Quantitative Risk Assessment of Susceptible and Ciprofloxacin-Resistant Salmonella from Retail Pork in Chiang Mai Province in Northern Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pork Samples
2.2. Enumeration of Salmonella
2.3. Antimicrobial Susceptibility Testing (AST)
2.4. Determination of Antimicrobial Resistance Genes
2.5. Risk Assessment Models
2.5.1. Exposure Assessment
- Probabilistic prevalence variable
- 2.
- Thermal inactivation model
- 3.
- Concentration variable
- 4.
- Consumption variable (CP)
- 5.
- Dose of Salmonella ingested
- 6.
- Probability of exposure (PE)
2.5.2. Hazard Characterization
- Probability of illness (PI)
- 2.
- Probability of mortality (PM)
- 3.
- Probability of mortality given illness (PMI)
2.5.3. Risk Characterization
2.6. Statistical Analysis
3. Results
3.1. Exposure Assessment
3.2. Hazard Characterization
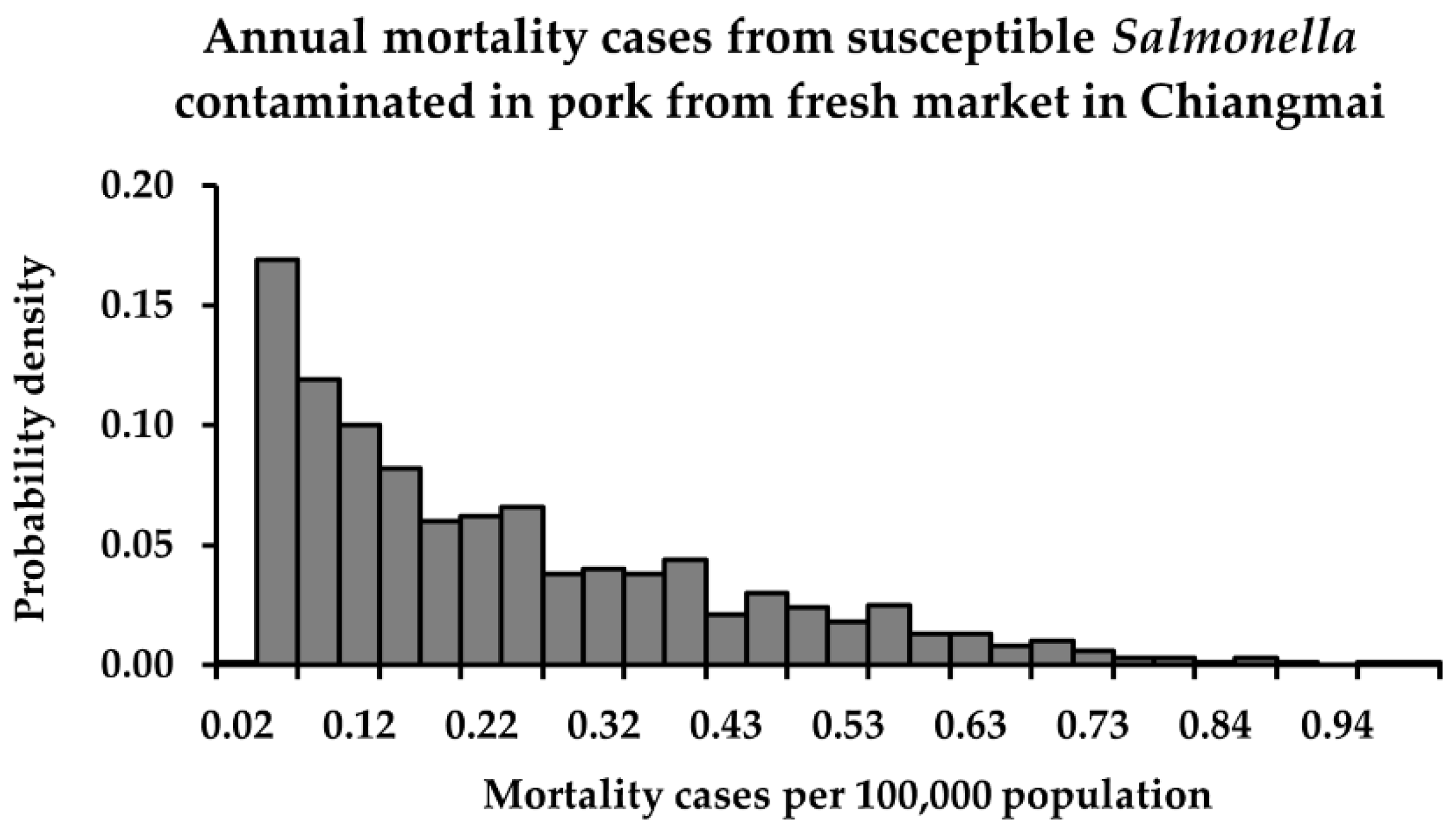
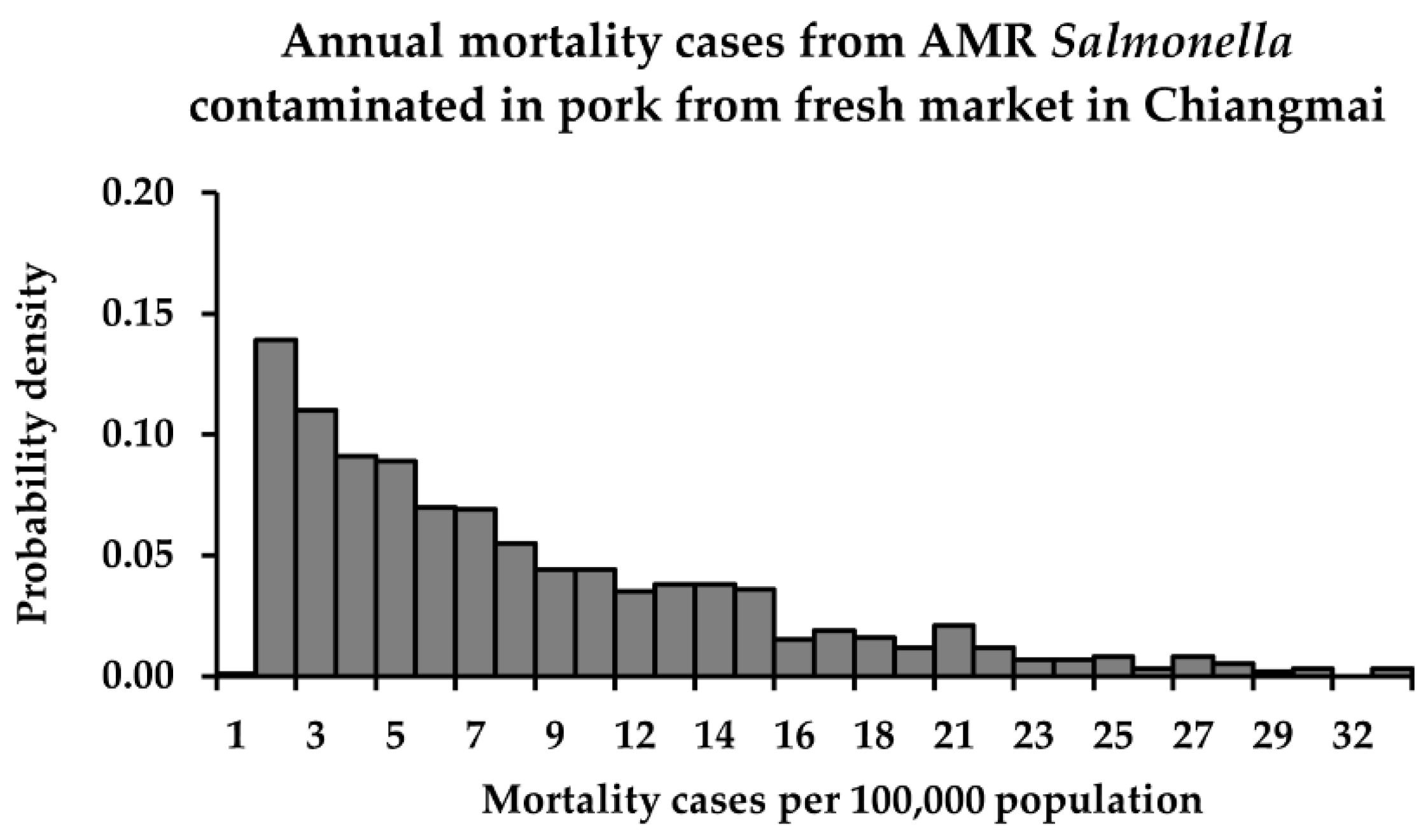
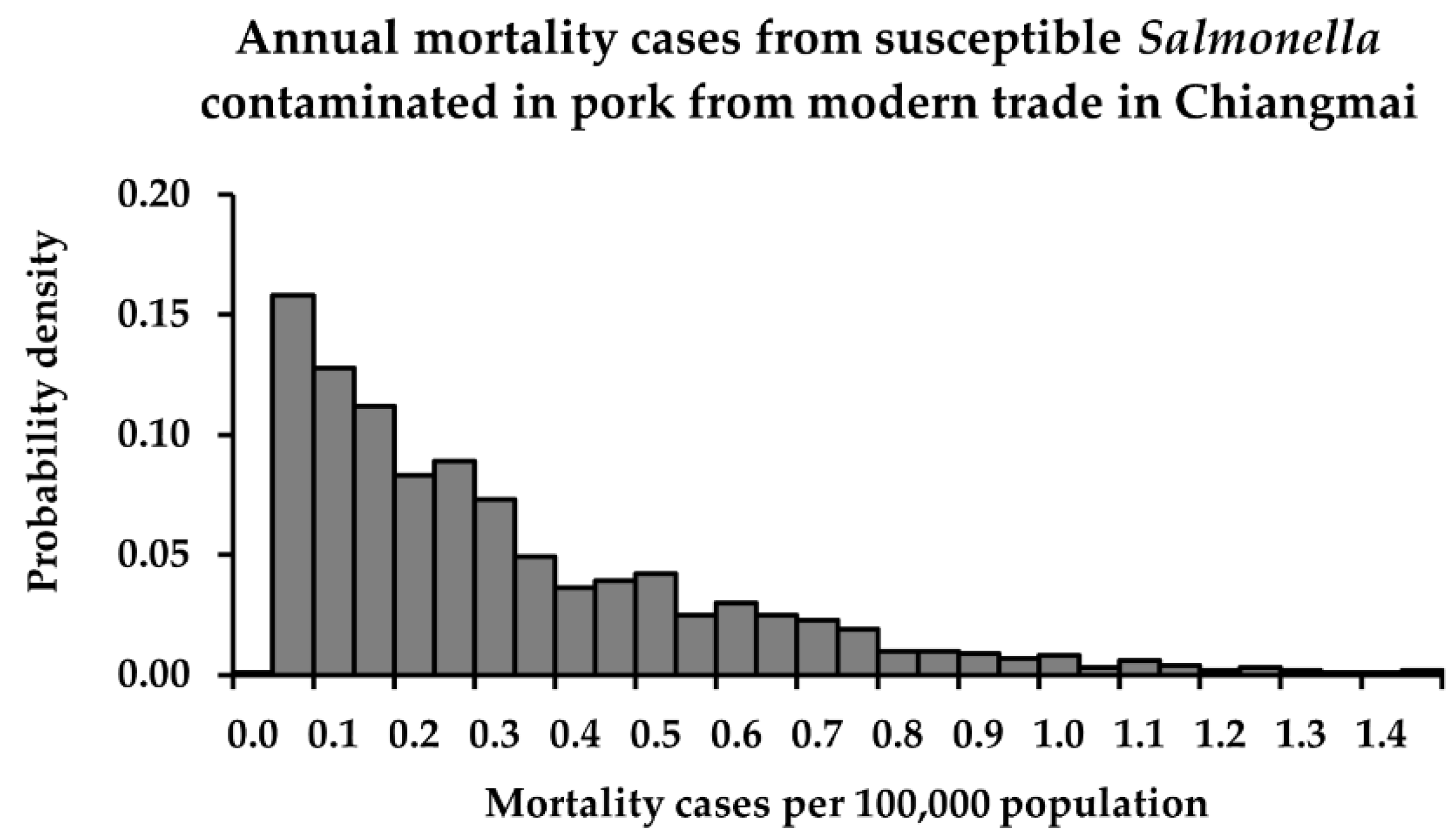
3.3. Risk Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Snary, E.; McEwen, S. Antimicrobial Resistance Risk Assessment. In Guide to Antimicrobial Use in Animals; Guardabassi, L., Jensen, L.B., Kruse, H., Eds.; Blackwell Publishing, Ltd.: Oxford, UK, 2008; pp. 27–43. [Google Scholar] [CrossRef]
- Booton, R.D.; Meeyai, A.; Alhusein, N.; Buller, H.; Feil, E.; Lambert, H.; Mongkolsuk, S.; Pitchforth, E.; Reyher, K.K.; Sakcamduang, W.; et al. One Health drivers of antibacterial resistance: Quantifying the relative impacts of human, animal and environmental use and transmission. One Health 2021, 12, 8. [Google Scholar] [CrossRef]
