Use of Large-Scale Genomics to Identify the Role of Animals and Foods as Potential Sources of Extraintestinal Pathogenic Escherichia coli That Cause Human Illness
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Strain Characterization
2.3. Virulence Group
3. Results
3.1. Characterization of IPEC and non-IPEC Strains
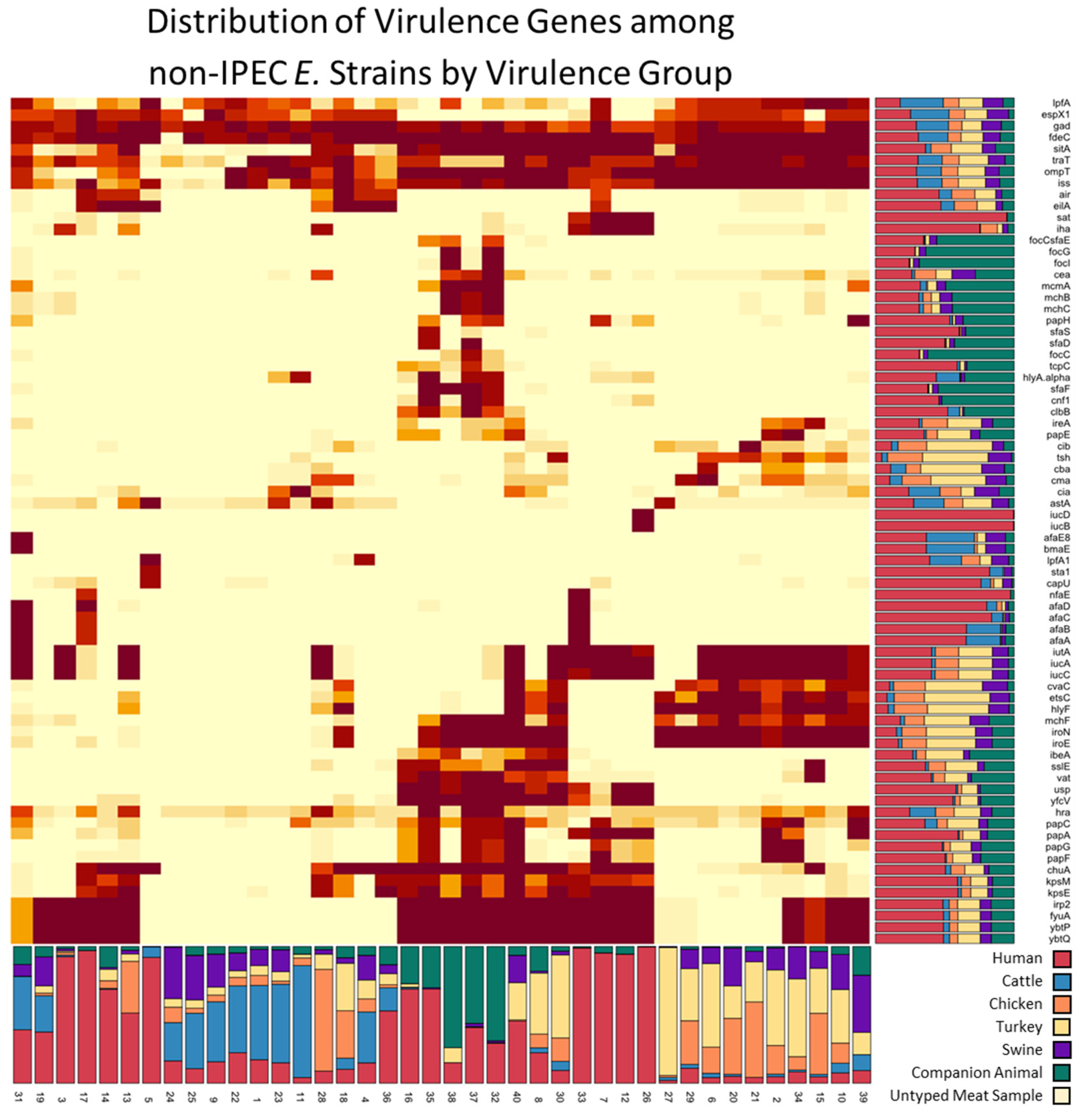
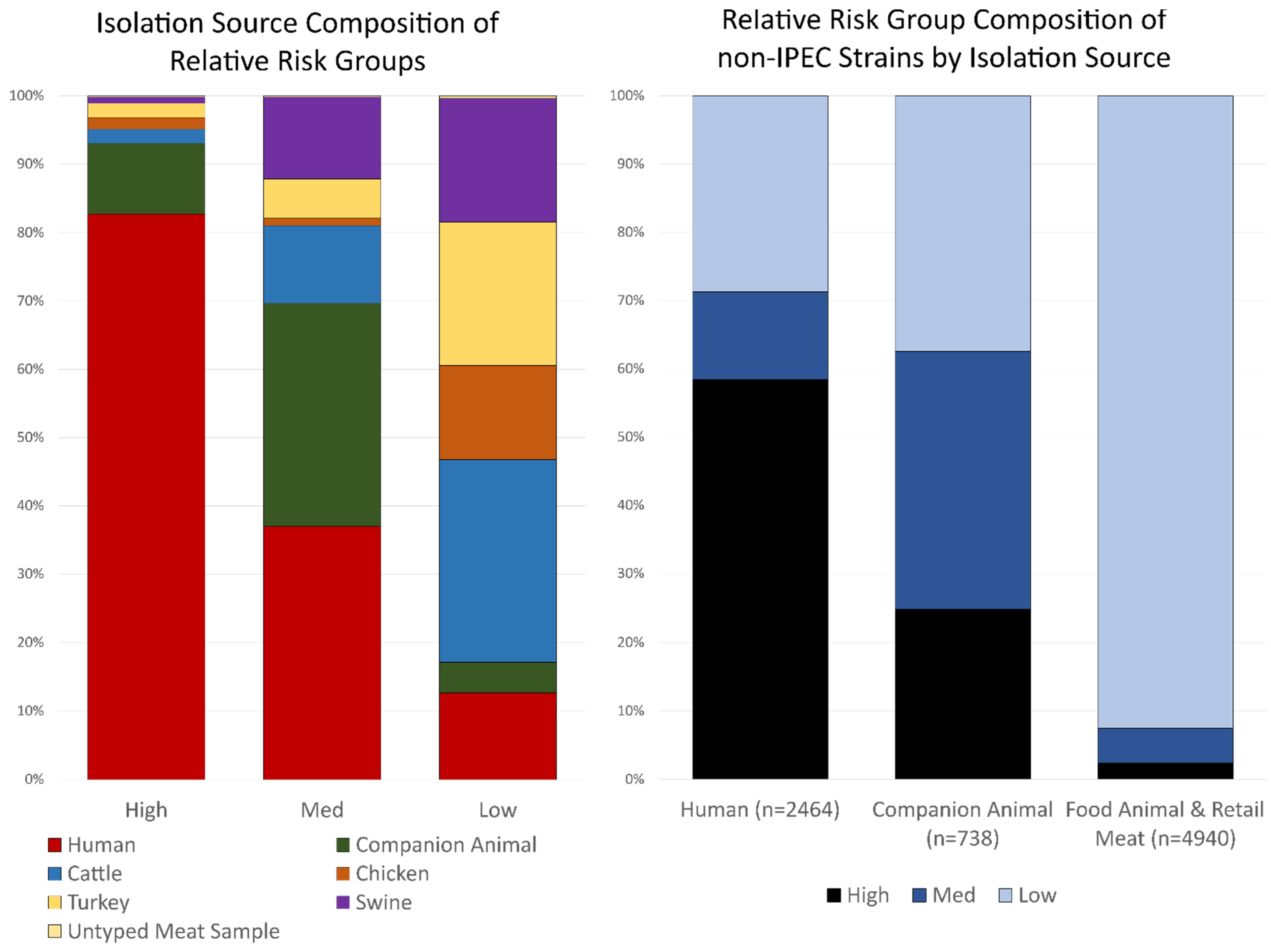
3.2. Isolation Source Composition of Virulence Groups
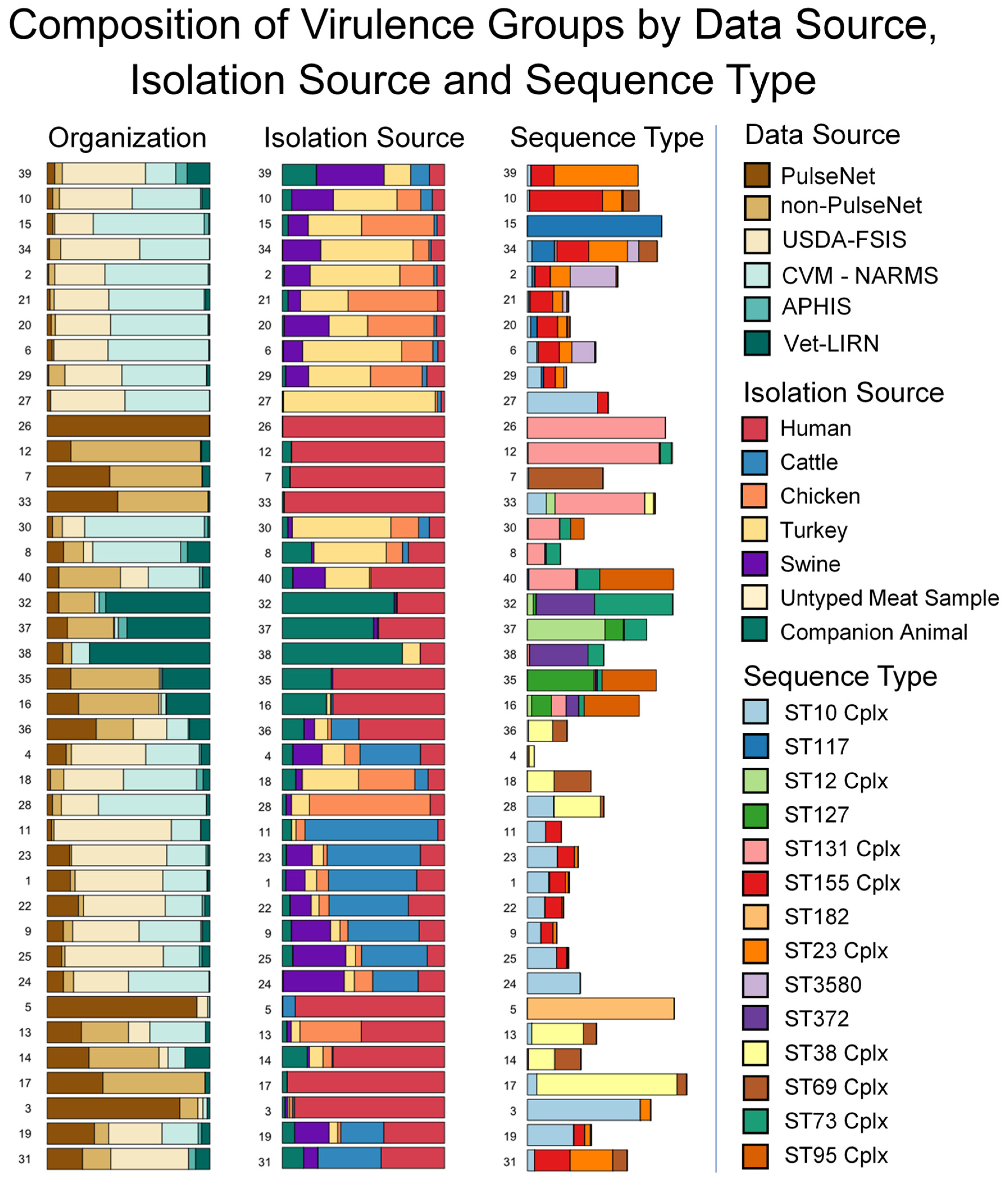
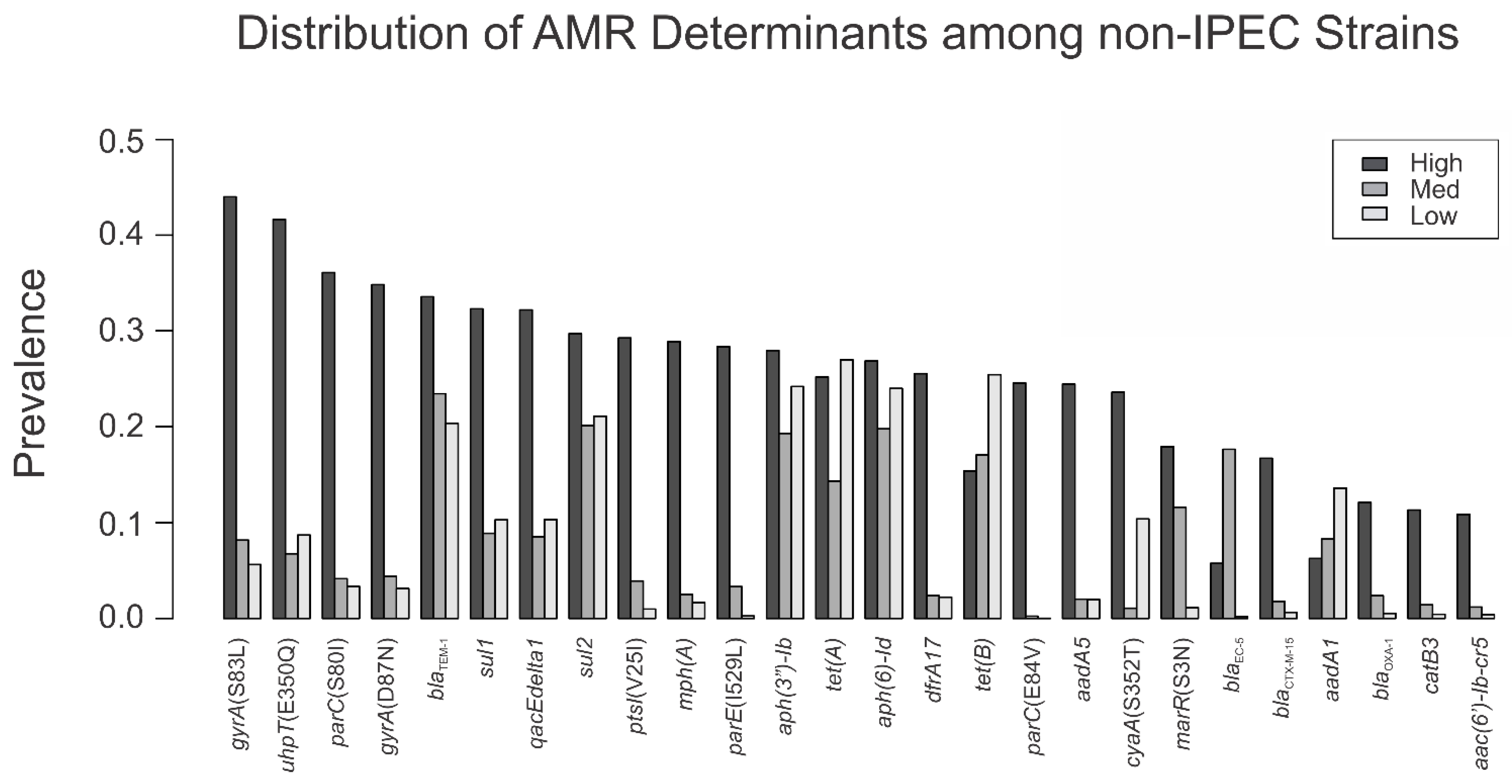
3.3. Sequence Type and AMR Gene Composition of Virulence Groups
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ishii, S.; Ksoll, W.B.; Hicks, R.E.; Sadowsky, M.J. Presence and Growth of Naturalized Escherichia coli in Temperate Soils from Lake Superior Watersheds. Appl. Environ. Microbiol. 