Potential Application of Living Microorganisms in the Detoxification of Heavy Metals
Abstract
1. Introduction
2. Heavy Metal Poisoning: Health Risks and Mechanisms of Toxicity
3. The Conventional Therapeutics for Heavy Metal Poisoning and Their Limitations
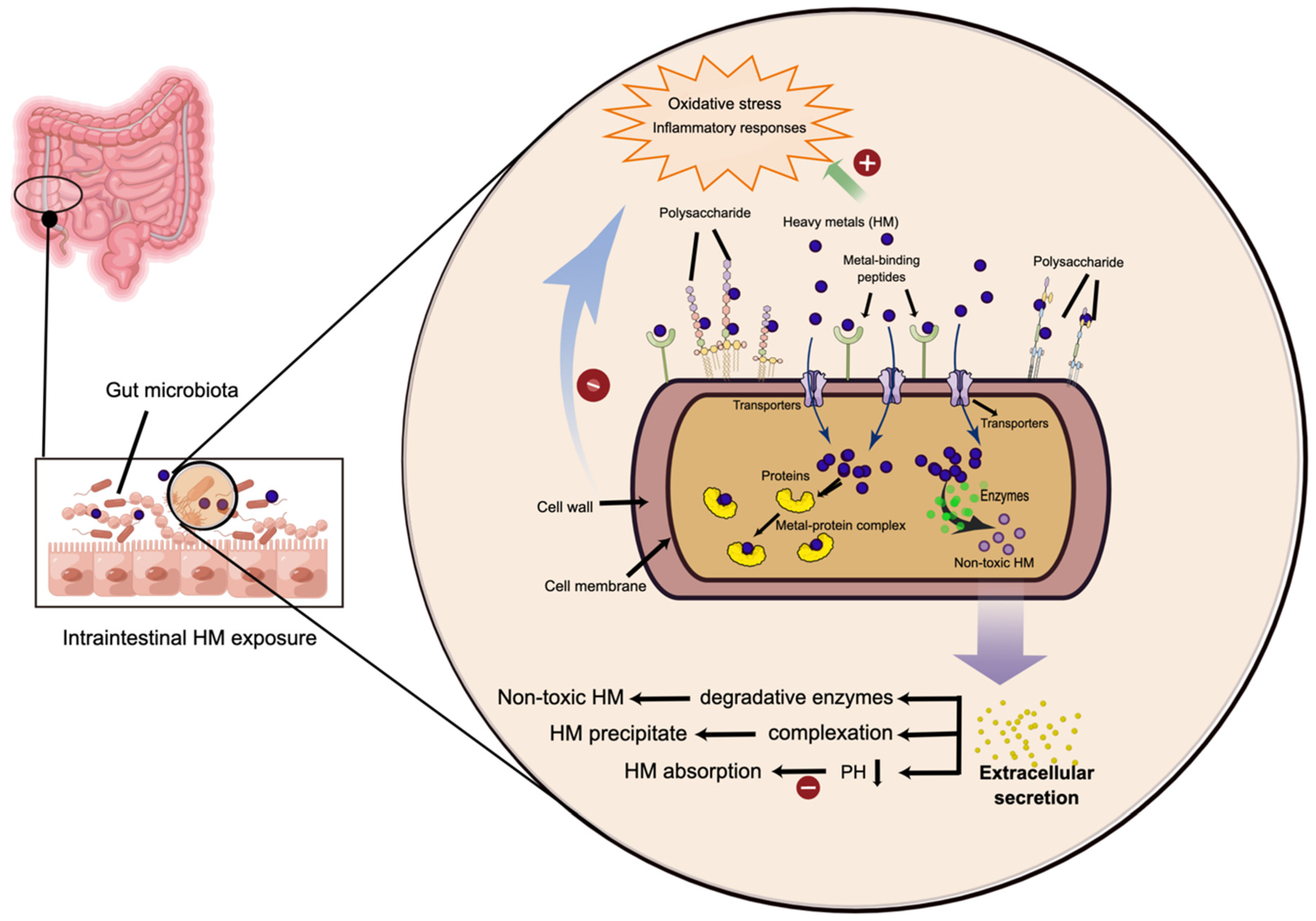
4. The Gut Microbiota: A Vital Mediator in the Heavy Metal-Induced Toxicity
5. Probiotic-Based Protective Strategies against Heavy Metal-Induced Gut Dysbiosis
