Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables—Jiangshui
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Preparation of L. Plantarum WYH
2.3. Antibiotic Susceptibility Test
2.4. Detecting Antibiotic Resistance Genes
2.5. Study on Acid Resistance and Bile Salt Tolerance
2.6. Auto-Aggregation and Co-Aggregation Ability
2.7. Hemolytic Assay
2.8. In Vivo Toxicity Experiments (Acute Oral Toxicity and Repeated Dose 28-Day Oral Toxicity Study)
2.9. Statistical Analysis
3. Results and Discussion
3.1. Tolerance to Acid and Bile Salts
3.2. Auto-Aggregation and Co-Aggregation
3.3. Hemolysis Activity
3.4. Antibiotics Resistance Analysis
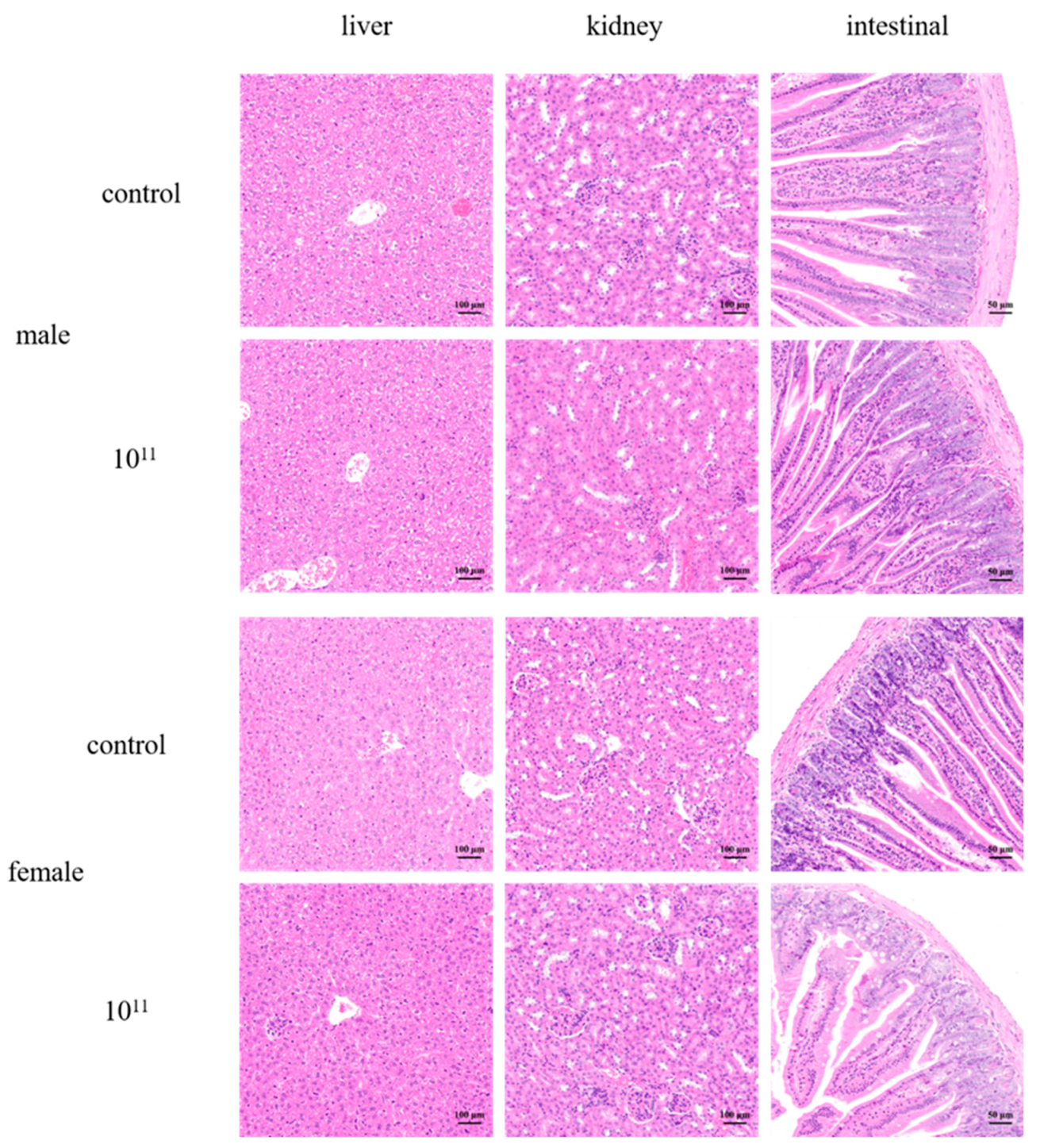
3.5. Acute Oral Toxicity Studies in Mice
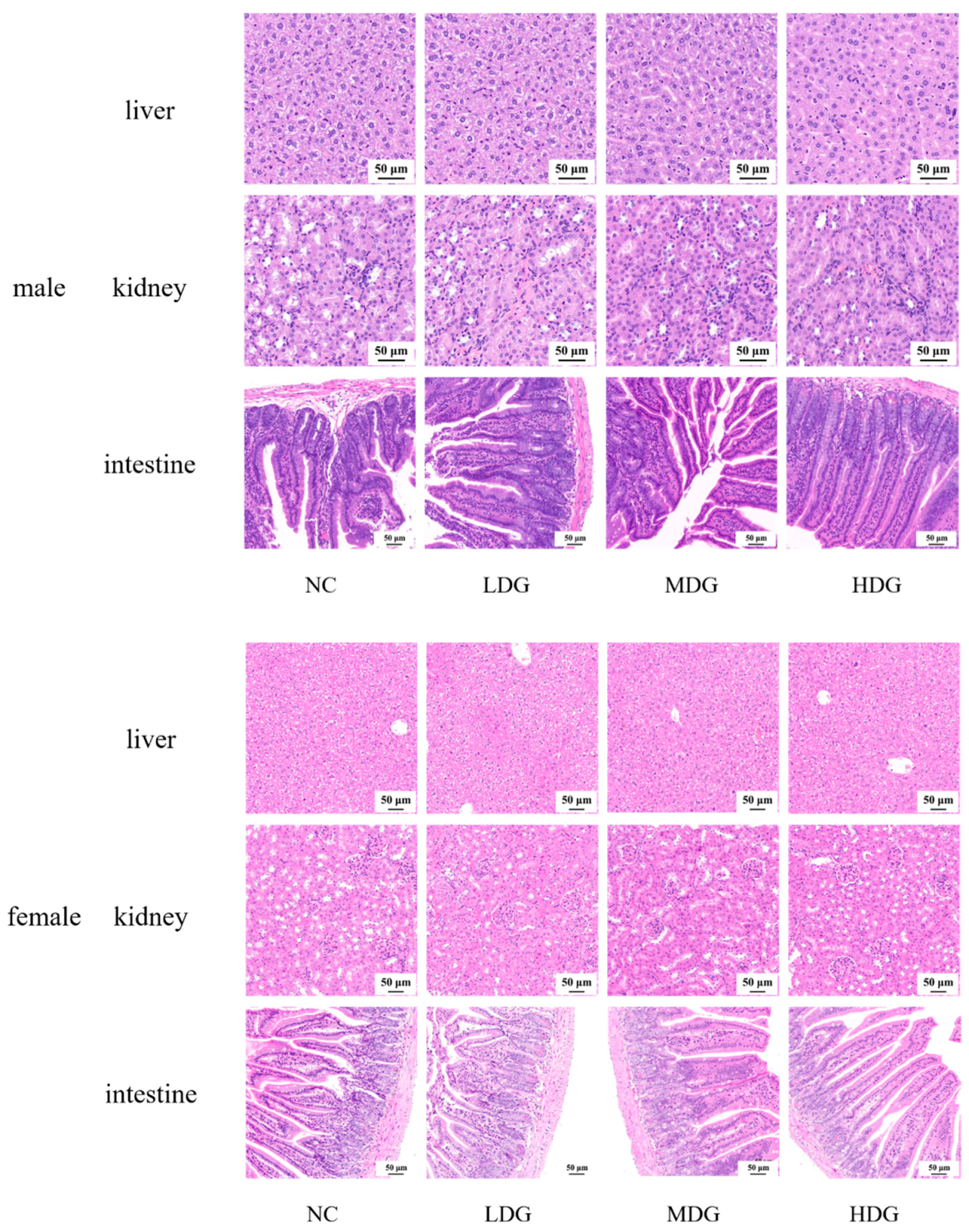
3.6. 28-Day Oral Toxicity Studies in Mice
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parker, R.B. Probiotics, the other half of the antibiotic story. Anim. Nutr. Health 1974, 29, 4–8. [Google Scholar]
- Hotel, A. Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria–Joint FAO/WHO Expert Consultation. Prevention 2001, 5, 1–10. [Google Scholar]
- Sakandar, H.A.; Zhang, H. Trends in probiotic(s)-fermented milks and their in vivo functionality: A review. Trends Food Sci. Technol. 2021, 110, 55–65. [Google Scholar] [CrossRef]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D.; et al. Shaping the future of probiotics and prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Chae, S.A.; Bang, W.Y.; Lee, M.; Ban, O.H.; Kim, S.J.; Jung, Y.H.; Yang, J. Anti-inflammatory potential of Lactiplantibacillus plantarum IDCC 3501 and its safety evaluation. Braz. J. Microbiol. 2021, 52, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Hua, X.; Zhao, W.; Liu, D.; Zhang, J.; Zhang, W.; Chen, W.; Yang, R. Protective effects of Lactobacillus plantarum CCFM436 against acute manganese toxicity in mice. Food Biosci. 2020, 35, 100583. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, L.; Duan, H.; Zhao, R.; Xiao, Y.; Guo, M.; Zhao, J.; Zhang, H.; Chen, W.; Tian, F. The protection of Lactiplantibacillus plantarum CCFM8661 against benzopyrene-induced toxicity via regulation of the gut microbiota. Front. Immunol. 2021, 12, 736129. [Google Scholar] [CrossRef]