2006, 72, 612–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leimbach, A.; Hacker, J.; Dobrindt, U. E. coli as an all-rounder: The thin line between commensalism and pathogenicity. Curr. Top Microbiol. Immunol. 2013, 358, 3–32. [Google Scholar] [PubMed]
- Johnson, J.R.; Russo, T.A. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli”. J. Lab Clin. Med. 2002, 139, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Johnson, J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000, 181, 1753–1754. [Google Scholar] [CrossRef] [Green Version]
- Lindstedt, B.A.; Finton, M.D.; Porcellato, D.; Brandal, L.T. High frequency of hybrid Escherichia coli strains with combined Intestinal Pathogenic Escherichia coli (IPEC) and Extraintestinal Pathogenic Escherichia coli (ExPEC) virulence factors isolated from human faecal samples. BMC Infect Dis. 2018, 18, 544. [Google Scholar] [CrossRef]
- Kohler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Salipante, S.J.; Roach, D.J.; Kitzman, J.O.; Snyder, M.W.; Stackhouse, B.; Butler-Wu, S.M.; Lee, C.; Cookson, B.T.; Shendure, J. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 2015, 25, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Boll, E.J.; Overballe-Petersen, S.; Hasman, H.; Roer, L.; Ng, K.; Scheutz, F.; Hammerum, A.M.; Dungu, A.; Hansen, F.; Johannesen, T.B. Emergence of enteroaggregative Escherichia coli within the ST131 lineage as a cause of extraintestinal infections. mBio 2020, 11, e00353-20. [Google Scholar] [CrossRef]
- Malberg, T.A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob Agents Chemother 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11, 1756287219832172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Borges, C.A.; Tarlton, N.J.; Riley, L.W. Escherichia coli from Commercial Broiler and Backyard Chickens Share Sequence