Spectroscopic Data for the Rapid Assessment of Microbiological Quality of Chicken Burgers
Abstract
:1. Introduction
2. Materials and Methods
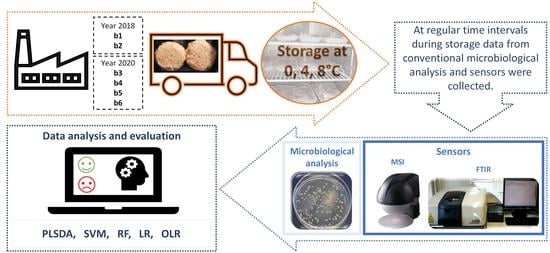
2.1. Purchase and Storage Conditions of Chicken Burgers
2.2. Microbiological Analysis and pH Measurement
2.3. Spectroscopy-Based Sensors
2.3.1. FTIR Measurements
2.3.2. MSI Acquisition
2.4. Data Analysis
- (i)
- ‘mixOmics’ [42] for PLSDA method [43], which is a linear classification model. It was employed for the reduction of dimensions and for the prediction of sample classes. In the current study the number of PLS components and the prediction distances (i.e., mahalanobis, centroids, maximum distance) were defined based on the classification error rate (leave-one-out cross-validation) of the training set.
- (ii)
- (iii)
- ‘e1071’ [44] was used for tuning the parameters (i.e., the number of trees to grow, the number of variables randomly sampled as candidates at each split) calculating the accuracy rate of 10-fold cross-validation for the training set and random forest [47] for implementing RF, which is an ensemble learning method for classification.
- (iv)
- ‘pls’ package [48] was used for extracting the PLS latent variables for the dimensionality reduction of the FTIR data and, subsequently, the ‘nnet’ [49] package was used to fit LR on the transformed (towards lower dimensions—due to memory issues caused by the large number of dimensions, i.e., >934m number of wavenumbers) FTIR data and on the raw MSI data. The number of LVs selected was defined as the knee-point of the curve of the variance explained vs. number of LVs.
- (v)
- ‘pls’ was applied on FTIR and MSI. The number of components was defined as the maximum number of components exhibiting at least 0.5% variance explained from the previous component. Subsequently, the MASS [49] package was used to fit OLR on the transformed FTIR and MSI data. The only difference between (iv) and (v) is that, in the second case, the Y-variable is handled as an ordinal categorical variable, defining the order of the three classes considered, i.e., in the continuum, “satisfactory”: 4–7 log CFU/g, “acceptable”: 7–8 log CFU/g, and “unacceptable”: >8 log CFU/g.
3. Results and Discussion
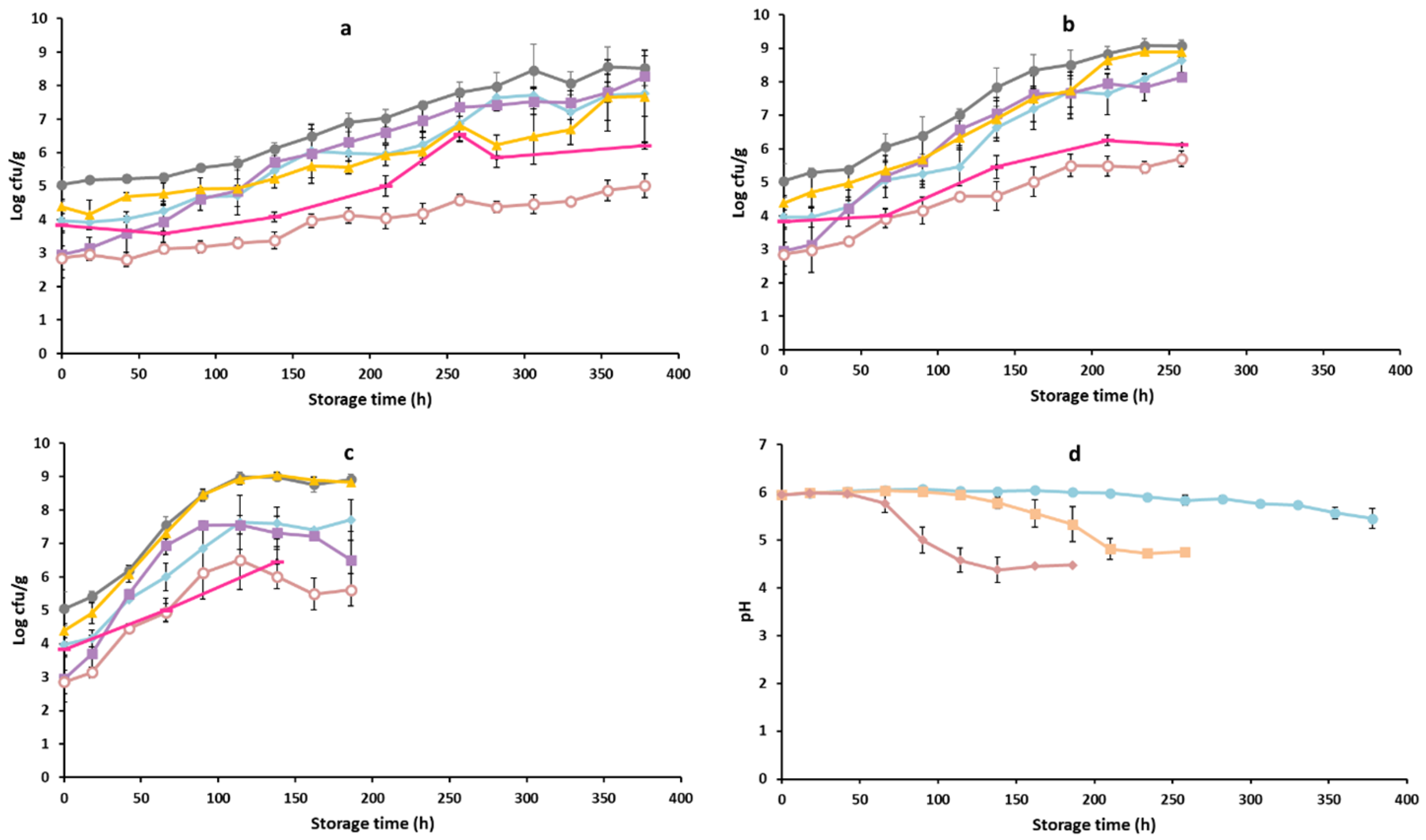
3.1. Microbiological Analysis and pH Measurements
3.2. FTIR Data
3.3. MSI Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. Food and Agricultural Organisation of the United Nations. In Meat Market Review—March 2021; FAO: Rome, Italy, 2020; p. 15. [Google Scholar]
- Nychas, G.-J.; Sims, E.; Tsakanikas, P.; Mohareb, F. Data science in the food industry. Annu. Rev. Biomed. Data Sci. 2021, 4, 341–367. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Jessep, W.; Sutton, J.J.; Hicks, T.M.; Loeffen, M.; Farouk, M.; Ward, J.F.; Bain, W.E.; Craigie, C.R.; Fraser-Miller, S.J.; et al. Evaluating low- mid- and high-level fusion strategies for combining Raman and infrared spectroscopy for quality assessment of red meat. Food Chem. 2021, 361, 130154. [Google Scholar] [CrossRef] [PubMed]