- Phumart, P.; Phodha, T.; Thamlikitkul, V.; Riewpaiboon, A.; Prakongsai, P.; Limwattananon, S. Health and economic impacts of antimicrobial resistant infections in Thailand: A Preliminary Study. J. Health Syst. Res. 2012, 6, 360. [Google Scholar]
- Codex Alimentarius Commission (CAC). Guidelines for Risk Analysis of Foodborne Antimicrobial Resistance (CXG 77-2011); FAO: Rome, Italy, 2011; p. 28. [Google Scholar]
- FAO; WHO. Principles and Guidelines for the Conduct of Microbiological Risk Assessment (CAC/GL-30), 2nd ed.; FAO: Rome, Italy; WHO: Geneva, Switzerland, 1999; p. 10. [Google Scholar]
- Khantasup, K.; Tungwongjulaniam, C.; Theerawat, R.; Lamaisri, T.; Piyalikit, K.; Nuengjamnong, C.; Nuanualsuwan, S. Cross-sectional risk assessment of zoonotic Streptococcus suis in pork and swine blood in Nakhon Sawan Province in northern Thailand. Zoonoses Public Health 2022, 69, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.; Kaneene, J.B.; Padungtod, P.; Hirokawa, K.; Zeno, C. Prevalence of Salmonella and E. coli, and their resistance to antimicrobial agents, in farming communities in northern Thailand. Southeast Asian J. Trop. Med. Public Health 2002, 33 (Suppl. S3), 126. [Google Scholar]
- Padungtod, P.; Kaneene, J.B. Salmonella in food animals and humans in northern Thailand. Int. J. Food Microbiol. 2006, 108, 354. [Google Scholar] [CrossRef]
- Sanpong, P.; Theeragool, G.; Wajjwalku, W.; Amavisit, P. Characterization of multiple-antimicrobial resistant Salmonella isolated from pig farms in Thailand. Agric. Nat. Resour. 2010, 44, 651. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 327. [Google Scholar] [CrossRef]
- Poomchuchit, S.; Kerdsin, A.; Chopjitt, P.; Boueroy, P.; Hatrongjit, R.; Akeda, Y.; Tomono, K.; Nuanualsuwan, S.; Hamada, S. Fluoroquinolone resistance in non-typhoidal Salmonella enterica isolated from slaughtered pigs in Thailand. J. Med. Microbiol. 2021, 70, 9. [Google Scholar] [CrossRef] [PubMed]