Types, Antimicrobial Resistance Profiles, and Resistance Genes with Human Extraintestinal Pathogenic Escherichia coli. Foodborne Pathog Dis. 2019, 16, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Buberg, M.L.; Mo, S.S.; Sekse, C.; Sunde, M.; Wasteson, Y.; Witsø, I.L. Population structure and uropathogenic potential of extended-spectrum cephalosporin-resistant Escherichia coli from retail chicken meat. BMC Microbiol. 2021, 21, 94. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.; García-Meniño, I.; Fernández, J.; García, V.; Mora, A. Chicken and turkey meat: Consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and high-risk lineages such as ST131. Int. J. Food Microbiol. 2020, 331, 108750. [Google Scholar] [CrossRef]
- Xia, X.; Meng, J.; Zhao, S.; Bodeis-Jones, S.; Gaines, S.A.; Ayers, S.L.; McDermott, P.F. Identification and antimicrobial resistance of extraintestinal pathogenic Escherichia coli from retail meats. J. Food Prot. 2011, 74, 38–44. [Google Scholar] [CrossRef]
- Yamaji, R.; Friedman, C.R.; Rubin, J.; Suh, J.; Thys, E.; McDermott, P.; Hung-Fan, M.; Riley, L.W. A population-based surveillance study of shared genotypes of Escherichia coli isolates from retail meat and suspected cases of urinary tract infections. Msphere 2018, 3, e00179-18. [Google Scholar] [CrossRef] [Green Version]
- Manges, A.R.; Smith, S.P.; Lau, B.J.; Nuval, C.J.; Eisenberg, J.N.; Dietrich, P.S.; Riley, L.W. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: A case–control study. Foodborne Pathog. Dis. 2007, 4, 419–431. [Google Scholar] [CrossRef] [Green Version]
- Robinson, T.P.; Bu, D.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- FDA. NARMS Now. 2021. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data (accessed on 28 January 2021).