- Dixit, Y.; Hitchman, S.; Hicks, T.M.; Lim, P.; Wong, C.K.; Holibar, L.; Gordon, K.C.; Loeffen, M.; Farouk, M.M.; Craigie, C.R.; et al. Non-invasive spectroscopic and imaging systems for prediction of beef quality in a meat processing pilot plant. Meat Sci. 2021, 181, 108410. [Google Scholar] [CrossRef]
- Wang, W.; Peng, Y.; Sun, H.; Zheng, X.; Wei, W. Spectral detection techniques for non-destructively monitoring the quality, safety, and classification of fresh red meat. Food Anal. Methods 2018, 11, 2707–2730. [Google Scholar] [CrossRef]
- Fu, X.; Chen, J. A review of hyperspectral imaging for chicken meat safety and quality evaluation: Application, hardware, and software. Compr. Rev. Food Sci. Food Saf. 2019, 18, 535–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbin, D.F.; Badaró, A.T.; Honorato, D.C.B.; Ida, E.Y.; Shimokomaki, M. Identification of turkey meat and processed products using near infrared spectroscopy. Food Control 2020, 107, 106816. [Google Scholar] [CrossRef]
- Yang, Y.; Zhuang, H.; Yoon, S.-C.; Wang, W.; Jiang, H.; Jia, B.; Li, C. Quality assessment of intact chicken breast fillets using factor analysis with VIS/NIR spectroscopy. Food Anal. Methods 2018, 11, 1356–1366. [Google Scholar] [CrossRef]
- Vasconcelos, H.; Saraiva, C.; de Almeida, J.M. Evaluation of the spoilage of raw chicken breast fillets using Fourier transform infrared spectroscopy in tandem with chemometrics. Food Bioprocess Technol. 2014, 7, 2330–2341. [Google Scholar] [CrossRef]
- Barbin, D.F.; Kaminishikawahara, C.M.; Soares, A.L.; Mizubuti, I.Y.; Grespan, M.; Shimokomaki, M.; Hirooka, E.Y. Prediction of chicken quality attributes by near infrared spectroscopy. Food Chem. 2015, 168, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhuang, H.; Yoon, S.-C.; Wang, W.; Jiang, H.; Jia, B. Rapid classification of intact chicken breast fillets by predicting principal component score of quality traits with visible/near-infrared spectroscopy. Food Chem. 2018, 244, 184–189. [Google Scholar] [CrossRef]
- Feng, Y.-Z.; Sun, D.-W. Determination of total viable count (TVC) in chicken breast fillets by near-infrared hyperspectral imaging and spectroscopic transforms. Talanta 2013, 105, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Zając, A.; Dymińska, L.; Lorenc, J.; Hanuza, J. Fourier transform infrared and Raman spectroscopy studies of the time-dependent changes in chicken meat as a tool for recording spoilage processes. Food Anal. Methods 2017, 10, 640–648. [Google Scholar] [CrossRef]
