Anti-Spoilage Activity and Exopolysaccharides Production by Selected Lactic Acid Bacteria
Abstract
:1. Introduction
2. Material and Methods
2.1. Lactic Acid Bacteria Strains
2.2. Antimould Activity Screening
2.3. Antibacterial Activity Screening
2.4. Screening for EPSs Production
2.4.1. EPS Extraction Procedure
2.4.2. EPSs Characterization
ATR-FTIR Spectroscopy
ESI-ION TRAP LC MS/MS (LCMSIT) Analysis
2.5. Statistical Analysis
3. Results and Discussion
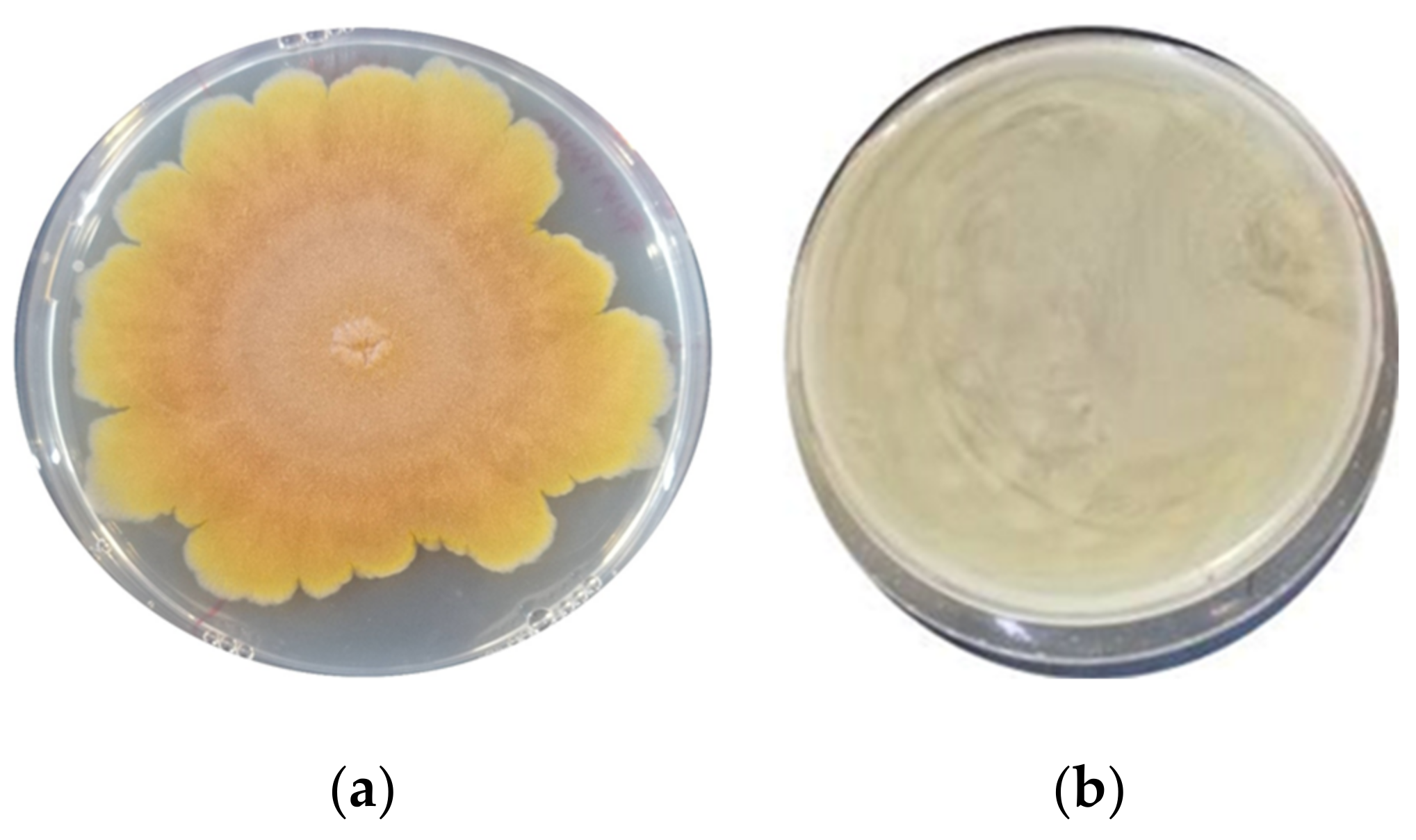
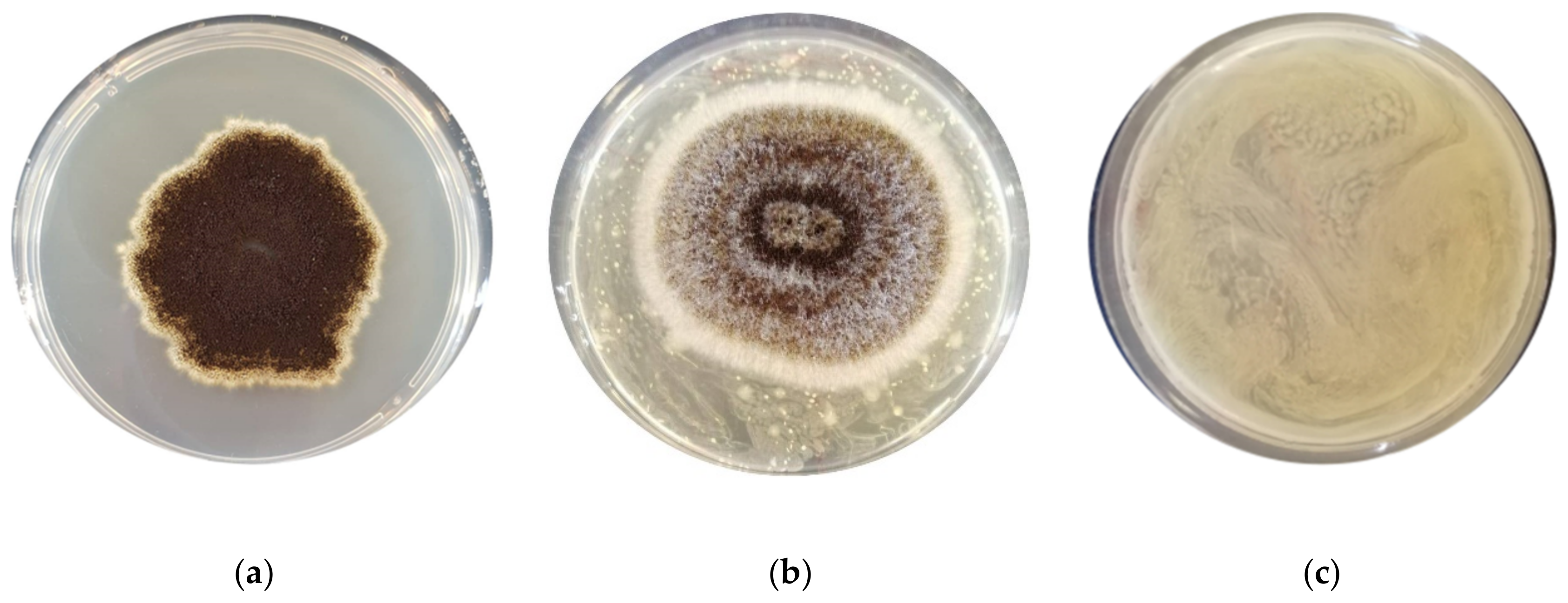
3.1. Assessment of the Antimould Activity
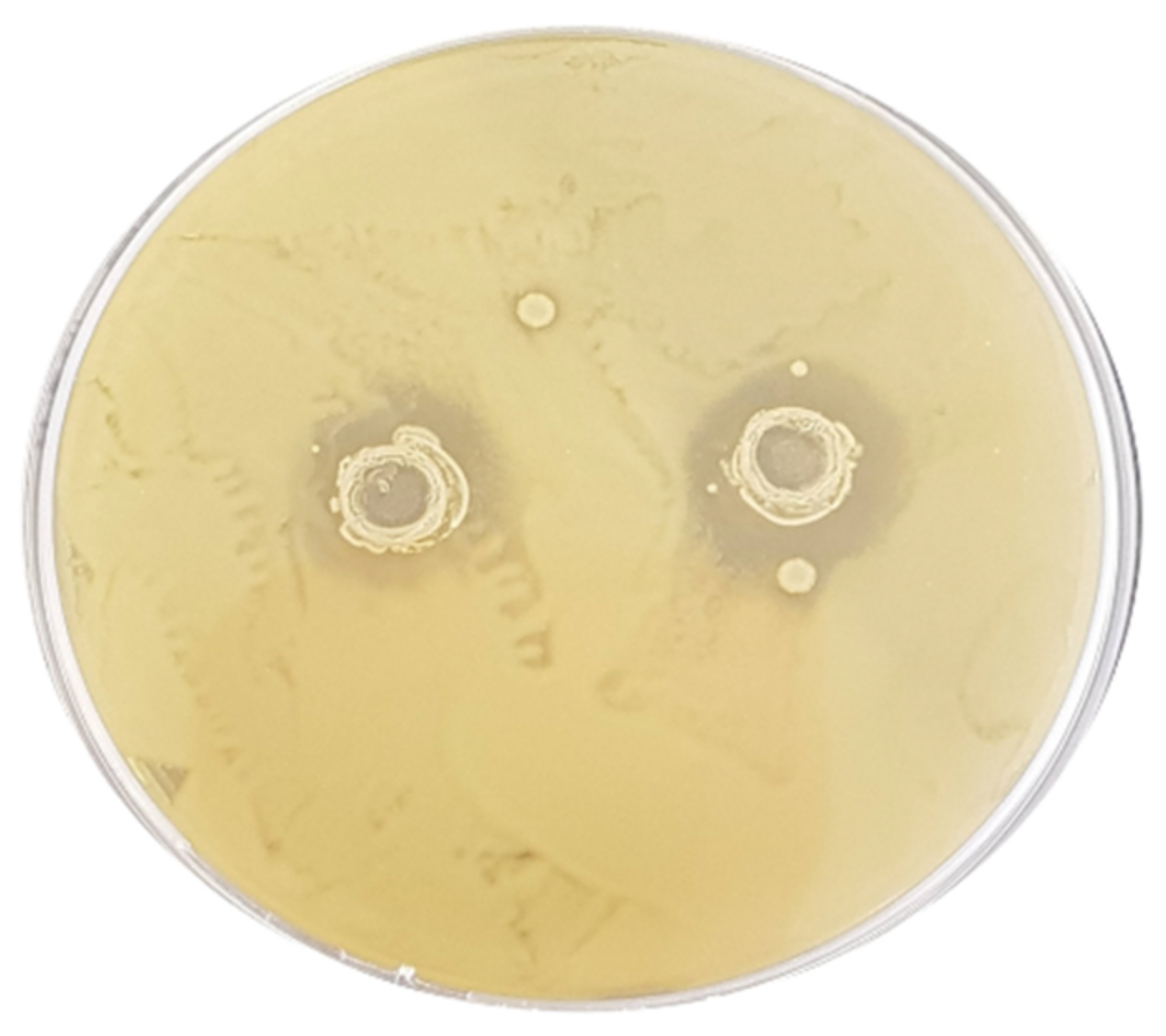
3.2. Assessment of the Antibacterial Activity
3.3. Characterization of the EPSs Produced by LAB Strains
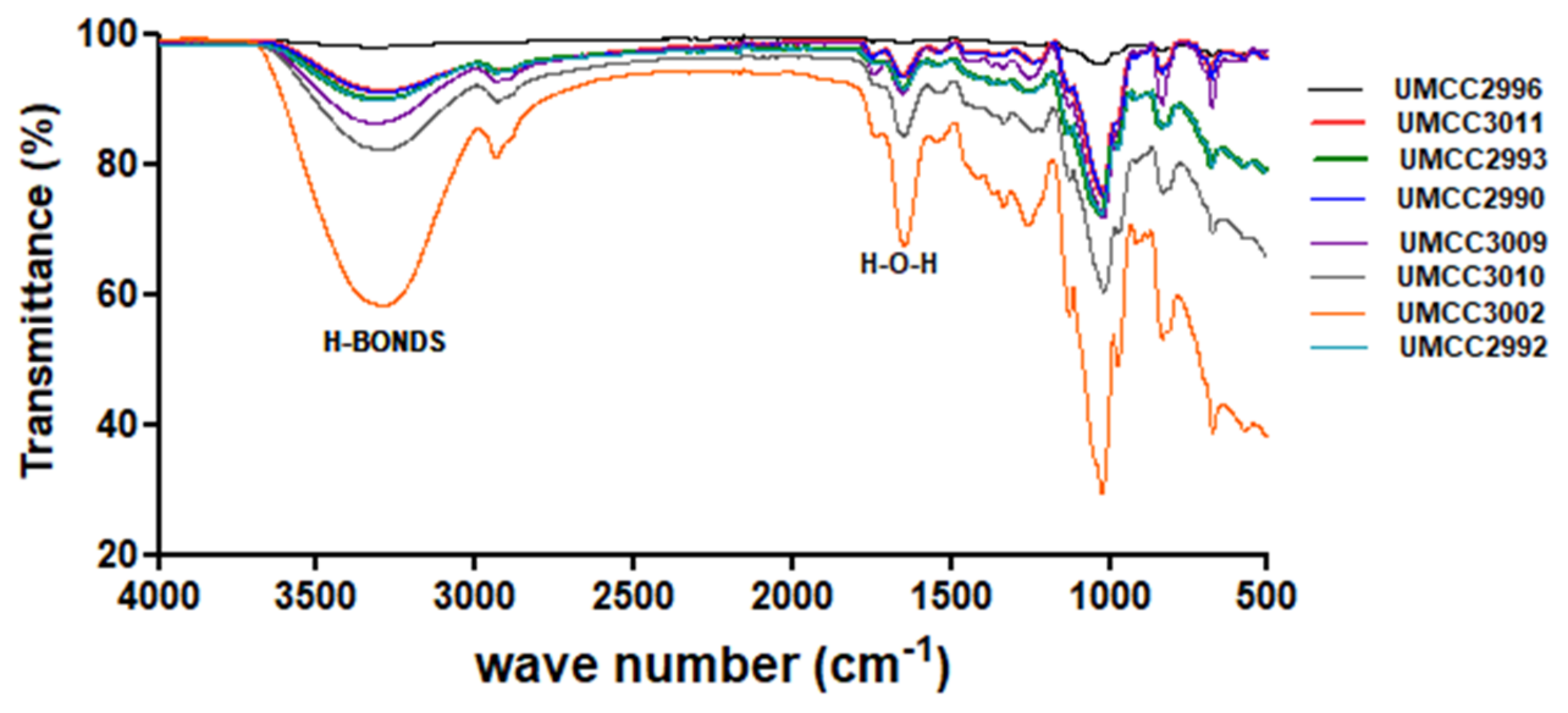
3.3.1. ATR-FTIR Analysis
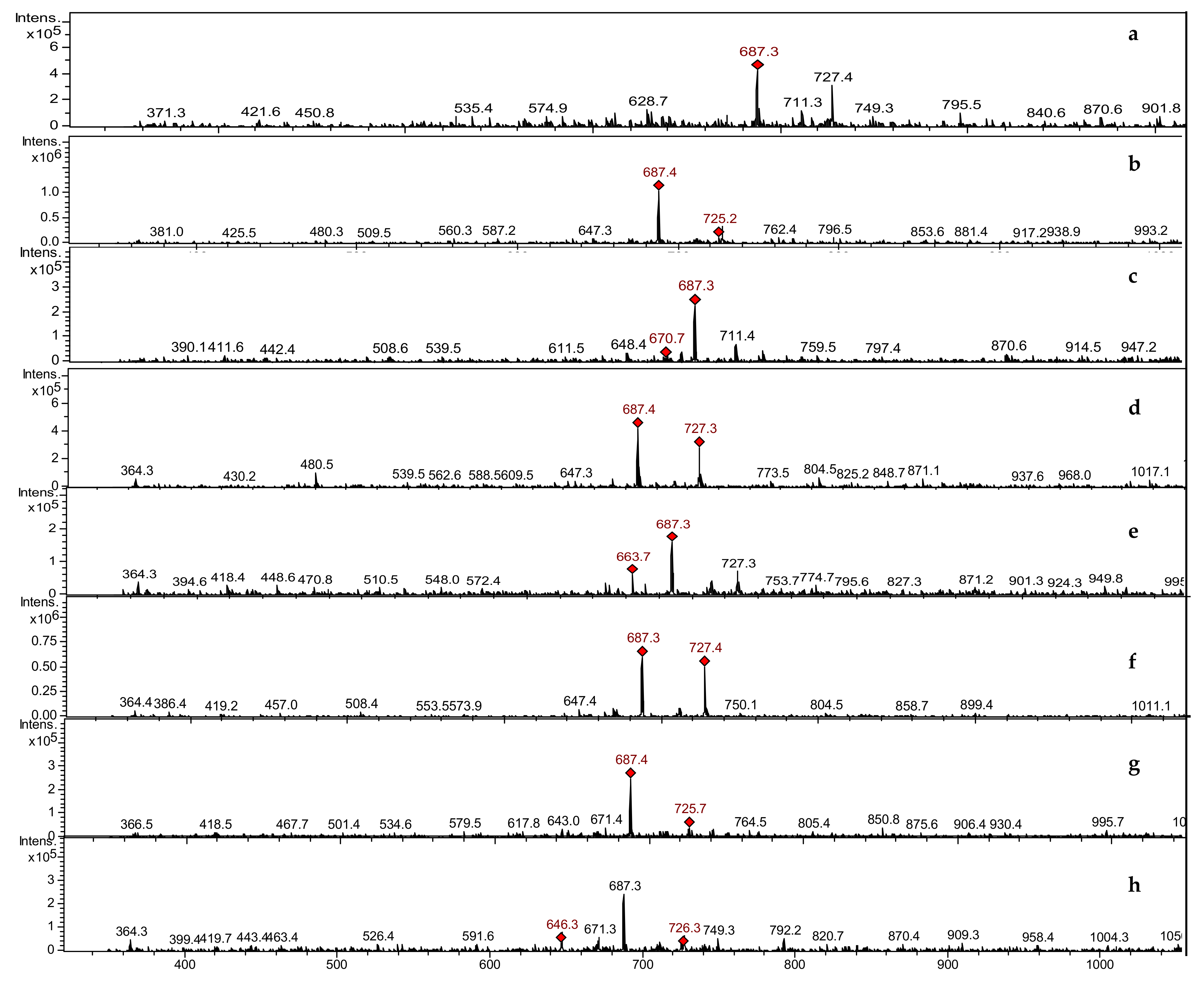
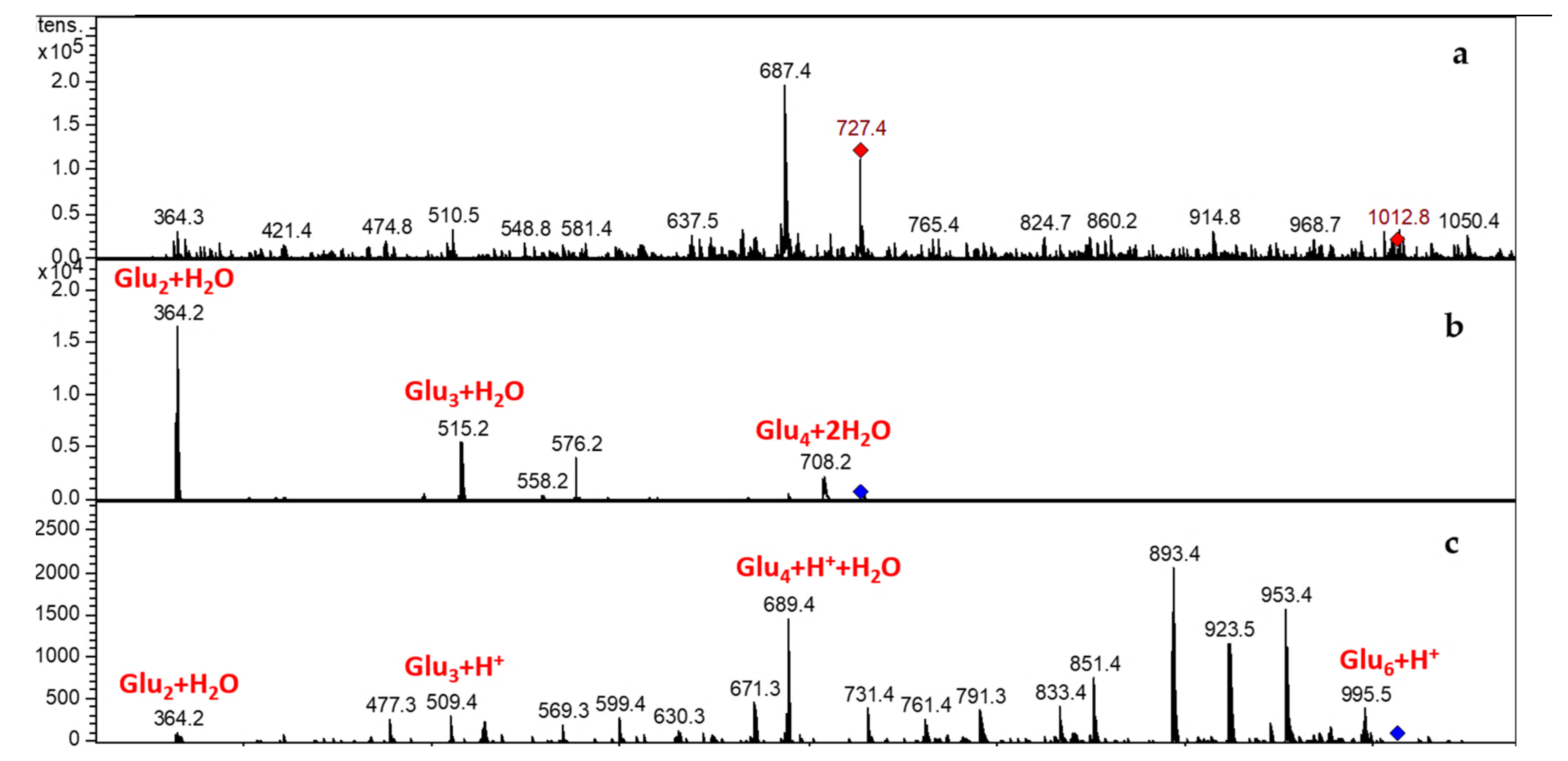
3.3.2. LCMSIT Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Vero, L.; Iosca, G.; Gullo, M.; Pulvirenti, A. Functional and healthy features of conventional and non-conventional sourdoughs. Appl. Sci. 2021, 11, 3694. [Google Scholar] [CrossRef]
- Gadhoumi, H.; Gullo, M.; De Vero, L.; Martinez-Rojas, E.; Saidani Tounsi, M.; Hayouni, E.A. Design of a new fermented beverage from medicinal plants and organic sugarcane molasses via lactic fermentation. Appl. Sci. 2021, 11, 6089. [Google Scholar] [CrossRef]
- Linares, D.M.; Gómez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C. Lactic Acid Bacteria and Bifidobacteria with Potential to Design Natural Biofunctional Health-Promoting Dairy Foods. Front. Microbiol. 2017, 8, 846. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health benefits of lactic acid bacteria (Lab) fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef] [PubMed]
- Al-Sahlany, S.T.G.; Niamah, A.K. Bacterial viability, antioxidant stability, antimutagenicity and sensory properties of onion types fermentation by using probiotic starter during storage. Nutr. Food Sci. 2022. [Google Scholar] [CrossRef]
- Cao, H.; Meng, D.; Zhang, W.; Ye, T.; Yuan, M.; Yu, J.; Wu, X.; Li, Y.; Yin, F.; Fu, C.; et al. Growth inhibition of Fusarium graminearum and deoxynivalenol detoxification by lactic acid bacteria and their application in sourdough bread. Int. J. Food Sci. Technol. 2020, 6, 2304–2314. [Google Scholar] [CrossRef]