- Kidsley, A.K.; O’Dea, M.; Saputra, S.; Jordan, D.; Johnson, J.R.; Gordon, D.M.; Turni, C.; Djordjevic, S.P.; Abraham, S.; Trott, D.J. Genomic analysis of phylogenetic group B2 extraintestinal pathogenic E. coli causing infections in dogs in Australia. Vet. Microbiol. 2020, 248, 108783. [Google Scholar] [CrossRef] [PubMed]
- Paul, R. State of the globe: Rising antimicrobial resistance of pathogens in urinary tract infection. J. Glob. Infect. Dis. 2018, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Naylor, N.R.; Pouwels, K.B.; Hope, R.; Green, N.; Henderson, K.L.; Knight, G.M.; Atun, R.; Robotham, J.V.; Deeny, S.R. The health and cost burden of antibiotic resistant and susceptible Escherichia coli bacteraemia in the English hospital setting: A national retrospective cohort study. PLoS ONE 2019, 14, e0221944. [Google Scholar] [CrossRef] [PubMed]
- Wasiński, B. Extra-intestinal pathogenic Escherichia coli–threat connected with food-borne infections. Ann. Agric. Environ. Med. 2019, 26, 532–537. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [Green Version]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Venkatesan, M.M.; Buysse, J.M.; Kopecko, D.J. Use of Shigella flexneri ipaC and ipaH gene sequences for the general identification of Shigella spp. and enteroinvasive Escherichia coli. J. Clin. Microbiol. 1989, 27, 2687–2691. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. Available online: https://www.R-Project.org/ (accessed on 28 January 2021).
- Weihs, C.; Ligges, U.; Luebke, K.; Raabe, N. klaR analyzing German business cycles. In Data Analysis and Decision Support; Springer: Berlin/Heidelberg, Germany, 2005; pp. 335–343. [Google Scholar] [CrossRef]
- Yuan, C.; Yang, H. Research on K-value selection method of K-means clustering algorithm. J. Multidiscip. Sci. J. 2019, 2, 226–235. [Google Scholar] [CrossRef] [Green Version]
- Spurbeck, R.R.; Dinh, P.C., Jr.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Signorell, A.; Aho, K.; Alfons, A.; Anderegg, N.; Aragon, T.; Arachchige, C.; Arppe, A.; Baddeley, A.; Barton, K.; Bolker, B. DescTools: Tools for Descriptive Statistics. R Package Version 0.99. 44. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://cran.r-project.org/web/packages/DescTools/index.html (accessed on 28 January 2021).
- Poolman, J.T.; Wacker, M. Extraintestinal pathogenic Escherichia coli, a common human pathogen: Challenges for vaccine development and progress in the field. J. Infect. Dis. 2016, 213, 6–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitout, J.D. Extraintestinal pathogenic Escherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti Infect Ther. 2012, 10, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, E.; Oprea, S.F.; Donabedian, S.M.; Perri, M.; Bozigar, P.; Bartlett, P.; Zervos, M.J. Epidemiology of antimicrobial resistance in enterococci of animal origin. J. Antimicrob Chemother 2005, 55, 127–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, W. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 2000, 1 (Suppl. 16), S19–S24. [Google Scholar] [CrossRef]