6. Potential Application of Engineered Bacteria in the Detoxification of Heavy Metals
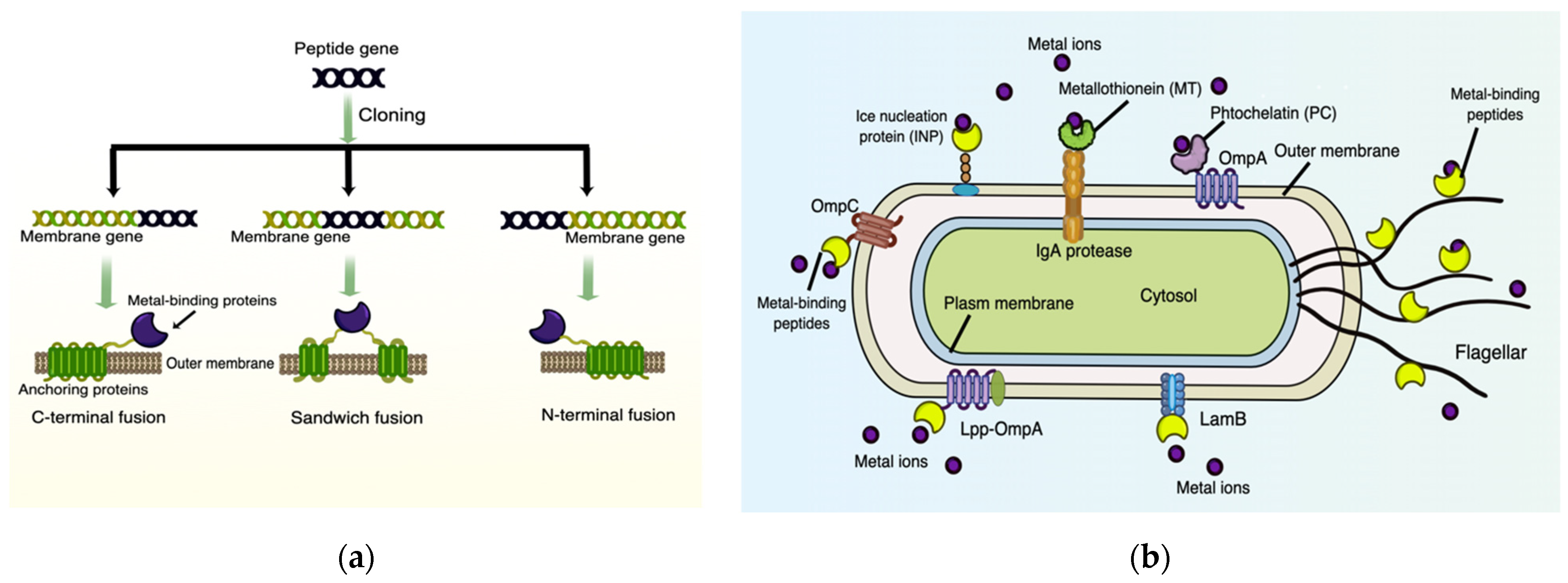
6.1. Surface-Displayed Proteins/Peptides for Heavy Metal Biosorption
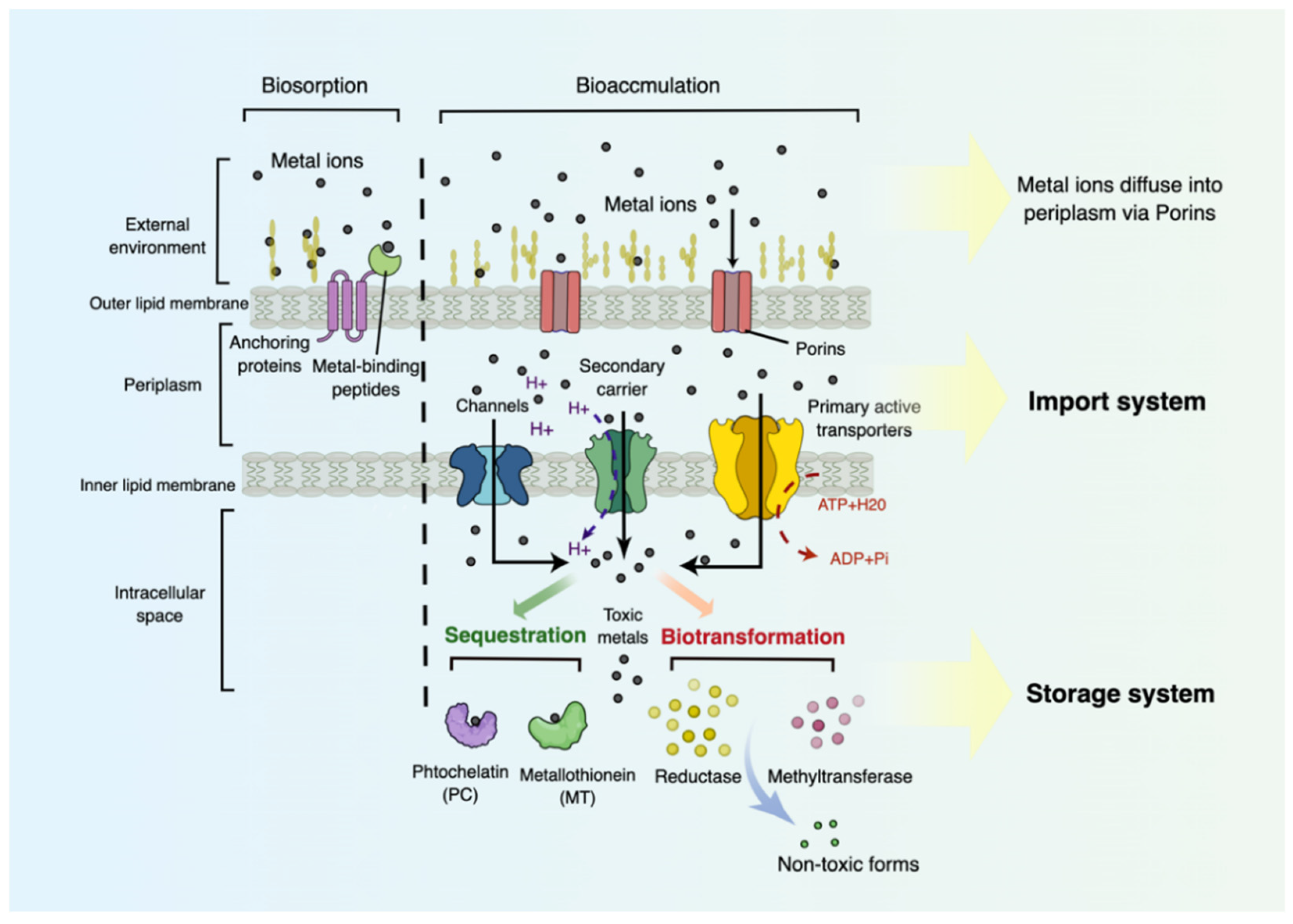
6.2. Transport and Storage Systems for Heavy Metal Bioaccumulation
6.3. In Vivo Attempts for Heavy Metal Detoxification by Engineered Strains
7. Current Limitations and Future Prospects of GEMs
8. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christoforidis, A.; Stamatis, N. Heavy metal contamination in street dust and roadside soil along the major national road in Kavala’s region, Greece. Geoderma 2009, 151, 257–263. [Google Scholar] [CrossRef]
- Chen, H.-W. Gallium, indium, and arsenic pollution of groundwater from a semiconductor manufacturing area of Taiwan. Bull. Environ. Contam. Toxicol. 2006, 77, 289–296. [Google Scholar] [CrossRef]
- Department of Industrial Statistics NBoSoCD. China Industrial Economy Statistical Yearbook; China Statistic Press: Beijing, China, 2011.