- Zannini, E.; Pontonio, E.; Waters, D.M.; Arendt, E.K. Applications of microbial fermentations for production of gluten-free products and perspectives. Appl. Microbiol. Biotechnol. 2012, 93, 473–485. [Google Scholar] [CrossRef]
- Bartkiene, E.; Lele, V.; Ruzauskas, M.; Domig, K.J.; Starkute, V.; Zavistanaviciute, P.; Bartkevics, V.; Pugajeva, I.; Klupsaite, D.; Juodeikiene, G.; et al. Lactic acid bacteria isolation from spontaneous sourdough and their characterization including antimicrobial and antifungal properties evaluation. Microorganisms 2020, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Hajinia, F.; Sadeghi, A.; Sadeghi Mahoonak, A. The use of antifungal oat-sourdough lactic acid bacteria to improve safety and technological functionalities of the supplemented wheat bread. J. Food Saf. 2020, 41, e12873. [Google Scholar] [CrossRef]
- Nasrollahzadeh, A.; Mokhtari, S.; Khomeiri, M.; Saris, P.E.J. Antifungal Preservation of Food by Lactic Acid Bacteria. Foods 2022, 11, 395. [Google Scholar] [CrossRef]
- Axel, C.; Brosnan, B.; Zannini, E.; Peyer, L.C.; Furey, A.; Coffey, A.; Arendt, E.K. Antifungal activities of three different Lactobacillus species and their production of antifungal carboxylic acids in wheat sourdough. Appl. Microbiol. Biotechnol. 2016, 100, 1701–1711. [Google Scholar] [CrossRef]
- Capozzi, V.; Fragasso, M.; Romaniello, R.; Berbegal, C.; Russo, P.; Spano, G. Spontaneous food fermentations and potential risks for human health. Fermentation 2017, 3, 49. [Google Scholar] [CrossRef]
- Abdalla, A.K.; Ayyash, M.M.; Olaimat, A.N.; Osaili, T.M.; Al-Nabulsi, A.A.; Shah, N.P.; Holley, R. Exopolysaccharides as Antimicrobial Agents: Mechanism and Spectrum of Activity. Front. Microbiol. 2021, 12, 664395. [Google Scholar] [CrossRef]
- La China, S.; De Vero, L.; Anguluri, K.; Brugnoli, M.; Mamlouk, D.; Gullo, M. Kombucha Tea as a Reservoir of Cellulose Producing Bacteria: Assessing Diversity among Komagataeibacter Isolates. Appl. Sci. 2021, 11, 1595. [Google Scholar] [CrossRef]
- Anguluri, K.; La China, S.; Brugnoli, M.; De Vero, L.; Pulvirenti, A.; Cassanelli, S.; Gullo, M. Candidate Acetic acid bacteria strains for levan production. Polymers 2022, 14, 2000. [Google Scholar] [CrossRef]