- Angkititrakul, S.; Chomvarin, C.; Chaita, T.; Kanistanon, K.; Waethewutajarn, S. Epidemiology of antimicrobial resistance in Salmonella isolated from pork, chicken meat and humans in Thailand. Southeast Asian J. Trop. Med. Public Health 2005, 36, 1515. [Google Scholar]
- Holmberg, S.D.; Solomon, S.L.; Blake, P.A. Health and economic impacts of antimicrobial resistance. Rev. Infect. Dis. 1987, 9, 1078. [Google Scholar] [CrossRef]
- Molbak, K.; Baggesen, D.L.; Aarestrup, F.M.; Ebbesen, J.M.; Engberg, J.; Frydendahl, K.; Gerner-Smidt, P.; Petersen, A.M.; Wegener, H.C. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N. Engl. J. Med. 1999, 341, 1425. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.A.; Lawson, A.J.; Lindsay, E.A.; Ward, L.R.; Wright, P.A.; Bolton, F.J.; Wareing, D.R.; Corkish, J.D.; Davies, R.H.; Threlfall, E.J. Decreased susceptibility to ciprofloxacin in outbreak-associated multiresistant Salmonella typhimurium DT104. Vet. Rec. 2000, 147, 395–396. [Google Scholar] [CrossRef] [PubMed]
- Meakins, S.; Fisher, I.S.; Berghold, C.; Gerner-Smidt, P.; Tschape, H.; Cormican, M.; Luzzi, I.; Schneider, F.; Wannett, W.; Coia, J.; et al. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000–2004: A report from the Enter-net International Surveillance Network. Microb. Drug Resist. 2008, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Threlfall, E.J.; Ward, L.R.; Skinner, J.A.; Graham, A. Antimicrobial drug resistance in non-typhoidal Salmonellas from humans in England and Wales in 1999: Decrease in multiple resistance in Salmonella enterica serotypes Typhimurium, Virchow, and Hadar. Microb. Drug Resist. 2000, 6, 325. [Google Scholar] [CrossRef] [PubMed]