- Magalhães, S.; Goodfellow, B.J.; Nunes, A. FTIR spectroscopy in biomedical research: How to get the most out of its potential. Appl. Spectrosc. Rev. 2021, 56, 869–907. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.-J.E. Data mining derived from food analyses using non-invasive/non-destructive analytical techniques; determination of food authenticity, quality & safety in tandem with computer science disciplines. Trends Food Sci. Technol. 2016, 50, 11–25. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Pavlidis, D.; Nychas, G.-J.E. High throughput multispectral image processing with applications in food science. PLoS ONE 2015, 10, e0140122. [Google Scholar] [CrossRef] [PubMed]
- Argyri, A.A.; Panagou, E.Z.; Tarantilis, P.A.; Polysiou, M.; Nychas, G.-J.E. Rapid qualitative and quantitative detection of beef fillets spoilage based on fourier transform infrared spectroscopy data and artificial neural networks. Sens. Actuators B Chem. 2010, 145, 146–154. [Google Scholar] [CrossRef] [Green Version]
- Candoğan, K.; Altuntas, E.G.; İğci, N. Authentication and quality assessment of meat products by fourier-transform infrared (FTIR) spectroscopy. Food Eng. Rev. 2020, 13, 66–91. [Google Scholar] [CrossRef]
- Manthou, E.; Karnavas, A.; Fengou, L.-C.; Bakali, A.; Lianou, A.; Tsakanikas, P.; Nychas, G.-J.E. Spectroscopy and imaging technologies coupled with machine learning for the assessment of the microbiological spoilage associated to ready-to-eat leafy vegetables. Int. J. Food Microbiol. 2022, 361, 109458. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, C.; Vasconcelos, H.; de Almeida, J.M. A chemometrics approach applied to fourier transform infrared spectroscopy (FTIR) for monitoring the spoilage of fresh salmon (Salmo salar) stored under modified atmospheres. Int. J. Food Microbiol. 2017, 241, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Panagou, E.Z.; Papadopoulou, O.; Carstensen, J.M.; Nychas, G.-J.E. Potential of multispectral imaging technology for rapid and non-destructive determination of the microbiological quality of beef filets during aerobic storage. Int. J. Food Microbiol. 2014, 174, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tsakanikas, P.; Pavlidis, D.; Panagou, E.; Nychas, G.-J. Exploiting multispectral imaging for non-invasive contamination assessment and mapping of meat samples. Talanta 2016, 161, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Barbin, D.F.; ElMasry, G.; Sun, D.-W.; Allen, P.; Morsy, N. Non-destructive assessment of microbial contamination in porcine meat using nir hyperspectral imaging. Innov. Food Sci. Emerg. Technol. 2013, 17, 180–191. [Google Scholar] [CrossRef]
- Liu, Y.; Pu, H.; Sun, D.W. Hyperspectral imaging technique for evaluating food quality and safety during various processes: A review of recent applications. Trends Food Sci. Technol. 2017, 69, 25–35. [Google Scholar] [CrossRef]