- Das, D.; Chatterjee, A.; Mandal, B.K.; Samanta, G.; Chakraborti, D.; Chanda, B. Arsenic in ground water in six districts of West Bengal, India: The biggest arsenic calamity in the world. Part 2. Arsenic concentration in drinking water, hair, nails, urine, skin-scale and liver tissue (biopsy) of the affected people. Analyst 1995, 120, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Hassan, T.; Majid, S. Heavy metal toxicity and their harmful effects on living organisms—A review. Int. J. Med. Sci. Diagn. Res. 2019, 3, 106–122. [Google Scholar]
- Flora, S. Metal poisoning: Threat and management. Al Ameen J. Med. Sci. 2009, 2, 4–26. [Google Scholar]
- Lentini, P.; Zanoli, L.; Granata, A.; Signorelli, S.S.; Castellino, P.; Dell’Aquila, R. Kidney and heavy metals-The role of environmental exposure. Mol. Med. Rep. 2017, 15, 3413–3419. [Google Scholar] [CrossRef] [PubMed]
- Amadi, C.N.; Offor, S.J.; Frazzoli, C.; Orisakwe, O.E. Natural antidotes and management of metal toxicity. Environ. Sci. Pollut. Res. Int. 2019, 26, 18032–18052. [Google Scholar] [CrossRef]
- Kim, J.J.; Kim, Y.S.; Kumar, V. Heavy metal toxicity: An update of chelating therapeutic strategies. J. Trace Elem. Med. Biol. 2019, 54, 226–231. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Rosenfeld, C.S. Gut Dysbiosis in Animals Due to Environmental Chemical Exposures. Front. Cell Infect Microbiol. 2017, 7, 396. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Dauros-Singorenko, P.; Hong, J.; Vanholsbeeck, F.; Phillips, A.; Swift, S. The role of host molecules in communication with the resident and pathogenic microbiota: A review. Med. Microecol. 2020, 4, 100005. [Google Scholar] [CrossRef]
- Yu, L.; Qiao, N.; Li, T.; Yu, R.; Zhai, Q.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Dietary supplementation with probiotics regulates gut microbiota structure and function in Nile tilapia exposed to aluminum. PeerJ 2019, 7, e6963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Kholodkevich, S.; Sharov, A.; Chen, C.; Feng, Y.; Ren, N.; Sun, K. Effects of cadmium on intestinal histology and microbiota in freshwater crayfish (Procambarus clarkii). Chemosphere 2020, 242, 125105. [Google Scholar] [CrossRef]
- Zhou, C.; Xu, P.; Huang, C.; Liu, G.; Chen, S.; Hu, G.; Li, G.; Liu, P.; Guo, X. Effects of subchronic exposure of mercuric chloride on intestinal histology and microbiota in the cecum of chicken. Ecotoxicol. Environ. Saf. 2020, 188, 109920. [Google Scholar] [CrossRef] [PubMed]
- Eggers, S.; Safdar, N.; Sethi, A.K.; Suen, G.; Peppard, P.E.; Kates, A.E.; Skarlupka, J.H.; Kanarek, M.; Malecki, K.M. Urinary lead concentration and composition of the adult gut microbiota in a cross-sectional population-based sample. Environ. Int. 2019, 133, 105122. [Google Scholar] [CrossRef]
- Lu, K.; Abo, R.P.; Schlieper, K.A.; Graffam, M.E.; Levine, S.; Wishnok, J.S.; Swenberg, J.A.; Tannenbaum, S.R.; Fox, J.G. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: An integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 2014, 122, 284–291. [Google Scholar] [CrossRef]
- Feng, S.; Liu, Y.; Huang, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Influence of oral administration of Akkermansia muciniphila on the tissue distribution and gut microbiota composition of acute and chronic cadmium exposure mice. FEMS Microbiol. Lett. 2019, 366, fnz160. [Google Scholar] [CrossRef]
- Giri, S.S.; Jun, J.W.; Yun, S.; Kim, H.J.; Kim, S.G.; Kang, J.W.; Kim, S.W.; Han, S.J.; Park, S.C.; Sukumaran, V. Characterisation of lactic acid bacteria isolated from the gut of Cyprinus carpio that may be effective against lead toxicity. Probiotics Antimicrob. Proteins 2019, 11, 65–73. [Google Scholar] [CrossRef]
