The Ability of Shiga Toxin-Producing Escherichia coli to Grow in Raw Cow’s Milk Stored at Low Temperatures
Abstract
1. Introduction
2. Materials and Methods
2.1. Culturing Conditions
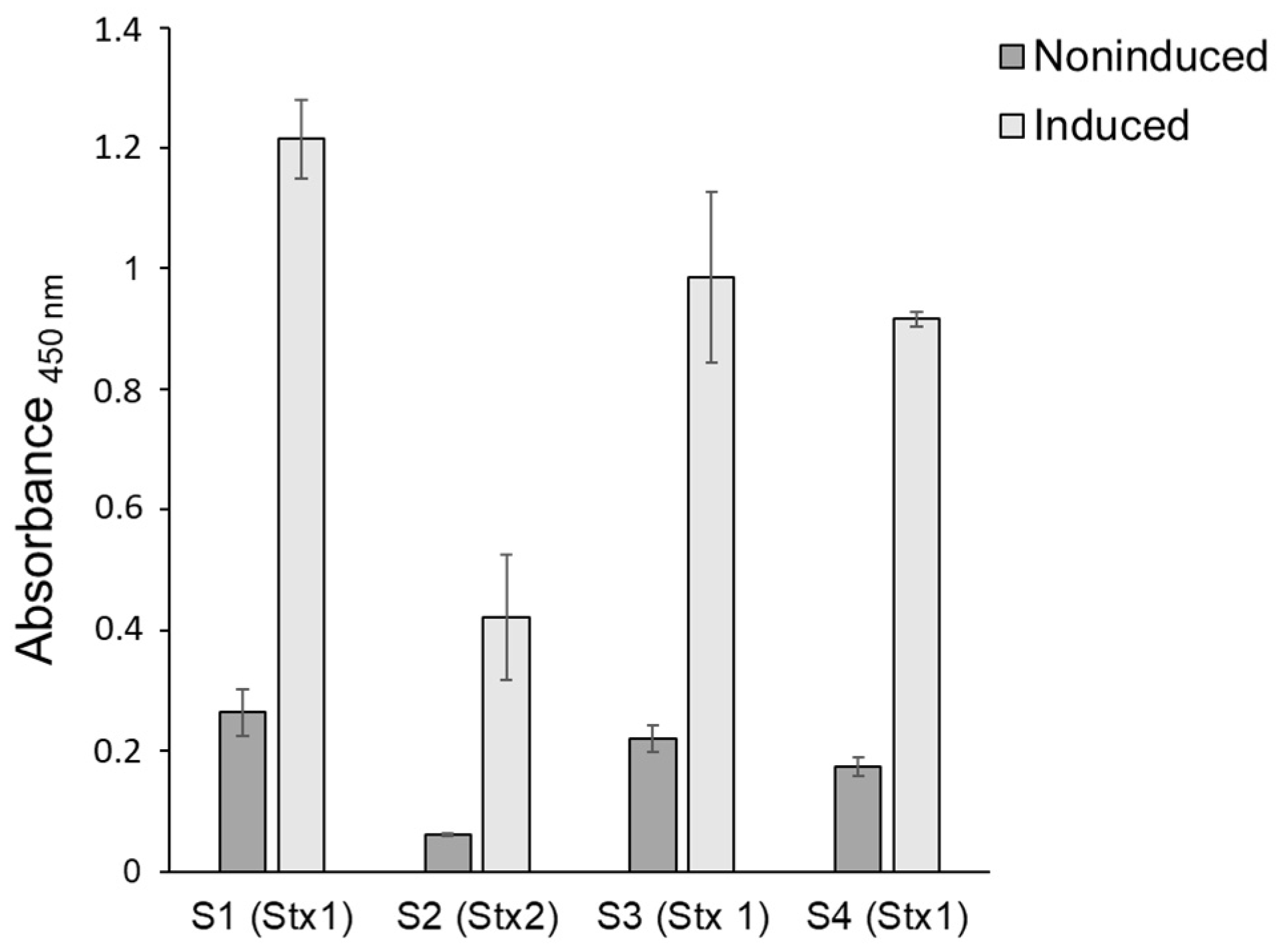
2.2. Stx Production
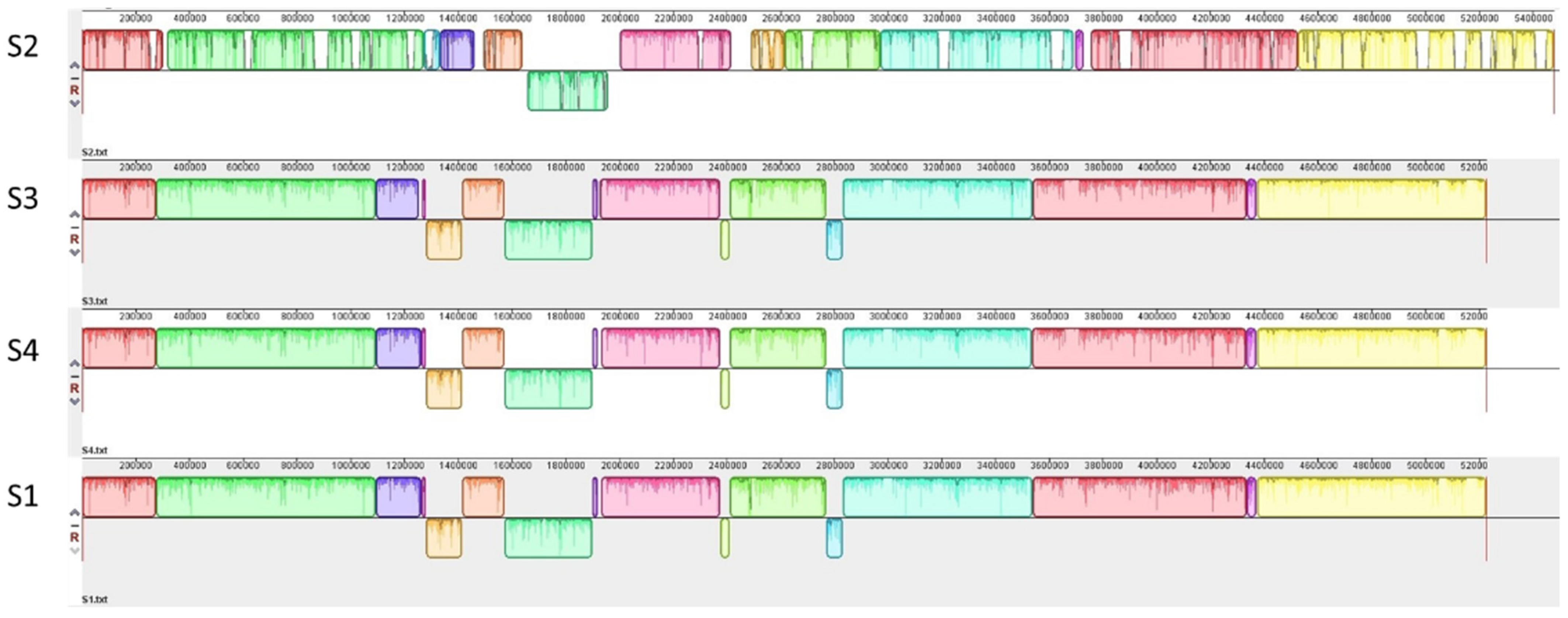
2.3. Genome Sequence Analyses
2.4. Statistics
3. Results
3.1. Genetic Characterization of STEC Isolates from Raw Milk
3.2. Stx Production
3.3. Growth Characteristics of STEC Isolates in Raw Milk at Different Storage Temperatures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karmali, M.A. Factors in the emergence of serious human infections associated with highly pathogenic strains of shiga toxin-producing Escherichia coli. Int. J. Med. Microbiol. 2018, 308, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Joseph, A.; Cointe, A.; Mariani-Kurkdjian, P.; Rafat, C.; Hertig, A. Shiga toxin-associated hemolytic uremic syndrome: A narrative review. Toxins 2020, 12, 67. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority & European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef]
- Gould, L.H.; Mody, R.K.; Ong, K.L.; Clogher, P.; Cronquist, A.B.; Garman, K.N.; Latrop, S.; Medus, C.; Spina, N.L.; Webb, T.H. Increased recognition of non-O157 Shiga toxin–producing Escherichia coli infections in the United States during 2000–2010: Epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 2013, 10, 453–460. [Google Scholar] [CrossRef]
- Hedican, E.B.; Medus, C.; Besser, J.M.; Juni, B.A.; Koziol, B.; Taylor, C.; Smith, K.E. Characteristics of O157 versus non-O157 Shiga toxin-producing Escherichia coli infections in Minnesota, 2000–2006. Clin. Infect. Dis. 2009, 49, 358–364. [Google Scholar] [CrossRef]
- Hadler, J.L.; Clogher, P.; Hurd, S.; Phan, Q.; Mandour, M.; Bemis, K.; Marcus, R. Ten-year trends and risk factors for non-O157 Shiga toxin–producing Escherichia coli found through Shiga toxin testing, Connecticut, 2000–2009. Clin. Infect. Dis. 2011, 53, 269–276. [Google Scholar] [CrossRef]
- Tuttle, J.; Gomez, T.; Doyle, M.; Wells, J.; Zhao, T.; Tauxe, R.; Griffin, P.M. Lessons from a large outbreak of Escherichia coli O157:H7 infections: Insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 1999, 122, 185–192. [Google Scholar] [CrossRef]
- Sperandio, V.; Nguyen, Y. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 90. [Google Scholar] [CrossRef]
- Llarena, A.-K.; Aspholm, M.; O’Sullivan, K.; Wêgrzyn, G.; Lindbäck, T. Replication region analysis reveals non-lambdoid shiga toxin converting bacteriophages. Front. Microbiol. 2021, 12, 640945. [Google Scholar] [CrossRef]