- Chen, Q.; Hui, Z.; Zhao, J.; Ouyang, Q. Evaluation of chicken freshness using a low-cost colorimetric sensor array with AdaBoost–OLDA classification algorithm. LWT—Food Sci. Technol. 2014, 57, 502–507. [Google Scholar] [CrossRef]
- Wojnowski, W.; Majchrzak, T.; Dymerski, T.; Gębicki, J.; Namieśnik, J. Poultry meat freshness evaluation using electronic nose technology and ultra-fast gas chromatography. Monatshefte Für Chem.—Chem. Mon. 2017, 148, 1631–1637. [Google Scholar] [CrossRef] [Green Version]
- ur Rahman, U.; Sahar, A.; Pasha, I.; ur Rahman, S.; Ishaq, A. Assessing the capability of Fourier transform infrared spectroscopy in tandem with chemometric analysis for predicting poultry meat spoilage. PeerJ 2018, 6, e5376. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Carvelo, A.M.; González-Casado, A.; Bagur-González, M.G.; Cuadros-Rodríguez, L. Alternative data mining/machine learning methods for the analytical evaluation of food quality and authenticity—A review. Food Res. Int. 2019, 122, 25–39. [Google Scholar] [CrossRef]
- Alexandrakis, D.; Downey, G.; Scannell, A.G.M. Rapid non-destructive detection of spoilage of intact chicken breast muscle using near-infrared and Fourier transform mid-infrared spectroscopy and multivariate statistics. Food Bioprocess Technol. 2012, 5, 338–347. [Google Scholar] [CrossRef]
- Lin, M.; Al-Holy, M.; Mousavi-Hesary, M.; Al-Qadiri, H.; Cavinato, A.G.; Rasco, B.A. Rapid and quantitative detection of the microbial spoilage in chicken meat by diffuse reflectance spectroscopy (600–1100 nm). Lett. Appl. Microbiol. 2004, 39, 148–155. [Google Scholar] [CrossRef]
- Spyrelli, E.D.; Doulgeraki, A.I.; Argyri, A.A.; Tassou, C.C.; Panagou, E.Z.; Nychas, G.-J.E. Implementation of MultiSpectral Imaging (MSI) for microbiological quality assessment of poultry products. Microorganisms 2020, 8, 552. [Google Scholar] [CrossRef] [Green Version]
- Tsakanikas, P.; Karnavas, A.; Panagou, E.Z.; Nychas, G.-J.E. Machine learning workflow for raw food spectroscopic classification in a future industry. Sci. Rep. 2020, 11, 11212. [Google Scholar] [CrossRef] [PubMed]
- Moschopoulou, E.; Moatsou, G.; Syrokou, M.K.; Paramithiotis, S.; Drosinos, E.H. Food Quality Changes during Shelf Life. In Food Quality and Shelf Life; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–31. [Google Scholar] [CrossRef]
- Nychas, G.-J.E.; Skandamis, P.N.; Tassou, C.C.; Koutsoumanis, K.P. Meat Spoilage during Distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef] [PubMed]
- USDA (Food Safety and Inspection Service U.S. Department of Agriculture). Freezing and Food Safety. Available online: https://www.fsis.usda.gov/food-safety/safe-food-handling-and-preparation/food-safety-basics/freezing-and-food-safety (accessed on 3 August 2022).