- Zhou, Y.; Cui, Y.; Qu, X. Exopolysaccharides of lactic acid bacteria: Structure, bioactivity and associations: A review. Carbohydr. Polym. 2019, 207, 317–332. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Biosynthesis to Health-Promoting Properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Korcz, E.; Varga, L. Exopolysaccharides from lactic acid bacteria: Techno-functional application in the food industry. Trends Food Sci. Technol. 2021, 110, 375–384. [Google Scholar] [CrossRef]
- Angelin, J.; Kavitha, M. Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 2020, 162, 853–865. [Google Scholar] [CrossRef]
- Castro-Bravo, N.; Wells, J.M.; Margolles, A.; Ruas-Madiedo, P. Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment. Front. Microbiol. 2018, 9, 2426. [Google Scholar] [CrossRef] [Green Version]
- Nehal, F.; Sahnoun, M.; Smaoui, S.; Jaouadi, B.; Bejar, S.; Mohammed, S. Characterization, high production and antimicrobial activity of exopolysaccharides from Lactococcus lactis F-mou. Microb. Pathog. 2019, 132, 10–19. [Google Scholar] [CrossRef]
- Dertli, E.; Mayer, M.J.; Narbad, A. Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol. 2015, 15, 22. [Google Scholar] [CrossRef] [Green Version]
- Crowley, S.; Mahony, J.; van Sinderen, D. Current Perspectives on Antifungal Lactic Acid Bacteria as Natural Bio-Preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Axel, C.; Röcker, B.; Brosnan, B.; Zannini, E.; Furey, A.; Coffey, A.; Arendt, E.K. Application of Lactobacillus amylovorus DSM19280 in gluten-free sourdough bread to improve the microbial shelf life. Food Microbiol. 2015, 47, 36–44. [Google Scholar] [CrossRef]
- De Vero, L.; Iosca, G.; La China, S.; Licciardello, F.; Gullo, M.; Pulvirenti, A. Yeasts and lactic acid bacteria for panettone production: An assessment of candidate strains. Microorganisms 2021, 9, 1093. [Google Scholar] [CrossRef]
- Iosca, G.; Febo, C.; Haghighi, H.; De Vero, L.; Gullo, M.; Pulvirenti, A. Selection of Promising Exopolysaccharide-Producing Starter Cultures for Gluten-Free Sourdough. Biol. Life Sci. Forum 2021, 6, 38. [Google Scholar] [CrossRef]
- Hassan, Y.I.; Bullerman, L.B. Antifungal activity of Lactobacillus paracasei ssp. tolerans isolated from a sourdough bread culture. Int. J. Food Microbiol. 2008, 121, 112–115. [Google Scholar] [CrossRef]