- Fagerlund, A.; Aspholm, M.; Węgrzyn, G.; Lindbäck, T. High diversity in the regulatory region of Shiga toxin encoding bacteriophages. BMC Genom. 2022, 23, 230. [Google Scholar] [CrossRef] [PubMed]
- Bednarz, M.; Halliday, J.A.; Herman, C.; Golding, I. Revisiting bistability in the lysis/lysogeny circuit of bacteriophage lambda. PLoS ONE 2014, 9, e100876. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R.; Hendrix, R.W. Bacteriophage lambda: Early pioneer and still relevant. Virology 2015, 479, 310–330. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Skinner, S.O.; Zong, C.; Sippy, J.; Feiss, M.; Golding, I. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 2010, 141, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kohler, B.; Oswald, E.; Beutin, L.; Karch, H.; Morabito, S.; Caprioli, A.; Suerbaum, S.; Schmidt, H. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 2002, 40, 4486–4492. [Google Scholar] [CrossRef]
- Yang, X.; Sun, H.; Fan, R.; Fu, S.; Zhang, J.; Matussek, A.; Xiong, Y.; Bai, X. Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci. Rep. 2020, 10, 3275. [Google Scholar] [CrossRef]
- De Rauw, K.; Buyl, R.; Jacquinet, S.; Piérard, D. Risk determinants for the development of typical haemolytic uremic syndrome in Belgium and proposition of a new virulence typing algorithm for Shiga toxin-producing Escherichia coli. Epidemiol. Infect. 2019, 147, E6. [Google Scholar] [CrossRef]
- European Food Safety Authority & European Centre for Disease Prevention Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. EFSA J. 2011, 9, 2090. [Google Scholar] [CrossRef]
- Bosilevac, J.M.; Koohmaraie, M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 2011, 77, 2103–2112. [Google Scholar] [CrossRef]
- European Food Safety Authority. Scientific Opinion of the Panel on Biological Hazards (BIOHAZ)-Monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic VTEC types. EFSA J. 2007, 5, 579. [Google Scholar] [CrossRef]
- Gillespie, I.; Adak, G.; O’brien, S.; Bolton, F. Milkborne general outbreaks of infectious intestinal disease, England and Wales, 1992–2000. Epidemiol. Infect. 2003, 130, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Ballem, A.; Gonçalves, S.; Garcia-Meniño, I.; Flament-Simon, S.C.; Blanco, J.E.; Fernandes, C.; Saavedra, M.J.; Pinto, C.; Oliveira, H.; Blanco, J.; et al. Prevalence and serotypes of Shiga toxin-producing Escherichia coli (STEC) in dairy cattle from Northern Portugal. PLoS ONE 2021, 15, e0244713. [Google Scholar] [CrossRef] [PubMed]