- Argyri, A.A.; Jarvis, R.M.; Wedge, D.; Xu, Y.; Panagou, E.Z.; Goodacre, R.; Nychas, G.-J.E. A Comparison of Raman and FT-IR spectroscopy for the prediction of meat spoilage. Food Control 2013, 29, 461–470. [Google Scholar] [CrossRef]
- Carstensen, J.M.; Hansen, J.F. Apparatus and a Method of Recording an Image of an Object. Patent Family EP1051660, 15 November 2003. [Google Scholar]
- Spyrelli, E.D.; Ozcan, O.; Mohareb, F.; Panagou, E.Z.; Nychas, G.-J.E. Spoilage assessment of chicken breast fillets by means of Fourier transform infrared spectroscopy and multispectral image analysis. Curr. Res. Food Sci. 2021, 4, 121–131. [Google Scholar] [CrossRef]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: https://www.R-Project.org/ (accessed on 3 August 2022).
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 3 August 2022).
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. MixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: e1071); R Package Version 1.7-4; TU Wien: Vienna, Austria, 2020. [Google Scholar]
- Luts, J.; Ojeda, F.; Van de Plas, R.; De Moor, B.; Van Huffel, S.; Suykens, J.A.K. Tutorial on support vector machine-based methods for classification problems in chemometrics. Anal. Chim. Acta 2010, 665, 129–145. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomforest. R News 2002, 2, 5. [Google Scholar]
- Mevik, B.-H.; Wehrens, R.; Liland, K.H. PLS: Partial Least Squares and Principal Component Regression. R Package Version 2.7-3. 2020. Available online: https://CRAN.R-project.org/package=pls (accessed on 3 August 2022).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S; Springer: New York, NY, USA, 2002. [Google Scholar]
- Álvarez-Astorga, M.; Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, C. Microbiological quality of retail chicken by-products in Spain. Meat Sci. 2002, 62, 45–50. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Han, Y.Q.; Cao, J.X.; Xu, X.L.; Zhou, G.H.; Zhang, W.Y. The spoilage of air-packaged broiler meat during storage at normal and fluctuating storage temperatures. Poult. Sci. 2012, 91, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Ercolini, D.; Villani, F.; Nychas, G.-J.E. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012, 157, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Nychas, G.-J.E.; Tassou, C.C. Spoilage processes and proteolysis in chicken as detected by HPLC. J. Sci. Food Agric. 1997, 74, 199–208. [Google Scholar] [CrossRef]
- Ye, X.; Iino, K.; Zhang, S. Monitoring of bacterial contamination on chicken meat surface using a novel narrowband spectral index derived from hyperspectral imagery data. Meat Sci. 2016, 122, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Dourou, D.; Grounta, A.; Argyri, A.A.; Froutis, G.; Tsakanikas, P.; Nychas, G.-J.E.; Doulgeraki, A.I.; Chorianopoulos, N.G.; Tassou, C.C. Rapid microbial quality assessment of chicken liver inoculated or not with Salmonella using FTIR spectroscopy and machine learning. Front. Microbiol. 2021, 11, 623788. [Google Scholar] [CrossRef]