- Varma, J.K.; Greene, K.D.; Ovitt, J.; Barrett, T.J.; Medalla, F.; Angulo, F.J. Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984–2002. Emerg. Infect. Dis. 2005, 11, 946. [Google Scholar] [CrossRef] [PubMed]
- Crook, P.D.; Aguilera, J.F.; Threlfall, E.J.; O’Brien, S.J.; Sigmundsdottir, G.; Wilson, D.; Fisher, I.S.; Ammon, A.; Briem, H.; Cowden, J.M.; et al. A European outbreak of Salmonella enterica serotype Typhimurium definitive phage type 204b in 2000. Clin. Microbiol. Infect. 2003, 9, 845. [Google Scholar] [CrossRef]
- Horby, P.W.; O’Brien, S.J.; Adak, G.K.; Graham, C.; Hawker, J.I.; Hunter, P.; Lane, C.; Lawson, A.J.; Mitchell, R.T.; Reacher, M.H.; et al. A national outbreak of multi-resistant Salmonella enterica serovar Typhimurium definitive phage type DT 104 associated with consumption of lettuce. Epidemiol. Infect. 2003, 130, 178. [Google Scholar] [CrossRef]
- Bertrand, S.; Weill, F.X.; Cloeckaert, A.; Vrints, M.; Mairiaux, E.; Praud, K.; Dierick, K.; Wildemauve, C.; Godard, C.; Butaye, P.; et al. Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 2006, 44, 2903. [Google Scholar] [CrossRef]
- Cloeckaert, A.; Praud, K.; Doublet, B.; Bertini, A.; Carattoli, A.; Butaye, P.; Imberechts, H.; Bertrand, S.; Collard, J.M.; Arlet, G.; et al. Dissemination of an extended-spectrum-beta-lactamase blaTEM-52 gene-carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob. Agents Chemother. 2007, 51, 1875. [Google Scholar] [CrossRef]
- Caffrey, N.; Invik, J.; Waldner, C.; Ramsay, D.; Checkley, S. Risk assessments evaluating foodborne antimicrobial resistance in humans: A scoping review. J. Microb. Risk Anal. 2019, 11, 46. [Google Scholar] [CrossRef]