- Venegas-Vargas, C.; Henderson, S.; Khare, A.; Mosci, R.E.; Lehnert, J.D.; Singh, P.; Ouellette, L.M.; Norby, B.; Funk, J.A.; Rust, S. Factors associated with Shiga toxin-producing Escherichia coli shedding by dairy and beef cattle. Appl. Environ. Microbiol. 2016, 82, 5049–5056. [Google Scholar] [CrossRef] [PubMed]
- Jaakkonen, A.; Castro, H.; Hallanvuo, S.; Ranta, J.; Rossi, M.; Isidro, J.; Lindström, M.; Hakkinen, M. Longitudinal Study of Shiga Toxin-Producing Escherichia coli and Campylobacter jejuni on Finnish Dairy Farms and in Raw Milk. Appl. Environ. Microbiol. 2019, 85, e02910-18. [Google Scholar] [CrossRef]
- Idland, L.; Granquist, E.G.; Aspholm, M.; Lindbäck, T. The prevalence of Campylobacter spp., Listeria monocytogenes and Shiga toxin-producing Escherichia coli in Norwegian dairy cattle farms: A comparison between free stall and tie stall housing systems. J. Appl. Microbiol. 2022, 132, 3959–3972. [Google Scholar] [CrossRef]
- Geue, L.; Segura-Alvarez, M.; Conraths, F.; Kuczius, T.; Bockemühl, J.; Karch, H.; Gallien, P. A long-term study on the prevalence of Shiga toxin-producing Escherichia coli (STEC) on four German cattle farms. Epidemiol. Infect. 2002, 129, 173–185. [Google Scholar] [CrossRef]
- D’aoust, J.-Y.; Park, C.; Szabo, R.; Todd, E.; Emmons, D.; McKellar, R. Thermal inactivation of Campylobacter species, Yersinia enterocolitica, and hemorrhagic Escherichia coli 0157: H7 in fluid milk. J. Dairy Sci. 1988, 71, 3230–3236. [Google Scholar] [CrossRef]
- Leclair, R.M.; McLean, S.K.; Dunn, L.A.; Meyer, D.; Palombo, E.A. Investigating the effects of time and temperature on the growth of Escherichia coli O157: H7 and Listeria monocytogenes in raw cow’s milk based on simulated consumer food handling practices. Int. J. Environ. Res. Public Health 2019, 16, 2691. [Google Scholar] [CrossRef]
- Kauppi, K.; Tatini, S.; Harrell, F.; Feng, P. Influence of substrate and low temperature on growth and survival of verotoxigenic Escherichia coli. Food Microbiol. 1996, 13, 397–405. [Google Scholar] [CrossRef]
- International Commission on Microbiological Specifications for Foods. Microorganisms in Foods 5: Characteristics of Microbial Pathogens, 1st ed.; Kluwer Academic/Plenum Publishers: London, UK, 1996. [Google Scholar]
- Wang, G.; Zhao, T.; Doyle, M.P. Survival and growth of Escherichia coli O157: H7 in unpasteurized and pasteurized milk. J. Food Prot. 1997, 60, 610–613. [Google Scholar] [CrossRef]
- Dumitrașcu, L.; Nicolau, A.I.; Neagu, C.; Didier, P.; Maître, I.; Nguyen-The, C.; Skuland, S.E.; Møretrø, T.; Langsrud, S.; Truninger, M. Time-temperature profiles and Listeria monocytogenes presence in refrigerators from households with vulnerable consumers. Food Control 2020, 111, 107078. [Google Scholar] [CrossRef]