- Chen, Y.; Dai, G. Antifungal activity of plant extracts against Colletotrichum lagenarium, the causal agent of anthracnose in cucumber. J. Sci. Food Agric. 2012, 92, 1937–1943. [Google Scholar] [CrossRef]
- Arrioja-Bretón, D.; Mani-López, E.; Palou, E.; López-Malo, A. Antimicrobial activity and storage stability of cell-free supernatants from lactic acid bacteria and their applications with fresh beef. Food Control 2020, 115, 107286. [Google Scholar] [CrossRef]
- Ruiz Rodríguez, L.; Vera Pingitore, E.; Rollan, G.; Cocconcelli, P.; Fontana, C.; Saavedra, L.; Vignolo, G.; Hebert, E.M. Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented quinoa sourdoughs. J. Appl. Microbiol. 2016, 120, 1289–1301. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, P.T.; Nguyen, T.T.; Bui, D.C.; Hong, P.T.; Hoang, Q.K.; Nguyen, H.T. Exopolysaccharide production by lactic acid bacteria: The manipulation of environmental stresses for industrial applications. AIMS Microbiol. 2020, 6, 451–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ahmed, Z.; Feng, W.; Li, C.; Song, S. Physicochemical properties of exopolysaccharide produced by Lactobacillus kefiranofaciens ZW3 isolated from Tibet kefir. Int. J. Biol. Macromol. 2008, 43, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Vaaje-kolstad, G.; Westereng, B.; Bunsæ, A.C.; Stenstrøm, Y.; Mackenzie, A.; Sørlie, M.; Horn, S.J.; Eijsink, V.G.H. Cleavage of cellulose by a cbm33 protein. Protein Sci. 2011, 20, 1479–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallagnol, A.M.; Bustos, A.Y.; Martos, G.I.; de Valdez, G.F.; Gerez, C.L. Antifungal and antimycotoxigenic effect of Lactobacillus plantarum CRL 778 at different water activity values. Rev. Argent. Microbiol. 2019, 51, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Nguyen, T.T.H.; Humayun, S.; Park, S.H.; Oh, H.; Lim, S.; Mok, I.K.; Li, Y.; Pal, K.; Kim, D. Characteristics of sourdough bread fermented with Pediococcus pentosaceus and Saccharomyces cerevisiae and its bio-preservative effect against Aspergillus flavus. Food Chem. 2021, 345, 128787. [Google Scholar] [CrossRef]