- Alban, L.; Olsen, A.M.; Nielsen, B.; Sorensen, R.; Jessen, B. Qualitative and quantitative risk assessment for human salmonellosis due to multi-resistant Salmonella Typhimurium DT104 from consumption of Danish dry-cured pork sausages. Prev. Vet. Med. 2002, 52, 265. [Google Scholar] [CrossRef]
- Doménech, E.; Jiménez-Belenguer, A.; Pérez, R.; Ferrús, M.A.; Escriche, I. Risk characterization of antimicrobial resistance of Salmonella in meat products. J. Food Control 2015, 57, 23. [Google Scholar] [CrossRef]
- Collineau, L.; Chapman, B.; Bao, X.; Sivapathasundaram, B.; Carson, C.A.; Fazil, A.; Reid-Smith, R.J.; Smith, B.A. A farm-to-fork quantitative risk assessment model for Salmonella Heidelberg resistant to third-generation cephalosporins in broiler chickens in Canada. Int. J. Food Microbiol. 2020, 330, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, B.; Xu, D.; Wu, C.; Li, J.; Zheng, Y. Antimicrobial Resistance Risk Assessment Models and Database System for Animal-Derived Pathogens. Antibiotics 2020, 9, 829. [Google Scholar] [CrossRef] [PubMed]
- Andrews, W.H.; Jacobson, A.; Hammack, T. Chapter 5: Salmonella. In Bacteriological Analytical Manual (BAM); U.S. Food and Drug Administration: Silver Spring, MD, USA, 2018. [Google Scholar]
- Haas, C.N.; Rose, J.B.; Gerba, C.P. Quantitative Microbial Risk Assessment; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1999; p. 449. [Google Scholar]
- FAO; WHO. Risk Assessments of Salmonella in Eggs and Broiler Chickens; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2002; p. 302. [Google Scholar]
- M07-A11; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 10th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018.