- Chiquette, J. The role of probiotics in promoting dairy production. In Proceedings of the 30th Western Nutrition Conference, Winnipeg, MB, Canada, 23–24 September 2009; p. 2. [Google Scholar]
- Ranadheera, C.S.; Naumovski, N.; Ajlouni, S. Non-bovine milk products as emerging probiotic carriers: Recent developments and innovations. Curr. Opin. Food Sci. 2018, 22, 109–114. [Google Scholar] [CrossRef]
- Zhai, Q.; Tian, F.; Zhao, J.; Zhang, H.; Narbad, A.; Chen, W. Oral administration of probiotics inhibits absorption of the heavy metal cadmium by protecting the intestinal barrier. Appl. Environ. Microbiol. 2016, 82, 4429–4440. [Google Scholar] [CrossRef]
- Zhai, Q.; Liu, Y.; Wang, C.; Qu, D.; Zhao, J.; Zhang, H.; Tian, F.; Chen, W. Lactobacillus plantarum CCFM8661 modulates bile acid enterohepatic circulation and increases lead excretion in mice. Food Funct. 2019, 10, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Dey, U.; Chatterjee, S.; Mondal, N.K. Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol. Rep. 2016, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Holmström, S.J. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Lehr, C.R.; Yuan, C.; Le, X.C.; McDermott, T.R.; Rosen, B.P. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc. Natl. Acad. Sci. USA 2009, 106, 5213–5217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chen, X.; Sheng, H.; Shen, X.; Sun, X.; Yan, Y.; Wang, J.; Yuan, Q. Engineering probiotics as living diagnostics and therapeutics for improving human health. Microb. Cell Fact 2020, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Mazhar, S.F.; Afzal, M.; Almatroudi, A.; Munir, S.; Ashfaq, U.A.; Rasool, M.; Raza, H.; Munir, H.M.W.; Rajoka, M.S.R.; Khurshid, M. The Prospects for the Therapeutic Implications of Genetically Engineered Probiotics. J. Food Qual. 2020, 2020, 9676452. [Google Scholar] [CrossRef]
- Riglar, D.T.; Silver, P.A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 2018, 16, 214–225. [Google Scholar] [CrossRef]
- Takiishi, T.; Cook, D.P.; Korf, H.; Sebastiani, G.; Mancarella, F.; Cunha, J.P.M.C.M.; Wasserfall, C.; Casares, N.; Lasarte, J.J.; Steidler, L. Reversal of diabetes in NOD mice by clinical-grade proinsulin and IL-10–secreting lactococcus lactis in combination with low-dose anti-CD3 depends on the induction of Foxp3-positive T cells. Diabetes 2017, 66, 448–459. [Google Scholar] [CrossRef]
- Isabella, V.M.; Ha, B.N.; Castillo, M.J.; Lubkowicz, D.J.; Rowe, S.E.; Millet, Y.A.; Anderson, C.L.; Li, N.; Fisher, A.B.; West, K.A. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018, 36, 857–864. [Google Scholar] [CrossRef]
- Braat, H.; Rottiers, P.; Hommes, D.W.; Huyghebaert, N.; Remaut, E.; Remon, J.P.; Van Deventer, S.J.; Neirynck, S.; Peppelenbosch, M.P.; Steidler, L. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 2006, 4, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.H.; Nguyen, V.H.; Jiang, S.-N.; Park, S.-H.; Tan, W.; Hong, S.H.; Shin, M.G.; Chung, I.-J.; Hong, Y.; Bom, H.-S. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017, 9, eaak9537. [Google Scholar] [CrossRef] [PubMed]
- Järup, L. Hazards of heavy metal contamination. Br. Med. Bull. 2003, 68, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Jain, J.; Gauba, P. Heavy metal toxicity-implications on metabolism and health. Int. J. Pharma Bio Sci. 2017, 8, 452–460. [Google Scholar] [CrossRef]
- Sharma, P.; Purchase, D.; Chandra, R. Residual pollutants in treated pulp paper mill wastewater and their phytotoxicity and cytotoxicity in Allium cepa. Environ. Geochem. Health 2021, 43, 2143–2164. [Google Scholar] [CrossRef]