- Valerio, F.; Favilla, M.; De Bellis, P.; Sisto, A.; de Candia, S.; Lavermicocca, P. Antifungal activity of strains of lactic acid bacteria isolated from a semolina ecosystem against Penicillium roqueforti, Aspergillus niger and Endomyces fibuliger contaminating bakery products. Syst. Appl. Microbiol. 2009, 32, 438–448. [Google Scholar] [CrossRef]
- Corsetti, A.; Gobbetti, M.; Rossi, J.; Damiani, P. Antimould activity of sourdough lactic acid bacteria: Identification of a mixture of organic acids produced by Lactobacillus sanfrancisco CB1. Appl. Microbiol. Biotechnol. 1998, 50, 253–256. [Google Scholar] [CrossRef]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- Rocha, J.M.; Malcata, F.X. Microbial Ecology Dynamics in Portuguese Broa Sourdough. J. Food Qual. 2016, 39, 634–648. [Google Scholar] [CrossRef]
- Ndagano, D.; Lamoureux, T.; Dortu, C.; Vandermoten, S.; Thonart, P. Antifungal Activity of 2 Lactic Acid Bacteria of the Weissella Genus Isolated from Food. J. Food Sci. 2011, 76, M305–M311. [Google Scholar] [CrossRef]
- Black, B.A.; Zannini, E.; Curtis, J.M.; Gänzle, M.G. Antifungal hydroxy fatty acids produced during sourdough fermentation: Microbial and enzymatic pathways, and antifungal activity in bread. Appl. Environ. Microbiol. 2013, 79, 1866–1873. [Google Scholar] [CrossRef] [Green Version]
- Divyashree, S.; Anjali, P.G.; Somashekaraiah, R.; Sreenivasa, M.Y. Probiotic properties of Lactobacillus casei–MYSRD 108 and Lactobacillus plantarum-MYSRD 71 with potential antimicrobial activity against Salmonella paratyphi. Biotechnol. Rep. 2021, 32, e00672. [Google Scholar] [CrossRef]
- Thao, T.T.P.; Thoa, L.T.K.; Ngoc, L.M.T.; Lan, T.T.P.; Phuong, T.V.; Truong, H.T.H.; Khoo, K.S.; Manickam, S.; Hoa, T.T.; Tram, N.D.Q.; et al. Characterization halotolerant lactic acid bacteria Pediococcus pentosaceus HN10 and in vivo evaluation for bacterial pathogens inhibition. Chem. Eng. Process.-Process Intensif. 2021, 168, 108576. [Google Scholar] [CrossRef]
- Pato, U.; Yusuf, Y.; Fitriani, S.; Fauzi, D.A. Antibacterial Activity of Bacteriocin from Pediococcus pentosaceus Strain 2397 and Application as Biopreservative for Fishballs. Philipp J. Sci. 2022, 151, 713–725. [Google Scholar]
- Koo, O.K.; Kim, S.M.; Kang, S.H. Antimicrobial potential of Leuconostoc species against E. coli O157:H7 in ground meat. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 831–838. [Google Scholar] [CrossRef]