- M100-S30; Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2020.
- Ciesielczuk, H.; Hornsey, M.; Choi, V.; Woodford, N.; Wareham, D.W. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 2013, 62, 1827. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, H.; Liu, Y.; Zhou, X.; Wang, J.; Liu, T.; Beier, R.C.; Hou, X. Characterization of quinolone resistance in Salmonella enterica serovar Indiana from chickens in China. Poult. Sci. 2015, 94, 460. [Google Scholar] [CrossRef]
- Alali, W.Q.; Mann, D.A.; Beuchat, L.R. Viability of Salmonella and Listeria monocytogenes in delicatessen salads and hummus as affected by sodium content and storage temperature. J. Food Prot. 2012, 75, 1056. [Google Scholar] [CrossRef]
- Ruchusatsawat, K.; Nuengjamnong, C.; Tawatsin, A.; Thiemsing, L.; Kawidam, C.; Somboonna, N.; Nuanualsuwan, S. Quantitative Risk Assessments of Hepatitis A Virus and Hepatitis E Virus from Raw Oyster Consumption. Risk Anal. 2022, 42, 965. [Google Scholar] [CrossRef]
- Nuanualsuwan, S.; Songkasupa, T.; Boonpornprasert, P.; Suwankitwat, N.; Lohlamoh, W.; Nuengjamnong, C. Thermal Inactivation of African Swine Fever Virus in Swill. Front. Vet. Sci. 2022, 9, 8. [Google Scholar] [CrossRef]
- Bywater, R.J.; Casewell, M.W. An assessment of the impact of antibiotic resistance in different bacterial species and of the contribution of animal sources to resistance in human infections. J. Antimicrob. Chemother. 2000, 46, 645. [Google Scholar] [CrossRef]
- Presi, P.; Stark, K.D.; Stephan, R.; Breidenbach, E.; Frey, J.; Regula, G. Risk scoring for setting priorities in a monitoring of antimicrobial resistance in meat and meat products. Int. J. Food Microbiol. 2009, 130, 100. [Google Scholar] [CrossRef] [PubMed]
- Berends, B.R.; van den Bogaard, A.E.; Van Knapen, F.; Snijders, J.M. Human health hazards associated with the administration of antimicrobials to slaughter animals. Part II. An assessment of the risks of resistant bacteria in pigs and pork. Vet. Q. 2001, 23, 21. [Google Scholar] [CrossRef]
- Hald, T.; Lo Fo Wong, D.M.; Aarestrup, F.M. The attribution of human infections with antimicrobial resistant Salmonella bacteria in Denmark to sources of animal origin. Foodborne Pathog. Dis. 2007, 4, 326. [Google Scholar] [CrossRef]
- Sommer, H.M.; Aabo, S.; Christensen, B.B.; Saadby, P.; Nielsen, N.; Nørrung, B.; Wong, D.L.F. Risk Assessment of the Impact on Human Health Related to Multiresistant Salmonella Typhimurium DT104 from Slaughter Pigs; Institute of Food Safety and Nutrition: Mørkhøj, Denmark, 2003; p. 104.