- Amachawadi, R.; Giok, F.; Shi, X.; Soto, J.; Narayanan, S.K.; Tokach, M.D.; Apley, M.D.; Nagaraja, T.G. Antimicrobial resistance of Enterococcus faecium strains isolated from commercial probiotic products used in cattle and swine. J. Anim. Sci. 2018, 96, 912–920. [Google Scholar] [CrossRef]
- Siciliano, R.; Reale, A.; Mazzeo, M.; Morandi, S.; Silvetti, T.; Brasca, M. Paraprobiotics: A new perspective for functional foods and nutraceuticals. Nutrients 2021, 13, 1225. [Google Scholar] [CrossRef]
- Key, T.J.; Bradbury, K.E.; Perez-Cornago, A.; Sinha, R.; Tsilidis, K.K.; Tsugane, S. Diet, nutrition, and cancer risk: What do we know and what is the way forward? BMJ 2021, 9, 1224. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing CLSI Supplement M100S; Clinical and Laboratory Standards Institude: Wayne, PA, USA, 2018. [Google Scholar]
- Zarzecka, U.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Microorganisms from starter and protective cultures-Occurrence of antibiotic resistance and conjugal transfer of tet genes in vitro and during food fermentation. LWT 2022, 153, 112490. [Google Scholar] [CrossRef]
- Dlamini, Z.C.; Langa, R.L.S.; Aiyegoro, O.A.; Okoh, A.I. Safety evaluation and colonisation abilities of four lactic acid bacteria as future probiotics. Probiotics Antimicrob. Proteins 2019, 11, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Escamilla-Montes, R.; Luna-González, A.; Flores-Miranda, M.d.C.; Álvarez-Ruiz, P.; Fierro-Coronado, J.A.; Sánchez-Ortiz, A.C.; Ávila-Leal, J. Isolation and characterization of potential probiotic bacteria suitable for mollusk larvae cultures. Thai J. Vet. Med. 2015, 45, 11–21. [Google Scholar]
- GB15193.3-2014; Procedure and Methods of Food Safety Toxicological Assessment. China’s Ministry of Health 2014a: Beijing, China, 2014. (In Chinese)
- GB15193.22-2014; Procedure and Methods of Food Safety Toxicological Assessment. China’s Ministry of Health 2014b: Beijing, China, 2014. (In Chinese)
- OECD. Test No. 423: Acute Oral toxicity-Acute Toxic Class Method; OECD Guidelines for the Testing of Chemicals, Section 4; OECD Publishing: Paris, France, 2002. [Google Scholar] [CrossRef]