- Siroli, L.; Patrignani, F.; Serrazanetti, D.I.; Gardini, F.; Lanciotti, R. Innovative strategies based on the use of bio-control agents to improve the safety, shelf-life and quality of minimally processed fruits and vegetables. Trends Food Sci. Technol. 2015, 46, 302–310. [Google Scholar] [CrossRef]
- Cizeikiene, D.; Juodeikiene, G.; Paskevicius, A.; Bartkiene, E. Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 2013, 31, 539–545. [Google Scholar] [CrossRef]
- Siedler, S.; Balti, R.; Neves, A.R. Bioprotective mechanisms of lactic acid bacteria against fungal spoilage of food. Curr. Opin. Biotechnol. 2019, 56, 138–146. [Google Scholar] [CrossRef]
- Cirrincione, S.; Breuer, Y.; Mangiapane, E.; Mazzoli, R.; Pessione, E. ’Ropy’ phenotype, exopolysaccharides and metabolism: Study on food isolated potential probiotics LAB. Microbiol. Res. 2018, 214, 137–145. [Google Scholar] [CrossRef]
- Daba, G.M.; Elnahas, M.O.; Elkhateeb, W.A. Contributions of exopolysaccharides from lactic acid bacteria as biotechnological tools in food, pharmaceutical, and medical applications. Int. J. Biol. Macromol. 2021, 173, 79–89. [Google Scholar] [CrossRef]
- Al-Roomi, F.W.; Al-Sahlany, S.T.G. Identification and Characterization of Xanthan Gum Produced from Date Juice by a Local Isolate of Bacteria Xanthomonas campestris. Basrah J. Agric. Sci. 2022, 35, 35–49. [Google Scholar] [CrossRef]
- Hospodarova, V.; Singovszka, E.; Stevulova, N. Characterization of Cellulosic Fibers by FTIR Spectroscopy for Their Further Implementation to Building Materials. Am. J. Anal. Chem. 2018, 9, 303–310. [Google Scholar] [CrossRef] [Green Version]
- Serra, I.; Piccinini, D.; Paradisi, A.; Ciano, L.; Bellei, M.; Bortolotti, C.A.; Battistuzzi, G.; Sola, M.; Walton, P.H.; Di Rocco, G. Activity and substrate specificity of lytic polysaccharide monooxygenases: An ATR FTIR-based sensitive assay tested on a novel species from Pseudomonas putida. Protein Sci. 2022, 31, 591–601. [Google Scholar] [CrossRef]
- Cichosz, S.; Masek, A. IR study on cellulose with the varied moisture contents: Insight into the supramolecular structure. Materials 2020, 13, 4573. [Google Scholar] [CrossRef]