- Adiani, V.; Gupta, S.; Variyar, P.S. Microbial quality assessment of minimally processed pineapple using GCMS and FTIR in tandem with chemometrics. Sci. Rep. 2020, 10, 6203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonah, E.; Huang, X.; Aheto, J.H.; Yi, R.; Yu, S.; Tu, H. Comparison of variable selection algorithms on vis-nir hyperspectral imaging spectra for quantitative monitoring and visualization of bacterial foodborne pathogens in fresh pork muscles. Infrared Phys. Technol. 2020, 107, 103327. [Google Scholar] [CrossRef]
- Wang, Y.-D.; Li, X.-L.; Liu, Z.-X.; Zhang, X.-X.; Hu, J.; Lü, J.-H. Discrimination of foodborne pathogenic bacteria using synchrotron FTIR Microspectroscopy. Nucl. Sci. Tech. 2017, 28, 49. [Google Scholar] [CrossRef]
| True Class | ||||
|---|---|---|---|---|
| A | B | C | ||
| PLSDA | Precision (%) | 88.24 | 75.00 | 88.57 |
| Recall (%) | 100.00 | 66.67 | 88.57 | |
| F1-score | 0.94 | 0.71 | 0.89 | |
| Accuracy (%) | 85.29 | |||
| SVM | Precision (%) | 88.24 | 82.35 | 94.12 |
| Recall (%) | 100.00 | 77.78 | 91.43 | |
| F1-score | 0.94 | 0.80 | 0.93 | |
| Accuracy (%) | 89.71 | |||
| RF | Precision (%) | 82.35 | 70.59 | 85.29 |
| Recall (%) | 93.33 | 66.67 | 82.86 | |
| F1-score | 0.87 | 0.69 | 0.84 | |
| Accuracy (%) | 80.88 | |||
| LR | Precision (%) | 100.00 | 77.78 | 88.57 |
| Recall (%) | 100.00 | 77.78 | 88.57 | |
| F1-score | 1.00 | 0.78 | 0.89 | |
| Accuracy (%) | 88.24 | |||
| OLR | Precision (%) | 87.50 | 60.00 | 87.50 |
| Recall (%) | 93.33 | 66.67 | 80.00 | |
| F1-score | 0.90 | 0.63 | 0.84 | |
| Accuracy (%) | 79.41 | |||
| True Class | ||||
|---|---|---|---|---|
| A | B | C | ||
| PLSDA | Precision (%) | 82.35 | 78.57 | 88.89 |
| Recall (%) | 100.00 | 61.11 | 91.43 | |
| F1-score | 0.90 | 0.69 | 0.90 | |
| Accuracy (%) | 85.07 | |||
| SVM | Precision (%) | 92.86 | 73.33 | 84.21 |
| Recall (%) | 92.86 | 61.11 | 91.43 | |
| F1-score | 0.93 | 0.67 | 0.88 | |
| Accuracy (%) | 83.58 | |||
| RF | Precision (%) | 73.68 | 62.50 | 77.50 |
| Recall (%) | 100.00 | 27.78 | 88.57 | |
| F1-score | 0.85 | 0.38 | 0.83 | |
| Accuracy (%) | 74.63 | |||
| LR | Precision (%) | 87.50 | 66.67 | 87.88 |
| Recall (%) | 100.00 | 66.67 | 82.86 | |
| F1-score | 0.93 | 0.67 | 0.85 | |
| Accuracy (%) | 82.09 | |||
| OLR | Precision (%) | 81.25 | 71.43 | 86.49 |
| Recall (%) | 92.86 | 55.56 | 91.43 | |
| F1-score | 0.86 | 0.62 | 0.89 | |
| Accuracy (%) | 82.09 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fengou, L.-C.; Liu, Y.; Roumani, D.; Tsakanikas, P.; Nychas, G.-J.E. Spectroscopic Data for the Rapid Assessment of Microbiological Quality of Chicken Burgers. Foods 2022, 11, 2386. https://doi.org/10.3390/foods11162386
Fengou L-C, Liu Y, Roumani D, Tsakanikas P, Nychas G-JE. Spectroscopic Data for the Rapid Assessment of Microbiological Quality of Chicken Burgers. Foods. 2022; 11(16):2386. https://doi.org/10.3390/foods11162386
Chicago/Turabian StyleFengou, Lemonia-Christina, Yunge Liu, Danai Roumani, Panagiotis Tsakanikas, and George-John E. Nychas. 2022. "Spectroscopic Data for the Rapid Assessment of Microbiological Quality of Chicken Burgers" Foods 11, no. 16: 2386. https://doi.org/10.3390/foods11162386
APA StyleFengou, L.-C., Liu, Y., Roumani, D., Tsakanikas, P., & Nychas, G.-J. E. (2022). Spectroscopic Data for the Rapid Assessment of Microbiological Quality of Chicken Burgers. Foods, 11(16), 2386. https://doi.org/10.3390/foods11162386