- OECD. Test No. 407: Repeated Dose 28-Day Oral Toxicity Study in Rodents; OECD Guidelines for the Testing of Chemicals, Section 4; OECD Publishing: Paris, France, 2008. [Google Scholar] [CrossRef]
- Sanders, M.E.; Walker, D.C.; Walker, K.M.; Aoyama, K.; Klaenhammer, T.R. Performance of commercial cultures in fluid milk applications. J. Dairy Sci. 1996, 79, 943–955. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.A.G.; Tsakalidou, E.; Nychas, G.J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.A.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Li, A.; Iqbal, M.; Zhang, L.; Pan, H.; Liu, Z.; Li, J. Probiotic potential and safety assessment of Lactobacillus isolated from yaks. Microb. Pathog. 2020, 145, 104213. [Google Scholar] [CrossRef]
- Rocha-Ramírez, L.M.; Hernández-Chiñas, U.; Moreno-Guerrero, S.S.; Ramírez-Pacheco, A.; Eslava, C.A. Probiotic properties and immunomodulatory activity of Lactobacillus strains isolated from dairy products. Microorganisms 2021, 9, 825. [Google Scholar] [CrossRef]
- Xu, Y.; Tian, Y.; Cao, Y.; Li, J.; Guo, H.; Su, Y.; Tian, Y.; Wang, C.; Wang, T.; Zhang, L. Probiotic properties of Lactobacillus paracasei subsp. paracasei L1 and its growth performance-promotion in chicken by improving the intestinal microflora. Front. Physiol. 2019, 10, 937. [Google Scholar] [CrossRef]
- Pavunc, A.L.; Penava, L.; Ranilovic, J.; Novak, J.; Banic, M.; Butorac, K.; Petrovic, E.; Mihaljevic-Herman, V.; Bendelja, K.; Mlakar, A.S.; et al. Influence of dehydrated wheat/rice cereal matrices on probiotic activity of Bifidobacterium animalistdui ssp. lactis BB-12®. Food Technol. Biotechnol. 2019, 57, 147–158. [Google Scholar] [CrossRef]
- He, Q.; Li, J.; Ma, Y.; Chen, Q.; Chen, G. Probiotic potential and cholesterol-lowering capabilities of bacterial strains isolated from pericarpium citri reticulatae ‘chachiensis’. Microorganisms 2021, 9, 1224. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Liu, X.; Guo, W.; Cui, S.; Tang, X.; Zhao, J.; Zhang, H.; Mao, B.; Chen, W. A comprehensive assessment of the safety of Blautia producta dsm 2950. Microorganisms 2021, 9, 908. [Google Scholar] [CrossRef] [PubMed]
- FAO; WHO. Guidelines for the Evaluation of Probiotics in Food (Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food); Food Agric Organ: Rome, Italy; United Nations World Health Organ: Geneva, Switzerland, 2002; p. 21. [Google Scholar]