| # of Isolates | IPEC | Non-IPEC | |
|---|---|---|---|
| Human (PulseNet) | 30,862 | 29,651 | 1211 |
| Human (non-PulseNet) | 1268 | 15 | 1253 |
| Retail meats | 2433 | 31 | 2402 |
| Food animal cecal | 2717 | 179 | 2538 |
| Companion animals | 752 | 14 | 738 |
| Total | 38,032 | 29,890 | 8142 |
| Virulence Gene | IPEC | Non-IPEC | Non-IPEC/IPEC |
|---|---|---|---|
| tsh | 2.78% | 14.01% | 5.046 |
| etsC | 5.12% | 25.35% | 4.954 |
| clbB | 1.89% | 9.36% | 4.945 |
| vat | 3.55% | 17.56% | 4.941 |
| cnf1 | 1.55% | 7.61% | 4.911 |
| hlyF | 5.61% | 27.14% | 4.842 |
| f17A | 0.75% | 3.61% | 4.817 |
| iroE | 6.98% | 33.39% | 4.785 |
| sfaF | 1.59% | 7.58% | 4.759 |
| iroN | 7.06% | 33.54% | 4.751 |
| focI | 0.97% | 4.58% | 4.742 |
| papH | 1.59% | 7.53% | 4.721 |
| tcpC | 1.26% | 5.96% | 4.721 |
| f17G | 0.72% | 3.39% | 4.697 |
| focG | 0.88% | 4.09% | 4.636 |
| papA | 4.77% | 21.94% | 4.594 |
| usp | 4.99% | 22.92% | 4.592 |
| focC | 0.77% | 3.45% | 4.506 |
| papF | 3.45% | 15.45% | 4.478 |
| kpsM | 6.37% | 28.21% | 4.429 |
| papC | 5.88% | 25.88% | 4.402 |
| ibeA | 1.95% | 8.47% | 4.339 |
| sfaD | 0.88% | 3.75% | 4.246 |
| papE | 2.09% | 8.86% | 4.231 |
| sslE | 3.64% | 15.35% | 4.217 |
| cvaC | 4.87% | 20.40% | 4.189 |
| iucD | 0.12% | 0.49% | 4.138 |
| iucB | 0.12% | 0.50% | 4.081 |
| yfcV | 6.25% | 24.80% | 3.967 |
| nfaE | 0.50% | 1.98% | 3.951 |
| sfaS | 0.54% | 2.08% | 3.843 |
| sfaE | 0.21% | 0.81% | 3.785 |
| lngA | 0.25% | 0.92% | 3.733 |
| kpsE | 8.46% | 31.45% | 3.720 |
| air | 2.44% | 8.90% | 3.653 |
| afaE | 0.29% | 1.04% | 3.646 |
| ltcA | 0.41% | 1.49% | 3.586 |
| hlyE | 0.22% | 0.77% | 3.574 |
| eilA | 3.36% | 11.61% | 3.450 |
| eatA | 0.50% | 1.72% | 3.435 |
| mchF | 8.09% | 27.61% | 3.413 |
| sat | 3.05% | 9.90% | 3.241 |
| hra | 9.97% | 30.19% | 3.029 |
| iroD | 0.02% | 0.06% | 2.931 |
| afaB | 1.15% | 3.33% | 2.906 |
| afaA | 1.17% | 3.34% | 2.853 |
| cma | 5.63% | 14.75% | 2.618 |
| afaC | 1.01% | 2.58% | 2.541 |
| ccI | 0.13% | 0.32% | 2.407 |
| mchB | 3.59% | 8.65% | 2.406 |
| afaD | 1.64% | 3.93% | 2.398 |
| faeG | 0.02% | 0.05% | 2.345 |
| ireA | 3.99% | 9.31% | 2.333 |
| iroB | 0.02% | 0.04% | 2.261 |
| iroC | 0.02% | 0.04% | 2.261 |
| neuC | 4.04% | 8.29% | 2.054 |
| A | B1 | B2 | C | D | E | E or Clade I | F | G | Unknown | |
|---|---|---|---|---|---|---|---|---|---|---|
| High | 10.8 | 0.9 | 56.2 | 0.6 | 20.4 | 4.6 | 0.2 | 4.7 | 1.3 | 0.2 |
| Med | 20.2 | 17 | 59.3 | 2.4 | 0.5 | 0.4 | 0 | 0.1 | 0.1 | 0 |
| Low | 28.6 | 48.1 | 7.6 | 3.8 | 4 | 2.7 | 0 | 1.2 | 3.7 | 0.3 |