- European Food Safety Authority (EFSA). Scientific Opinion of the Panel on Biological Hazards on a request from the European Food Safety Authority on foodborne antimicrobial resistance as a biological hazard. EFSA 2008, 765, 87. [Google Scholar]
- Aslam, M.; Checkley, S.; Avery, B.; Chalmers, G.; Bohaychuk, V.; Gensler, G.; Reid-Smith, R.; Boerlin, P. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 2012, 32, 117. [Google Scholar] [CrossRef]
- Patchanee, P.; Tansiricharoenkul, K.; Buawiratlert, T.; Wiratsudakul, A.; Angchokchatchawal, K.; Yamsakul, P.; Yano, T.; Boonkhot, P.; Rojanasatien, S.; Tadee, P. Salmonella in pork retail outlets and dissemination of its pulsotypes through pig production chain in Chiang Mai and surrounding areas, Thailand. Prev. Vet. Med. 2016, 130, 105. [Google Scholar] [CrossRef]
- Prasertsee, T.; Chokesajjawatee, N.; Santiyanont, P.; Chuammitri, P.; Deeudom, M.; Tadee, P.; Patchanee, P. Quantification and rep-PCR characterization of Salmonella spp. in retail meats and hospital patients in Northern Thailand. Zoonoses Public Health 2019, 66, 309. [Google Scholar] [CrossRef]
- Sanguankiat, A.; Pinthong, R.; Padungtod, P.; Baumann, M.P.; Zessin, K.H.; Srikitjakarn, L.; Fries, R. A cross-sectional study of Salmonella in pork products in Chiang Mai, Thailand. Foodborne Pathog. Dis. 2010, 7, 878. [Google Scholar] [CrossRef]
- Tadee, P.; Boonkhot, P.; Pornruangwong, S.; Patchanee, P. Comparative phenotypic and genotypic characterization of Salmonella spp. in pig farms and slaughterhouses in two provinces in northern Thailand. PLoS ONE 2015, 10, 11. [Google Scholar] [CrossRef]
- Tadee, P.; Kumpapong, K.; Sinthuya, D.; Yamsakul, P.; Chokesajjawatee, N.; Nuanualsuwan, S.; Pornsukarom, S.; Molla, B.Z.; Gebreyes, W.A.; Patchanee, P. Distribution, quantitative load and characterization of Salmonella associated with swine farms in upper-northern Thailand. J. Vet. Sci. 2014, 15, 334. [Google Scholar] [CrossRef]
- European Commission. Microbiological Criteria for Foodstuffs: Commission Regulation (EC) No 2073/2005 on 2005; European Commission: Brussels, Belgium, 2005; pp. 1–26. [Google Scholar]
- Hald, T.; Duarte, A.R.; Stärk, K. Quantifying human exposure to antimicrobial resistance from animals and food. In Proceedings of the International Conference on Animal Health Surveillance, Wellington, New Zealand, 30 April–4 May 2017; p. 2. [Google Scholar]
- Biosecurity Australia. Generic Import Risk Analysis Report for Chicken Meat: Final Report; Biosecurity Australia: Canberra, Australia, 2008.
- Hurd, H.S.; Vaughn, M.B.; Holtkamp, D.; Dickson, J.; Warnick, L. Quantitative risk from fluoroquinolone-resistant Salmonella and Campylobacter due to treatment of dairy heifers with enrofloxacin for bovine respiratory disease. J. Foodborne Pathog. 2010, 7, 1322. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S.; Ruegg, P.L.; Bauman, D.E. Quantitative Risk Assessment of Antimicrobial-Resistant Foodborne Infections in Humans Due to Recombinant Bovine Somatotropin Usage in Dairy Cows. J. Food Prot. 2017, 80, 1116. [Google Scholar] [CrossRef] [PubMed]
- European Agency for the Evaluation of Medicinal Products. Antibiotic Resistance in the European Union Associated with Therapeutic Use of Veterinary Medicines: Report and Qualitative Risk Assessment by the Committee for Veterinary Medicinal Products; European Agency for the Evaluation of Medicinal Products: Amsterdam, The Netherlands, 1999.