- Roccato, A.; Uyttendaele, M.; Membré, J.-M. Analysis of domestic refrigerator temperatures and home storage time distributions for shelf-life studies and food safety risk assessment. Food Res. Int. 2017, 96, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Clausen, P.T.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Achtman, M.; Brown, D.; Chattaway, M.; Dallman, T.; Delahay, R.; Kornschober, C. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Creuzburg, K.; Recktenwald, J.R.; Kuhle, V.; Herold, S.; Hensel, M.; Schmidt, H. The Shiga toxin 1-converting bacteriophage BP-4795 encodes an NleA-like type III effector protein. J. Bacteriol. Parasitol. 2005, 187, 8494–8498. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Savarino, S.J.; McVeigh, A.; Watson, J.; Cravioto, A.; Molina, J.; Echeverria, P.; Bhan, M.K.; Levine, M.M.; Fasano, A. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 1996, 173, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, S.C.; Son, I.; Maounounen-Laasri, A.; Lin, A.; Fischer, M.; Kase, J.A. Prevalence of hemolysin genes and comparison of ehxA subtype patterns in Shiga toxin-producing Escherichia coli (STEC) and non-STEC strains from clinical, food, and animal sources. Appl. Environ. Microbiol. 2013, 79, 6301–6311. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, S.; Dubreuil, J.D. Presence of Escherichia coli carrying the EAST1 toxin gene in farm animals. Vet. Res. 2006, 37, 3–13. [Google Scholar] [CrossRef][Green Version]
- Hua, Y.; Zhang, J.; Jernberg, C.; Chromek, M.; Hansson, S.; Frykman, A.; Xiong, Y.; Wan, C.; Matussek, A.; Bai, X. Molecular Characterization of the Enterohemolysin Gene (ehxA) in Clinical Shiga Toxin-Producing Escherichia coli Isolates. Toxins 2021, 13, 71. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.; Dennis, J.; Wishart, D. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Melton-Celsa, A.R. Shiga toxin (Stx) classification, structure, and function. Microbiol. Spectr. 2014, 2, 6. [Google Scholar] [CrossRef]
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337. [Google Scholar] [CrossRef]
- L’Abée-Lund, T.M.; Jørgensen, H.J.; O’Sullivan, K.; Bohlin, J.; Ligård, G.; Granum, P.E.; Lindbäck, T. The highly virulent 2006 Norwegian EHEC O103: H25 outbreak strain is related to the 2011 German O104: H4 outbreak strain. PLoS ONE 2012, 7, e31413. [Google Scholar] [CrossRef]