- D’Aimmo, M.R.; Modesto, M.; Biavati, B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 2007, 115, 35–42. [Google Scholar] [CrossRef]
- Gotcheva, V.; Petrova, G.; Petkova, M.; Kuzmanova, Y.; Angelov, A. Molecular and in vitro assessment of some probiotic characteristics of amylolytic Lactobacillus plantarum strains from Bulgarian fermented products. Eng. Life Sci. 2018, 18, 820–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casado Muñoz, M.d.C.; Benomar, N.; Lerma, L.L.; Gálvez, A.; Abriouel, H. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int. J. Food Microbiol. 2014, 172, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Ogbolu, D.O.; Daini, O.A.; Ogunledun, A.; Terry Alli, A.O.; Olusoga-Ogbolu, F.F.; Webber, M.A. Effects of gyrA and parC mutations in quinolones resistant clinical gram-negative bacteria from Nigeria. Afr. J. Biomed. Res. 2012, 15, 97–104. [Google Scholar]
- Li, T.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, J. A critical review of antibiotic resistance in probiotic bacteria. Food Res. Int. 2020, 136, 109571. [Google Scholar] [CrossRef]
- Shao, Y.; Zhang, W.; Guo, H.; Pan, L.; Zhang, H.; Sun, T. Comparative studies on antibiotic resistance in Lactobacillus casei and Lactobacillus plantarum. Food Control. 2015, 50, 250–258. [Google Scholar] [CrossRef]
- Zhou, J.S.; Shu, Q.; Rutherfurd, K.J.; Prasad, J.; Gopal, P.K.; Gill, H.S. Acute oral toxicity and bacterial translocation studies on potentially probiotic strains of lactic acid bacteria. Food Chem. Toxicol. 2000, 38, 153–161. [Google Scholar] [CrossRef]
- Kim, J.; Han, M.; Jeon, W.K. Acute and subacute oral toxicity of mumefural, bioactive compound derived from processed fruit of Prunus mume sieb. Et zucc., in ICR mice. Nutrients 2020, 12, 1328. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Nasti, A.; Takeshita, Y.; Okumura, M.; Kitajima, S.; Honda, M.; Wada, T.; Nakamura, S.; Takamura, T.; Tamura, T.; et al. Eight-year longitudinal study of whole blood gene expression profiles in individuals undergoing long-term medical follow-up. Sci. Rep. 2021, 11, 16564. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kandhare, A.D.; Mukherjee, A.A.; Bodhankar, S.L. Acute and sub-chronic oral toxicity studies of hesperidin isolated from orange peel extract in Sprague Dawley rats. Regul. Toxicol. Pharmacol. 2019, 105, 77–85. [Google Scholar] [CrossRef]