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect Dis 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Hatrongjit, R.; Kerdsin, A.; Akeda, Y.; Hamada, S. Detection of plasmid-mediated colistin-resistant and carbapenem-resistant genes by multiplex PCR. MethodsX 2018, 5, 532–536. [Google Scholar] [CrossRef]
- Khanawapee, A.; Kerdsin, A.; Chopjitt, P.; Boueroy, P.; Hatrongjit, R.; Akeda, Y.; Tomono, K.; Nuanualsuwan, S.; Hamada, S. Distribution and Molecular Characterization of Escherichia coli Harboring mcr Genes Isolated from Slaughtered Pigs in Thailand. Microb. Drug Resist. 2021, 27, 971–979. [Google Scholar] [CrossRef]
| Retail | No. of Salmonella | Total | |
|---|---|---|---|
| Susceptible | QR | ||
| Fresh market | 30 | 28 | 58 (n =100) |
| Modern trade | 6 | 0 | 6 (n = 50) |
| Retail | PPROB (%) | Mean Concentration ± SD (log cfu/g) | ||||
|---|---|---|---|---|---|---|
| Salmonella spp. | Total | Salmonella spp. | Total * | |||
| Susceptible | QR | Susceptible | QR | |||
| Fresh market | 30.4 | 28.4 | 57.8 | 1.5 ± 0.8 | 2.1 ± 0.7 | 1.8 ± 0.8 |
| Modern trade | 13.5 | 1.9 | 6.9 | 1.9 ± 0.9 | 0 | 1.9 ± 0.9 |
| Retail | PE | PI | PMI | |||
|---|---|---|---|---|---|---|
| Salmonella spp. | Salmonella spp. | Salmonella | ||||
| Susceptible | QR | Susceptible | QR | Susceptible | QR | |
| Fresh market | 0.020 | 0.030 | 1.8 × 10−4 | 3.1 × 10−4 | 2.3 × 10−7 | 5.4 × 10−6 |
| Modern trade | 0.016 | 2 × 10−7 | 3.2 × 10−4 | 2.2 × 10−8 | 4.2 × 10−7 | 3.9 × 10−10 |
| Retail | Risk Estimate | Annual Cases * | |||
|---|---|---|---|---|---|
| Salmonella spp. | Salmonella spp. | ||||
| Susceptible | AMR | Susceptible | AMR | ||
| Fresh market | min | 5.3 × 10−13 | 8.8 × 10−11 | <1 | <1 |
| mean | 5.7 × 10−9 | 2.0 × 10−7 | <1 a | 7 b | |
| max | 2.7 × 10−8 | 8.8 × 10−7 | 1 | 32 | |
| Modern trade | min | 1.5 × 10−12 | 4.2 × 10−21 | <1 | <1 |
| mean | 7.9 × 10−9 | 7.4 × 10−17 | <1 c | <1 d | |
| max | 4.0 × 10−8 | 7.6 × 10−16 | 2 | <1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pulsrikarn, C.; Kedsin, A.; Boueroy, P.; Chopjitt, P.; Hatrongjit, R.; Chansiripornchai, P.; Suanpairintr, N.; Nuanualsuwan, S. Quantitative Risk Assessment of Susceptible and Ciprofloxacin-Resistant Salmonella from Retail Pork in Chiang Mai Province in Northern Thailand. Foods 2022, 11, 2942. https://doi.org/10.3390/foods11192942
Pulsrikarn C, Kedsin A, Boueroy P, Chopjitt P, Hatrongjit R, Chansiripornchai P, Suanpairintr N, Nuanualsuwan S. Quantitative Risk Assessment of Susceptible and Ciprofloxacin-Resistant Salmonella from Retail Pork in Chiang Mai Province in Northern Thailand. Foods. 2022; 11(19):2942. https://doi.org/10.3390/foods11192942
Chicago/Turabian StylePulsrikarn, Chaiwat, Anusak Kedsin, Parichart Boueroy, Peechanika Chopjitt, Rujirat Hatrongjit, Piyarat Chansiripornchai, Nipattra Suanpairintr, and Suphachai Nuanualsuwan. 2022. "Quantitative Risk Assessment of Susceptible and Ciprofloxacin-Resistant Salmonella from Retail Pork in Chiang Mai Province in Northern Thailand" Foods 11, no. 19: 2942. https://doi.org/10.3390/foods11192942
APA StylePulsrikarn, C., Kedsin, A., Boueroy, P., Chopjitt, P., Hatrongjit, R., Chansiripornchai, P., Suanpairintr, N., & Nuanualsuwan, S. (2022). Quantitative Risk Assessment of Susceptible and Ciprofloxacin-Resistant Salmonella from Retail Pork in Chiang Mai Province in Northern Thailand. Foods, 11(19), 2942. https://doi.org/10.3390/foods11192942