- Bolton, D.; Byrne, C.; Sheridan, J.; McDowell, D.; Blair, I. The survival characteristics of a non-toxigenic strain of Escherichia coli O157: H7. J. Appl. Microbiol. 1999, 86, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Doyle, M.P. Survival of enterohemorrhagic Escherichia coli O157: H7 in water. J. Food Prot. 1998, 61, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.; Lucchesi, P.M. Shiga toxins and stx phages: Highly diverse entities. Microbiology 2015, 161, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Brandal, L.T.; Wester, A.L.; Lange, H.; Løbersli, I.; Lindstedt, B.-A.; Vold, L.; Kapperud, G. Shiga toxin-producing Escherichia coli infections in Norway, 1992-2012: Characterization of isolates and identification of risk factors for haemolytic uremic syndrome. BMC Infect. Dis. 2015, 15, 324. [Google Scholar] [CrossRef]
- Muniesa, M.; Blanco, J.E.; De Simón, M.; Serra-Moreno, R.; Blanch, A.R.; Jofre, J. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 2004, 150, 2959–2971. [Google Scholar] [CrossRef]
- Beutin, L.; Steinrück, H.; Krause, G.; Steege, K.; Haby, S.; Hultsch, G.; Appel, B. Comparative evaluation of the Ridascreen® Verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J. Appl. Microbiol. 2007, 102, 630–639. [Google Scholar] [CrossRef]
- Itoh, Y.; Nagano, I.; Kunishima, M.; Ezaki, T. Laboratory investigation of enteroaggregative Escherichia coli O untypeable: H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 1997, 35, 2546–2550. [Google Scholar] [CrossRef]
- Sidhu, J.P.; Ahmed, W.; Hodgers, L.; Toze, S. Occurrence of virulence genes associated with diarrheagenic pathotypes in Escherichia coli isolates from surface water. Appl. Environ. Microbiol. 2013, 79, 328–335. [Google Scholar] [CrossRef]
- Heuvelink, A.; Bleumink, B.; Van Den Biggelaar, F.; Te Giffel, M.; Beumer, R.; De Boer, E. Occurrence and survival of verocytotoxin-producing Escherichia coli O157 in raw cow’s milk in The Netherlands. J. Food Prot. 1998, 61, 1597–1601. [Google Scholar] [CrossRef]
- Claeys, W.L.; Cardoen, S.; Daube, G.; De Block, J.; Dewettinck, K.; Dierick, K.; De Zutter, L.; Huyghebaert, A.; Imberechts, H.; Thiange, P. Raw or heated cow milk consumption: Review of risks and benefits. Food Control 2013, 31, 251–262. [Google Scholar] [CrossRef]
- European Food Safety Authority, panel on Biological Hazards. Scientific opinion on the public health risks related to the consumption of raw drinking milk. EFSA J. 2015, 13, 3940. [Google Scholar] [CrossRef]