- Jonoobi, M.; Harun, J.; Mathew, A.P.; Hussein, M.Z.B.; Oksman, K. Preparation of cellulose nanofibers with hydrophobic surface characteristics. Cellulose 2010, 17, 299–307. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Laws, A.; Gu, Y.; Marshall, V. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 2001, 19, 597–625. [Google Scholar] [CrossRef]
- Gonçalves, V.M.F.; Reis, A.; Domingues, M.R.M.; Lopes-da-Silva, J.A.; Fialho, A.M.; Moreira, L.M.; Sá-Correia, I.; Coimbra, M.A. Structural analysis of gellans produced by Sphingomonas elodea strains by electrospray tandem mass spectrometry. Carbohydr. Polym. 2009, 77, 10–19. [Google Scholar] [CrossRef]
- Wu, J.; Han, X.; Ye, M.; Li, Y.; Wang, X.; Zhong, Q. Exopolysaccharides synthesized by lactic acid bacteria: Biosynthesis pathway, structure-function relationship, structural modification and applicability. Crit. Rev. Food Sci. Nutr. 2022. [Google Scholar] [CrossRef]
| Species | Origin | |||
|---|---|---|---|---|
| UMCC ID | Other Denomination | Isolation Source | Region | |
| UMCC 2990 | BMA8 | Fructilactobacillus sanfranciscensis | sourdough type I | Emilia Romagna (Italy) |
| UMCC 2992 | BIA2 | Lentilactobacillus parabuchneri | dough for Panettone | Emilia Romagna (Italy) |
| UMCC 2993 | BIA6 | Leuconostoc citreum | dough for Panettone | Emilia Romagna (Italy) |
| UMCC 2996 | BFA1 | Lactiplantibacillus plantarum | dough for Panettone | Emilia Romagna (Italy) |
| UMCC 3002 | BIB7 | Furfurilactobacillus rossiae | dough for Panettone | Emilia Romagna (Italy) |
| UMCC 3011 | BFB3 | Leuconostoc citreum | dough for Panettone | Emilia Romagna (Italy) |
| UMCC 3009 | LC1RC13 | Lactiplantibacillus plantarum | gluten-free sourdough | Sicily (Italy) |
| UMCC 3010 | LL1C9 | Pediococcus pentosaceus | gluten-free sourdough | Sicily (Italy) |
| MOULDS | CONTROL | Leuc. citreum UMCC 3011 | Lacp. plantarum UMCC 2996 | P. pentosaceus UMCC 3010 | Lacp. plantarum UMCC 3009 | Leuc. citreum UMCC 2993 | Lenl. parabuchneri UMCC 2992 | Fl. sanfranciscensis UMCC 2990 | Furl. rossiae UMCC 3002 |
|---|---|---|---|---|---|---|---|---|---|
| F. graminearum ITEM 5356 | 32.2 ± 0.66 b | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a |
| A. flavus ITEM 7828 | 41.8 ± 0.32 c | 35.7 ± 0.38 b | 0 ± 0 a | 27.1 ± 0.93 b | 0 ± 0 a | 34.5 ± 0.34 bc | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a |