- McKelvey, W.; Gwynn, R.C.; Jeffery, N.; Kass, D.; Thorpe, L.E.; Garg, R.K.; Palmer, C.D.; Parsons, P.J. A biomonitoring study of lead, cadmium, and mercury in the blood of New York city adults. Environ. Health Perspect. 2007, 115, 1435–1441. [Google Scholar] [CrossRef]
- Sheehan, M.C.; Burke, T.A.; Navas-Acien, A.; Breysse, P.N.; McGready, J.; Fox, M.A. Global methylmercury exposure from seafood consumption and risk of developmental neurotoxicity: A systematic review. Bull. World Health Organ. 2014, 92, 254–269F. [Google Scholar] [CrossRef]
- Sall, M.L.; Diaw, A.K.D.; Gningue-Sall, D.; Efremova Aaron, S.; Aaron, J.-J. Toxic heavy metals: Impact on the environment and human health, and treatment with conducting organic polymers, a review. Environ. Sci. Pollut. Res. 2020, 27, 29927–29942. [Google Scholar] [CrossRef]
- Chung, J.-Y.; Yu, S.-D.; Hong, Y.-S. Environmental source of arsenic exposure. J. Prev. Med. Public Health 2014, 47, 253. [Google Scholar] [CrossRef]
- Centeno, J.A.; Tchounwou, P.B.; Patlolla, A.K.; Mullick, F.G.; Murakata, L.; Meza, E.; TodorTodorov, D.L.; Yedjou, C.G. Environmental pathology and health effects of arsenic poisoning. In Managing Arsenic in the Environment: From Soil to Human Health; CSIRO PUBLISHING: Collingwood, Australia, 2006; pp. 311–327. [Google Scholar]
- Tchounwou, P.B.; Centeno, J.A.; Patlolla, A.K. Arsenic toxicity, mutagenesis, and carcinogenesis–a health risk assessment and management approach. Mol. Cell. Biochem. 2004, 255, 47–55. [Google Scholar] [CrossRef]
- Ratnaike, R.N. Acute and chronic arsenic toxicity. Postgrad. Med. J. 2003, 79, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.-S.; Song, K.-H.; Chung, J.-Y. Health effects of chronic arsenic exposure. J. Prev. Med. Public Health 2014, 47, 245. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.J.; Graham, B.; Walker, A.M.; Tchounwou, P.B.; Rogers, C. The effects of arsenic trioxide on DNA synthesis and genotoxicity in human colon cancer cells. Int. J. Environ. Res. Public Health 2010, 7, 2018–2032. [Google Scholar] [CrossRef]
- Saha, J.; Dikshit, A.; Bandyopadhyay, M.; Saha, K. A review of arsenic poisoning and its effects on human health. Crit. Rev. Environ. Sci. Technol. 1999, 29, 281–313. [Google Scholar] [CrossRef]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Beryllium, Cadmium, Mercury, and Exposures in the Glass Manufacturing Industry; International Agency for Research on Cancer: Lyon, French, 1993. [Google Scholar]
- Shimada, H.; Yasutake, A.; Hirashima, T.; Takamure, Y.; Kitano, T.; Waalkes, M.P.; Imamura, Y. Strain difference of cadmium accumulation by liver slices of inbred Wistar-Imamichi and Fischer 344 rats. Toxicol. Vitr. 2008, 22, 338–343. [Google Scholar] [CrossRef]
- Bernhoft, R.A. Cadmium toxicity and treatment. Sci. World J. 2013, 2013, 394652. [Google Scholar] [CrossRef]
- Dewanjee, S.; Gangopadhyay, M.; Sahu, R.; Karmakar, S. Cadmium induced pathophysiology: Prophylactic role of edible jute (Corchorus olitorius) leaves with special emphasis on oxidative stress and mitochondrial involvement. Food Chem. Toxicol. 2013, 60, 188–198. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The effects of cadmium toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, V.; Paliwal, R. Lead toxicity, oxidative damage and health implications. A review. Int. J. Biotechnol. Mol. Biol. Res. 2011, 2, 215–221. [Google Scholar] [CrossRef]
- Roper, W.L.; Houk, V.; Falk, H.; Binder, S. Preventing Lead Poisoning in Young Children: A Statement by the Centers for Disease Control, October 1991; Centers for Disease Control: Atlanta, GA, USA, 1991.