- Marklinder, I.; Lindblad, M.; Eriksson, L.; Finnson, A.; Lindqvist, R. Home storage temperatures and consumer handling of refrigerated foods in Sweden. J. Food Prot. 2004, 67, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.W.; Redmond, E.C. Time-temperature profiling of United Kingdom consumers’ domestic refrigerators. J. Food Prot. 2016, 79, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Jackson, V.; Blair, I.; McDowell, D.; Cowan, C.; Bolton, D. Food safety knowledge of consumers and the microbiological and temperature status of their refrigerators. J. Food Prot. 2005, 68, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Pavlis, A.; Nychas, G.-J.E.; Xanthiakos, K. Probabilistic model for Listeria monocytogenes growth during distribution, retail storage, and domestic storage of pasteurized milk. Appl. Environ. Microbiol. 2010, 76, 2181–2191. [Google Scholar] [CrossRef]
- Zech, H.; Echtermeyer, C.; Wöhlbrand, L.; Blasius, B.; Rabus, R. Biological versus technical variability in 2-D DIGE experiments with environmental bacteria. Proteomics 2011, 11, 3380–3389. [Google Scholar] [CrossRef]



| S1 | S2 | S3 | S4 | |
|---|---|---|---|---|
| Source | Cattle feces (Farm B) | Cattle feces (Farm A) | Milk filter (Farm A) | Cattle feces (Farm A) |
| Year of isolation | 2019 (November) | 2020 (January) | 2020 (June) | 2020 (June) |
| Country | Norway | Norway | Norway | Norway |
| Pathotype | STEC | STEC | STEC | STEC |
| Serotype | ONT:H28 | O108:H25 | ONT:H28 | ONT:H28 |
| NCBI accession no | JANWGF000000000 | JANWGE000000000 | JANWGD000000000 | JANWGC000000000 |
| LEE operons | five | five | five | five |
| Intimin type | gamma | alpha | gamma | gamma |
| ehxA | yes | yes | yes | yes |
| astA ST toxin | yes | yes (2) | yes | yes |
| Stx type | Stx1a | Stx2a | Stx1a | Stx1a |
| Eru type | lambdoid | Eru1 | lambdoid | lambdoid |
| Stx phage CI clade | V | II | V | V |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Idland, L.; Bø-Granquist, E.G.; Aspholm, M.; Lindbäck, T. The Ability of Shiga Toxin-Producing Escherichia coli to Grow in Raw Cow’s Milk Stored at Low Temperatures. Foods 2022, 11, 3411. https://doi.org/10.3390/foods11213411
Idland L, Bø-Granquist EG, Aspholm M, Lindbäck T. The Ability of Shiga Toxin-Producing Escherichia coli to Grow in Raw Cow’s Milk Stored at Low Temperatures. Foods. 2022; 11(21):3411. https://doi.org/10.3390/foods11213411
Chicago/Turabian StyleIdland, Lene, Erik G. Bø-Granquist, Marina Aspholm, and Toril Lindbäck. 2022. "The Ability of Shiga Toxin-Producing Escherichia coli to Grow in Raw Cow’s Milk Stored at Low Temperatures" Foods 11, no. 21: 3411. https://doi.org/10.3390/foods11213411
APA StyleIdland, L., Bø-Granquist, E. G., Aspholm, M., & Lindbäck, T. (2022). The Ability of Shiga Toxin-Producing Escherichia coli to Grow in Raw Cow’s Milk Stored at Low Temperatures. Foods, 11(21), 3411. https://doi.org/10.3390/foods11213411