| P. paneum ITEM 13081 | 38.5± 0.53 ab | 35.5 ± 0.23 ab | 39.4 ± 0.47 b | 38.2 ± 0.21 ab | 39.4 ± 0.13 b | 37.0 ± 0.37 ab | 34.1 ± 0.19 a | 34.1 ± 0.19 a | 33.5 ± 0.40 a |
| A. niger ITEM 7090 | 31.8 ± 1.03 b | 0 ± 0 a | 33.2 ± 0.66 bc | 0 ± 0 a | 32.1 ± 0.53 b | 33.1 ± 0.38 bc | 0± 0 a | 39.4± 0.30 c | 0 ± 0 a |
| PATHOGEN STRAIN | Leuc. citreum UMCC 3011 | Lacp. plantarum UMCC 2996 | P. pentosaceus UMCC 3010 | Lacp. plantarum UMCC 3009 | Leuc. citreum UMCC 2993 | Lenl. parabuchneri UMCC 2992 | Fl. sanfranciscensis UMCC 2990 | Furl. rossiae UMCC 3002 |
|---|---|---|---|---|---|---|---|---|
| E. coli ATCC 43888 | 12.6 ± 0.17 Ad | 7.2 ± 0.27 Ac | 11.3 ± 0.24 Ad | 4.4 ± 0.16 Bb | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa |
| S. typhimurium ATCC 14028 | 13.4 ± 0.43 Ab | 13.4 ± 0.43 Bb | 11.3 ± 0.60 Ab | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa |
| L. monocytogenes ATCC 19115 | 12.3 ± 0.09 Aa | 11.8 ± 0.14 Ba | 18.0 ± 0.16 Bb | 10.9 ± 0.22 Ca | 10.7 ± 0.14 Ca | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa |
| C. jejuni ATCC 33250 | 13.1 ± 0.21 Ab | 14.6 ± 0.34 Bb | 15.6 ± 0.44 ABb | 11.6 ± 0.45 Cab | 8.2 ± 0.25 Ba | 0 ± 0 Aa | 0 ± 0 Aa | 0 ± 0 Aa |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iosca, G.; De Vero, L.; Di Rocco, G.; Perrone, G.; Gullo, M.; Pulvirenti, A. Anti-Spoilage Activity and Exopolysaccharides Production by Selected Lactic Acid Bacteria. Foods 2022, 11, 1914. https://doi.org/10.3390/foods11131914
Iosca G, De Vero L, Di Rocco G, Perrone G, Gullo M, Pulvirenti A. Anti-Spoilage Activity and Exopolysaccharides Production by Selected Lactic Acid Bacteria. Foods. 2022; 11(13):1914. https://doi.org/10.3390/foods11131914
Chicago/Turabian StyleIosca, Giovanna, Luciana De Vero, Giulia Di Rocco, Giancarlo Perrone, Maria Gullo, and Andrea Pulvirenti. 2022. "Anti-Spoilage Activity and Exopolysaccharides Production by Selected Lactic Acid Bacteria" Foods 11, no. 13: 1914. https://doi.org/10.3390/foods11131914
APA StyleIosca, G., De Vero, L., Di Rocco, G., Perrone, G., Gullo, M., & Pulvirenti, A. (2022). Anti-Spoilage Activity and Exopolysaccharides Production by Selected Lactic Acid Bacteria. Foods, 11(13), 1914. https://doi.org/10.3390/foods11131914