| Resistance Determinant | Human | Cattle | Chicken | Turkey | Swine | Untyped Meat Sample | Companion Animal |
|---|---|---|---|---|---|---|---|
| cyaA(S352T) | 15.1 | 2.5 | 32.6 | 14.9 | 5.4 | 43.5 | 8.9 |
| gyrA(S83L) | 37.2 | 2.3 | 2.2 | 1.7 | 3.5 | 4.3 | 15.5 |
| parC(S80I) | 28.9 | 0.5 | 0.3 | 0.8 | 2.3 | 0.0 | 12.4 |
| gyrA(D87N) | 27.7 | 0.4 | 0.6 | 0.8 | 2.1 | 0.0 | 12.2 |
| uhpT(E350Q) | 29.6 | 6.9 | 12.7 | 13.3 | 4.1 | 8.7 | 13.8 |
| mph(A) | 22.1 | 0.2 | 0.1 | 0.2 | 2.0 | 0.0 | 5.8 |
| dfrA17 | 19.6 | 0.6 | 0.3 | 1.2 | 2.1 | 0.0 | 7.5 |
| parE(I529L) | 19.3 | 0.0 | 0.1 | 0.8 | 1.9 | 0.0 | 3.9 |
| ptsI(V25I) | 20.1 | 0.1 | 1.8 | 2.7 | 2.0 | 4.3 | 4.0 |
| aadA5 | 18.5 | 0.5 | 0.1 | 1.3 | 1.4 | 0.0 | 7.4 |
| parC(E84V) | 16.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 |
| marR(S3N) | 13.3 | 0.7 | 1.4 | 1.0 | 0.2 | 4.3 | 14.1 |
| blaCTX-M-15 | 12.8 | 0.1 | 0.0 | 0.1 | 0.1 | 0.0 | 3.1 |
| blaOXA-1 | 9.6 | 0.0 | 0.0 | 0.4 | 0.0 | 0.0 | 2.6 |
| catB3 | 8.7 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 |
| aac(6′)-Ib-cr5 | 8.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 |
| sul1 | 28.0 | 2.8 | 20.5 | 14.8 | 5.8 | 21.7 | 7.3 |
| qacEdelta1 | 27.9 | 2.7 | 20.4 | 14.6 | 5.8 | 21.7 | 7.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harrison, L.; Tyson, G.H.; Strain, E.; Lindsey, R.L.; Strockbine, N.; Ceric, O.; Fortenberry, G.Z.; Harris, B.; Shaw, S.; Tillman, G.; et al. Use of Large-Scale Genomics to Identify the Role of Animals and Foods as Potential Sources of Extraintestinal Pathogenic Escherichia coli That Cause Human Illness. Foods 2022, 11, 1975. https://doi.org/10.3390/foods11131975
Harrison L, Tyson GH, Strain E, Lindsey RL, Strockbine N, Ceric O, Fortenberry GZ, Harris B, Shaw S, Tillman G, et al. Use of Large-Scale Genomics to Identify the Role of Animals and Foods as Potential Sources of Extraintestinal Pathogenic Escherichia coli That Cause Human Illness. Foods. 2022; 11(13):1975. https://doi.org/10.3390/foods11131975
Chicago/Turabian StyleHarrison, Lucas, Gregory H. Tyson, Errol Strain, Rebecca L. Lindsey, Nancy Strockbine, Olgica Ceric, Gamola Z. Fortenberry, Beth Harris, Sheryl Shaw, Glenn Tillman, and et al. 2022. "Use of Large-Scale Genomics to Identify the Role of Animals and Foods as Potential Sources of Extraintestinal Pathogenic Escherichia coli That Cause Human Illness" Foods 11, no. 13: 1975. https://doi.org/10.3390/foods11131975
APA StyleHarrison, L., Tyson, G. H., Strain, E., Lindsey, R. L., Strockbine, N., Ceric, O., Fortenberry, G. Z., Harris, B., Shaw, S., Tillman, G., Zhao, S., & Dessai, U. (2022). Use of Large-Scale Genomics to Identify the Role of Animals and Foods as Potential Sources of Extraintestinal Pathogenic Escherichia coli That Cause Human Illness. Foods, 11(13), 1975. https://doi.org/10.3390/foods11131975