- ATSDR. Case studies in environmental medicine—Lead toxicity. In Public Health Service, U.S. Department of Health and Human Services; Agency for Toxic Substances and Disease Registry: Atlanta, GA, USA, 2017. [Google Scholar]
- Pirkle, J.L.; Kaufmann, R.B.; Brody, D.J.; Hickman, T.; Gunter, E.W.; Paschal, D.C. Exposure of the US population to lead, 1991–1994. Environ. Health Perspect. 1998, 106, 745–750. [Google Scholar] [CrossRef]
- Kaul, B.; Sandhu, R.S.; Depratt, C.; Reyes, F. Follow-up screening of lead-poisoned children near an auto battery recycling plant, Haina, Dominican Republic. Environ. Health Perspect. 1999, 107, 917–920. [Google Scholar] [CrossRef]
- Factor-Litvak, P.; Slavkovich, V.; Liu, X.; Popovac, D.; Preteni, E.; Capuni-Paracka, S.; Hadzialjevic, S.; Lekic, V.; LoIacono, N.; Kline, J. Hyperproduction of erythropoietin in nonanemic lead-exposed children. Environ. Health Perspect. 1998, 106, 361–364. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Finkelstein, Y.; Markowitz, M.E.; Rosen, J.F. Low-level lead-induced neurotoxicity in children: An update on central nervous system effects. Brain Res. Rev. 1998, 27, 168–176. [Google Scholar] [CrossRef]
- Jan, A.T.; Azam, M.; Siddiqui, K.; Ali, A.; Choi, I.; Haq, Q.M. Heavy metals and human health: Mechanistic insight into toxicity and counter defense system of antioxidants. Int. J. Mol. Sci. 2015, 16, 29592–29630. [Google Scholar] [CrossRef] [PubMed]
- Brochin, R.; Leone, S.; Phillips, D.; Shepard, N.; Zisa, D.; Angerio, A. The cellular effect of lead poisioning and its clinical picture. Management 2014, 8, 1–8. [Google Scholar]
- Ab Latif Wani, A.A.; Usmani, J.A. Lead toxicity: A review. Interdiscip. Toxicol. 2015, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D. Mechanisms of lead-induced hypertension and cardiovascular disease. Am. J. Physiol.-Heart Circ. Physiol. 2008, 295, H454–H465. [Google Scholar] [CrossRef] [PubMed]
- Trasande, L.; Landrigan, P.J.; Schechter, C. Public health and economic consequences of methyl mercury toxicity to the developing brain. Environ. Health Perspect. 2005, 113, 590–596. [Google Scholar] [CrossRef]
- Ha, E.; Basu, N.; Bose-O’Reilly, S.; Dórea, J.G.; McSorley, E.; Sakamoto, M.; Chan, H.M. Current progress on understanding the impact of mercury on human health. Environ. Res. 2017, 152, 419–433. [Google Scholar] [CrossRef]
- Gauba, P.; Shakeel, M.; Gaur, S. Mercury Neurotoxicity: A review of case studies. Asian J. Multidiscip. Stud. 2015, 3, 9–16. [Google Scholar]
- Rice, K.M.; Walker, E.M., Jr.; Wu, M.; Gillette, C.; Blough, E.R. Environmental mercury and its toxic effects. J. Prev. Med. Public Health 2014, 47, 74. [Google Scholar] [CrossRef]
- Zefferino, R.; Piccoli, C.; Ricciardi, N.; Scrima, R.; Capitanio, N. Possible mechanisms of mercury toxicity and cancer promotion: Involvement of gap junction intercellular communications and inflammatory cytokines. Oxidative Med. Cell. Longev. 2017, 2017, 7028583. [Google Scholar] [CrossRef] [PubMed]
- Grant, D. Detoxification pathways in the liver. J. Inherit. Metab. Dis. 1991, 14, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Aaseth, J.; Crisponi, G.; Anderson, O. Chelation Therapy in the Treatment of Metal Intoxication; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Blanusa, M.; Varnai, V.M.; Piasek, M.; Kostial, K. Chelators as antidotes of metal toxicity: Therapeutic and experimental aspects. Curr. Med. Chem. 2005, 12, 2771–2794. [Google Scholar] [CrossRef]
- Cao, Y.; Skaug, M.A.; Andersen, O.; Aaseth, J. Chelation therapy in intoxications with mercury, lead and copper. J. Trace Elem. Med. Biol. 2015, 31, 188–192. [Google Scholar] [CrossRef]
