.; Zhang, X.; Li, Z.; Li, J.J.; Du, Y. Overexpression and Biochemical Characterization of an Endo-alpha-1,4-polygalacturonase from Aspergillus nidulans in Pichia pastoris. Int. J. Mol Sci. 2020, 21, 2100. [Google Scholar] [CrossRef]
- Yuan, P.; Meng, K.; Huang, H.; Shi, P.; Luo, H.; Yang, P.; Yao, B. A novel acidic and low-temperature-active endo-polygalacturonase from Penicillium sp. CGMCC 1669 with potential for application in apple juice clarification. Food Chem. 2011, 129, 1369–1375. [Google Scholar] [CrossRef]
- Amin, F.; Mohsin, A.; Bhatti, H.N.; Bilal, M. Production, thermodynamic characterization, and fruit juice quality improvement characteristics of an Exo-polygalacturonase from Penicillium janczewskii. Biochim. Biophys. Acta Proteins. Proteom. 2020, 1868, 140379. [Google Scholar] [CrossRef]
- Li, K.; Meng, K.; Pan, X.; Ma, R.; Yang, P.; Huang, H.; Yao, B.; Su, X. Two thermophilic fungal pectinases from Neosartorya fischeri P1: Gene cloning, expression, and biochemical characterization. J. Mol. Catal. B Enzym. 2015, 118, 70–78. [Google Scholar] [CrossRef]
- Rafique, N.; Tabassum, R.; Awan, M.S.; Orts, W.; Wong, D.W.S. Cloning and Expression of Pectobacterium carotovorum Endo-polygalacturonase Gene in Pichia pastoris for Production of Oligogalacturonates. Bioresources 2016, 11, 5204–5214. [Google Scholar] [CrossRef]
- Nobre, C.; Sousa, S.C.; Silva, S.P.; Pinheiro, A.C.; Coelho, E.; Vicente, A.A.; Gomes, A.M.P.; Coimbra, M.A.; Teixeira, J.A.; Rodrigues, L.R. In vitro digestibility and fermentability of fructo-oligosaccharides produced by Aspergillus ibericus. J. Funct. Foods. 2018, 46, 278–287. [Google Scholar] [CrossRef]
- Gawrońska, M.; Kowalik, M.; Makowski, M. Recent advances in medicinal chemistry of ampicillin: Derivatives, metal complexes, and sensing approaches. Trends Analyt. Chem. 2022, 155, 116691. [Google Scholar] [CrossRef]
- Wang, S.; Ding, S.; Meng, K.; Liu, X.; Wang, Y.; Wang, X.; Qin, X.; Luo, H.; Yao, B.; Huang, H.; et al. Preparation of methyl-esterified pectin oligosaccharides with antibacterial activity using fungus-derived bifunctional pectinase. J. Clean Prod. 2022, 333, 130110. [Google Scholar] [CrossRef]
- Hu, X.; Jiang, X.; Gong, J.; Hwang, H.; Liu, Y.; Guan, H. Antibacterial activity of lyase-depolymerized products of alginate. J. Appl. Phycol. 2005, 17, 57–60. [Google Scholar] [CrossRef]
- Guo, Z.; Wei, Y.; Zhang, Y.; Xu, Y.; Zheng, L.; Zhu, B.; Yao, Z. Carrageenan oligosaccharides: A comprehensive review of preparation, isolation, purification, structure, biological activities and applications. Algal Res. 2022, 61, 102593. [Google Scholar] [CrossRef]
- Wu, M.-C.; Li, H.-c.; Wu, P.-H.; Huang, P.-H.; Wang, Y.-T. Assessment of Oligogalacturonide from Citrus Pectin as a Potential Antibacterial Agent Against Foodborne Pathogens. J. Food Sci. 2014, 79, M1541–M1544. [Google Scholar] [CrossRef]
- Chen, X.; Liang, L.; Han, C. Borate suppresses the scavenging activity of gallic acid and plant polyphenol extracts on DPPH radical: A potential interference to DPPH assay. LWT-Food Sci. Technol. 2020, 131, 109769. [Google Scholar] [CrossRef]
- Yang, W.; Fortunati, E.; Dominici, F.; Giovanale, G.; Mazzaglia, A.; Balestra, G.M.; Kenny, J.M.; Puglia, D. Effect of cellulose and lignin on disintegration, antimicrobial and antioxidant properties of PLA active films. Int. J. Biol. Macromol. 2016, 89, 360–368. [Google Scholar] [CrossRef]
- Zhang, T.; Tian, Y.; Jiang, B.; Miao, M.; Mu, W. Purification, preliminary structural characterization and in vitro antioxidant activity of polysaccharides from Acanthus ilicifolius. LWT-Food Sci. Technol. 2014, 56, 9–14. [Google Scholar] [CrossRef]
- Zhang, T.; Shuai, M.; Ma, P.; Huang, J.; Sun, C.; Yao, X.; Chen, Z.; Min, X.; Yan, S. Purification, chemical analysis and antioxidative activity of polysaccharides from pH-modified citrus pectin after dialyzation. LWT-Food Sci. Technol. 2020, 128, 109513. [Google Scholar] [CrossRef]
- Ezzati, S.; Ayaseh, A.; Ghanbarzadeh, B.; Heshmati, M.K. Pectin from sunflower by-product: Optimization of ultrasound-assisted extraction, characterization, and functional analysis. Int. J. Biol. Macromol. 2020, 165 Pt A, 776–786. [Google Scholar] [CrossRef]
- Sabater, C.; Blanco-Doval, A.; Montilla, A.; Corzo, N. Optimisation of an enzymatic method to obtain modified artichoke pectin and pectic oligosaccharides using artificial neural network tools. In silico and in vitro assessment of the antioxidant activity. Food Hydrocoll. 2021, 110, 106161. [Google Scholar] [CrossRef]




| Antimicrobial Activities/MIC (mg/mL) | pePGA-POS |
|---|---|
| E. coli JM109 | 50 |
| B. subtilis 168 | 50 |
| S. aureus | 12.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, M.-J.; Wu, D.; Xu, Y.; Tao, X.-M.; Li, N.; Yu, X.-W. A Novel Endo-Polygalacturonase from Penicillium rolfsii with Prebiotics Production Potential: Cloning, Characterization and Application. Foods 2022, 11, 3469. https://doi.org/10.3390/foods11213469
Hao M-J, Wu D, Xu Y, Tao X-M, Li N, Yu X-W. A Novel Endo-Polygalacturonase from Penicillium rolfsii with Prebiotics Production Potential: Cloning, Characterization and Application. Foods. 2022; 11(21):3469. https://doi.org/10.3390/foods11213469
Chicago/Turabian StyleHao, Meng-Jie, Dan Wu, Yan Xu, Xiu-Mei Tao, Ning Li, and Xiao-Wei Yu. 2022. "A Novel Endo-Polygalacturonase from Penicillium rolfsii with Prebiotics Production Potential: Cloning, Characterization and Application" Foods 11, no. 21: 3469. https://doi.org/10.3390/foods11213469
APA StyleHao, M.-J., Wu, D., Xu, Y., Tao, X.-M., Li, N., & Yu, X.-W. (2022). A Novel Endo-Polygalacturonase from Penicillium rolfsii with Prebiotics Production Potential: Cloning, Characterization and Application. Foods, 11(21), 3469. https://doi.org/10.3390/foods11213469