- Gómez del Pulgar, E.M.; Benítez-Páez, A.; Sanz, Y. Safety assessment of Bacteroides uniformis CECT 7771, a symbiont of the gut microbiota in infants. Nutrients 2020, 12, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Luo, C.; Zhang, X.; Guo, M.; Sun, M.; Yu, H.; Chen, Q.; Yang, W.; Wang, M.; Zuo, S.; et al. Probing the impact of sulfur/selenium/carbon linkages on prodrug nanoassemblies for cancer therapy. Nat. Commun. 2019, 10, 3211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, W.; Hou, K.; Lin, J.; Song, C.; Zhou, C.; Huang, B.; Tong, X.; Wang, J.; Rhine, W.; et al. Air pollution exposure associates with increased risk of neonatal jaundice. Nat. Commun. 2019, 10, 3211. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Hu, D.; Gao, Y.; Ai, Y.; Luo, S.; Chen, S.; Wang, B.; Zhou, L.; Dong, Y.; Wang, Y. Safety assessment of subtilisin QK in rats. BMC Pharmacol. Toxicol. 2021, 22, 38. [Google Scholar] [CrossRef]


| Gene | Primer Sequences (5′ to 3′) | |
|---|---|---|
| 16s rRNA | 27F | AGAGTTTGATCMTGGCTCAG |
| 1492R | CGGTTACCTTGTTACGACTT | |
| tetM | F | GTGGACAAAGGTACAACGAG |
| R | CGGTAAAGTTCGTCACACAC | |
| tetK | F | TTAGGTGAAGGGTTAGGTCC |
| R | GCAAACTCATTCCAGAAGCA | |
| tetL | F | CATTTGGTCTTATTGGATCG |
| R | ATTACACTTCCGATTTCGG | |
| tetS | F | TGGAACGCCAGAGAGGTATT |
| R | ACATAGACAAGCCGTTGACC | |
| bla | F | CATARTTCCGATAATASMGCC |
| R | CGTSTTTAACTAAGTATSGY | |
| vanA | F | TTGCTCAGAGGAGCATGACG |
| R | TCGGGAAGTGCAATACCTGC | |
| vanB | F | TTATCTTCGGCGGTTGCTCG |
| R | GCCAATGTAATCAGGCTGTC | |
| vanX | F | TCGCGGTAGTCCCACCATTCGTT |
| R | AAATCATCGTTGACCTGCGTTAT | |
| aac | F | CCAAGAGCAATAAGGGCATA |
| R | CACTATCATAACCACTACCG | |
| aph | F | GCCGATGTGGATTGCGAAAA |
| R | GCTTGATCCCCAGTAAGTCA | |
| strA | F | CTTGGTGATAACGGCAATTC |
| R | CCAATCGCAGATAGAAGGC | |
| strB | F | ATCGTCAAGGGATTGAAACC |
| R | GGATCGTAGAACATATTGGC | |
| aadA | F | ATCCTTCGGCGCGATTTTG |
| R | GCAGCGCAATGACATTCTTG | |
| ant | F | ACTGGCTTAATCAATTTGGG |
| R | GCCTTTCCGCCACCTCACCG | |
| cat | F | CATARTTCCGATAATASMGCC |
| R | TTAGGTTATTGGGATAAGTTA | |
| gyrA | F | CAMCGKCGKATTCTTTACGGAATG |
| R | TTRTTGATATCRCGBAGCATTTC | |
| parC | F | GCYTCNGTATAACGCATMGCCG |
| R | GCYTCNGTATAACGCATMGCCG | |
| ermE | F | CATTTAACGACGAAACTGGC |
| R | GGAACATCTGTGGTATGGCG | |
| ermC | F | CAAACCCGTATTCCACGATT |
| R | ATCTTTGAAATCGGCTCAGG |
| Antibiotic | Inhibition Zone (mm) a | CLSI Classification b | ||
|---|---|---|---|---|
| Lactiplantibacillus plantarum | Staphylococcus aureus | Susceptible Zone (mm) | S-I-R | |
| TE (30 μg) | 18.24 ± 0.26 | 21.95 ± 0.09 | ≥19 | I |
| MEM (10 μg) | 29.67 ± 0.31 | 20.74 ± 0.12 | ≥16 | S |
| AML (10 μg) | 33.61 ± 0.23 | 17.45 ± 0.05 | ≥18 | S |
| AMP (10 μg) | NA | 16.12 ± 0.12 | ≥19 | R |
| VA (30 μg) | NA | NA | ≥15 | R |
| S (10 μg) | NA | 19.21 ± 0.05 | ≥14 | R |
| CIP (5 μg) | NA | 25.25 ± 0.18 | ≥21 | R |
| K (30 μg) | 10.59 ± 0.10 | 7.82 ± 0.01 | ≥18 | S |
| C (30 μg) | 20.61 ± 0.09 | 23.72 ± 0.13 | ≥18 | S |
| GEN (10 μg) | 17.13 ± 0.14 | 14.35 ± 0.14 | ≥10 | S |
| ERM (15 μg) | 27.96 ± 0.06 | NA | ≥23 | S |
| NEO (10 μg) | 17.22 ± 0.16 | 14.80 ± 0.02 | ≥16 | S |
| AZI (15 μg) | 19.15 ± 0.07 | 9.00 ± 0.08 | ≥18 | S |
| Hematological Parameters | 0 | 1011 | ||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| White blood cells (109/L) | 3.69 ± 0.16 | 3.94 ± 0.06 | 4.20 ± 0.28 | 4.72 ± 0.39 |
| Neutrophils (109/L) | 0.06 ± 0.01 | 0.25 ± 0.02 | 0.08 ± 0.01 | 0.45 ± 0.12 |
| Lymphocytes (109/L) | 3.27 ± 0.23 | 3.67 ± 0.07 | 3.90 ± 0.46 | 4.26 ± 0.30 |
| Monocytes (109/L) | ND | ND | ND | ND |
| Eosinophilic (109/L) | ND | ND | ND | ND |
| Basophilic (109/L) | 0.01 ± 0.005 | 0.02 ± 0.000 | 0.02 ± 0.010 | 0.02 ± 0.010 |
| Neutrophil percentage (%) | 11.23 ± 4.58 | 6.25 ± 0.64 | 7.60 ± 5.78 | 9.18 ± 2.32 |
| Lymphocytes percentage (%) | 88.25 ± 4.65 | 93.20 ± 0.56 | 91.95 ± 5.79 | 90.38 ± 2.34 |
| Monocytes percentage (%) | 0.08 ± 0.04 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.10 ± 0.06 |
| Eosinophilic percentage (%) | ND | 0.030 ± 0.030 | ND | ND |
| Basophilic percentage (%) | 0.45 ± 0.05 | 0.50 ± 0.11 | 0.43 ± 0.13 | 0.35 ± 0.06 |
| Red blood cells (109/L) | 7.99 ± 1.23 | 10.49 ± 0.21 | 9.27 ± 0.84 | 9.92 ± 0.38 |
| Hemoglobin (g/L) | 151.00 ± 9.43 | 160.75 ± 4.27 | 157.25 ± 4.55 | 159.50 ± 3.40 |
| Hematocrit (%) | 38.78 ± 5.49 | 49.08 ± 1.33 | 45.75 ± 4.02 | 47.73 ± 0.57 |
| Mean corpuscular volume (fl) | 48.93 ± 0.82 | 46.73 ± 0.42 | 49.40 ± 0.60 | 48.25 ± 1.43 |
| Mean corpuscular hemoglobin (pg) | 15.88 ± 0.34 | 15.33 ± 0.18 | 17.33 ± 1.41 | 16.13 ± 0.34 |
| Mean corpuscular hemoglobin concentration (g/L) | 324.50 ± 1.96 | 327.75 ± 2.53 | 350.25 ± 25.96 | 334.25 ± 3.35 |
| Red blood cell distribution width (SD) | 14.20 ± 0.090 | 15.30 ± 0.45 | 13.95 ± 0.33 | 13.45 ± 0.39 |
| Red blood cell volume distribution width (CV) | 26.98 ± 0.56 | 27.83 ± 1.02 | 26.80 ± 0.72 | 25.20 ± 0.91 * |
| Platelets (109/L) | 401.25 ± 65.39 | 320.25 ± 160.97 | 365.00 ± 203.01 | 403.00 ± 198.78 |
| Mean platelet volume (fl) | 7.13 ± 0.23 | 22.58 ± 15.81 | 7.20 ± 0.19 | 6.48 ± 0.19 |
| Platelet distribution width (%) | 15 ± 0.23 | 14.78 ± 0.11 | 14.85 ± 0.06 | 14.90 ± 0.11 |
| Plateletcrit (%) | 0.12 ± 0.04 | 0.26 ± 0.09 | 0.10 ± 0.02 | 0.25 ± 0.12 |
| NC | LDG | MDG | HDG | |
|---|---|---|---|---|
| Hematological parameters | ||||
| White blood cells (109/L) | 6.98 ± 0.70 | 8.88 ± 0.61 | 5.26 ± 0.24 | 5.72 ± 0.61 |
| Neutrophils (109/L) | 1.26 ± 0.28 | 1.18 ± 0.49 | 0.61 ± 0.22 | 0.68 ± 0.23 |
| Lymphocytes (109/L) | 5.65 ± 0.62 | 8.36 ± 0.78 | 4.34 ± 0.59 | 7.50 ± 2.74 |
| Monocytes (109/L) | 0.030 ± 0.010 | 0.030 ± 0.010 | 0.010 ± 0.00 | 0.010 ± 0.010 |
| Eosinophilic (109/L) | ND | ND | ND | ND |
| Basophilic (109/L) | 0.050 ± 0.010 | 0.070 ± 0.020 | 0.040 ± 0.010 | 0.040 ± 0.010 |
| Neutrophil percentage (%) | 17.95 ± 4.12 | 10.93 ± 3.37 | 13.55 ± 5.34 | 8.88 ± 2.62 |
| Lymphocytes percentage (%) | 80.95 ± 4.06 | 88.05 ± 3.14 | 85.55 ± 5.26 | 90.48 ± 2.60 |
| Monocytes percentage (%) | 0.40 ± 0.15 | 0.28 ± 0.11 | 0.15 ± 0.030 | 0.15 ± 0.050 |
| Eosinophilic percentage (%) | 0.030 ± 0.030 | ND | ND | ND |
| Basophilic percentage (%) | 0.68 ± 0.12 | 0.75 ± 0.27 | 0.75 ± 0.18 | 0.50 ± 0.070 |
| Red blood cells (109/L) | 9.68 ± 0.14 | 9.89 ± 0.18 | 8.44 ± 0.99 | 9.66 ± 0.20 |
| Hemoglobin (g/L) | 144.50 ± 1.32 | 147.25 ± 3.28 | 129.75 ± 15.94 | 143.25 ± 1.55 |
| Hematocrit (%) | 45.33 ± 0.54 | 46.85 ± 1.09 | 41.63 ± 4.85 | 46.00 ± 0.72 |
| Mean corpuscular volume (fl) | 46.80 ± 0.64 | 47.38 ± 0.66 | 49.38 ± 0.050 | 47.60 ± 0.39 |
| Mean corpuscular hemoglobi (pg) | 14.93 ± 0.21 | 14.90 ± 0.070 | 15.35 ± 0.22 | 14.85 ± 0.18 |
| Mean corpuscular hemoglobin concentration (g/L) | 319.25 ± 1.80 | 314.25 ± 4.33 | 310.75 ± 4.31 | 311.50 ± 1.66 |
| Red blood cell distribution width (SD) | 17.08 ± 0.86 | 18.70 ± 1.50 | 16.88 ± 0.65 | 17.08 ± 0.57 |
| Red blood cell volume distribution width (CV) | 33.00 ± 1.32 | 36.70 ± 3.26 | 33.88 ± 1.32 | 33.60 ± 1.33 |
| Platelets (109/L) | 885.75 ± 98.30 | 626.25 ± 126.72 | 614.75 ± 121.37 | 1058.25 ± 110.94 |
| Mean platelet volume (fl) | 5.63 ± 0.090 | 5.88 ± 0.24 | 6.20 ± 0.11 | 5.68 ± 0.14 |
| Platelet distribution width (%) | 15.20 ± 0.070 | 15.08 ± 0.13 | 15.28 ± 0.060 | 15.15 ± 0.030 |
| Plateletcrit (%) | 0.50 ± 0.050 | 0.36 ± 0.070 | 0.38 ± 0.080 | 0.60 ± 0.050 |
| Serum biochemistry | ||||
| ALT (U/L) | 61.68 ± 5.97 | 74.65 ± 2.77 | 64.14 ± 6.65 | 61.08 ± 3.48 |
| AST (U/L) | 139.28 ± 5.76 | 163.15 ± 14.68 | 151.10 ± 3.40 | 139.90 ± 10.12 |
| Glu (mmol/L) | 6.35 ± 0.49 | 6.05 ± 0.345 | 6.95 ± 0.50 | 7.05 ± 0.41 |
| Urea (mmol/L) | 18.68 ± 0.61 | 20.76 ± 1.04 | 18.26 ± 2.38 | 20.51 ± 1.34 |
| CREA (μmol/L) | 26.29 ± 1.09 | 21.64 ± 0.14 | 24.19 ± 2.39 | 26.08 ± 1.89 |
| CHO (mmol/L) | 2.26 ± 0.12 | 2.33 ± 0.17 | 2.65 ± 0.12 | 2.40 ± 0.085 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ou, D.; Ling, N.; Wang, X.; Zou, Y.; Dong, J.; Zhang, D.; Shen, Y.; Ye, Y. Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables—Jiangshui. Foods 2022, 11, 2177. https://doi.org/10.3390/foods11152177
Ou D, Ling N, Wang X, Zou Y, Dong J, Zhang D, Shen Y, Ye Y. Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables—Jiangshui. Foods. 2022; 11(15):2177. https://doi.org/10.3390/foods11152177
Chicago/Turabian StyleOu, Dexin, Na Ling, Xihao Wang, Yanyan Zou, Jingjing Dong, Danfeng Zhang, Yizhong Shen, and Yingwang Ye. 2022. "Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables—Jiangshui" Foods 11, no. 15: 2177. https://doi.org/10.3390/foods11152177
APA StyleOu, D., Ling, N., Wang, X., Zou, Y., Dong, J., Zhang, D., Shen, Y., & Ye, Y. (2022). Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables—Jiangshui. Foods, 11(15), 2177. https://doi.org/10.3390/foods11152177