- Bjørklund, G.; Mutter, J.; Aaseth, J. Metal chelators and neurotoxicity: Lead, mercury, and arsenic. Arch. Toxicol. 2017, 91, 3787–3797. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Li, X.; Yu, D.; Wang, Y.; Yuan, H.; Ning, X.; Rui, B.; Lei, Z.; Yuan, J.; Yan, J.; Li, M. The Intestinal Dysbiosis of Mothers with Gestational Diabetes Mellitus (GDM) and Its Impact on the Gut Microbiota of Their Newborns. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 3044534. [Google Scholar] [CrossRef]
- Lange, K.; Buerger, M.; Stallmach, A.; Bruns, T. Effects of antibiotics on gut microbiota. Dig. Dis. 2016, 34, 260–268. [Google Scholar] [CrossRef]
- Bielka, W.; Przezak, A.; Pawlik, A. The Role of the Gut Microbiota in the Pathogenesis of Diabetes. Int. J. Mol. Sci. 2022, 23, 480. [Google Scholar] [CrossRef]
- Duan, H.; Yu, L.; Tian, F.; Zhai, Q.; Fan, L.; Chen, W. Gut microbiota: A target for heavy metal toxicity and a probiotic protective strategy. Sci. Total Environ. 2020, 742, 140429. [Google Scholar] [CrossRef]
- Lin, X.; Zhao, J.; Zhang, W.; He, L.; Wang, L.; Chang, D.; Cui, L.; Gao, Y.; Li, B.; Chen, C. Acute oral methylmercury exposure perturbs the gut microbiome and alters gut-brain axis related metabolites in rats. Ecotoxicol. Environ. Saf. 2020, 190, 110130. [Google Scholar] [CrossRef]
- Wu, J.; Wen, X.W.; Faulk, C.; Boehnke, K.; Zhang, H.; Dolinoy, D.C.; Xi, C. Perinatal lead exposure alters gut microbiota composition and results in sex-specific bodyweight increases in adult mice. Toxicol. Sci. 2016, 151, 324–333. [Google Scholar] [CrossRef]
- Zhai, Q.; Li, T.; Yu, L.; Xiao, Y.; Feng, S.; Wu, J.; Zhao, J.; Zhang, H.; Chen, W. Effects of subchronic oral toxic metal exposure on the intestinal microbiota of mice. Sci. Bull. 2017, 62, 831–840. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Nookaew, I.; Petranovic, D.; Nielsen, J. Prospects for systems biology and modeling of the gut microbiome. Trends Biotechnol. 2011, 29, 251–258. [Google Scholar] [CrossRef]
- Sagar, N.M.; Cree, I.A.; Covington, J.A.; Arasaradnam, R.P. The interplay of the gut microbiome, bile acids, and volatile organic compounds. Gastroenterol. Res. Pract. 2015, 2015, 398585. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.; Bian, X.; Gao, B.; Tu, P.; Ru, H.; Lu, K. The effects of an environmentally relevant level of arsenic on the gut microbiome and its functional metagenome. Toxicol. Sci. 2017, 160, 193–204. [Google Scholar] [CrossRef]
- Wang, J.; Hu, W.; Yang, H.; Chen, F.; Shu, Y.; Zhang, G.; Liu, J.; Liu, Y.; Li, H.; Guo, L. Arsenic concentrations, diversity and co-occurrence patterns of bacterial and fungal communities in the feces of mice under sub-chronic arsenic exposure through food. Environ. Int. 2020, 138, 105600. [Google Scholar] [CrossRef] [PubMed]
- Farina, M.; Rocha, J.B.; Aschner, M. Mechanisms of methylmercury-induced neurotoxicity: Evidence from experimental studies. Life Sci. 2011, 89, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Gokulan, K.; Arnold, M.G.; Jensen, J.; Vanlandingham, M.; Twaddle, N.C.; Doerge, D.R.; Cerniglia, C.E.; Khare, S. Exposure to arsenite in CD-1 mice during juvenile and adult stages: Effects on intestinal microbiota and gut-associated immune status. MBio 2018, 9, e01418. [Google Scholar] [CrossRef] [PubMed]
- Dheer, R.; Patterson, J.; Dudash, M.; Stachler, E.N.; Bibby, K.J.; Stolz, D.B.; Shiva, S.; Wang, Z.; Hazen, S.L.; Barchowsky, A. Arsenic induces structural and compositional colonic microbiome change and promotes host nitrogen and amino acid metabolism. Toxicol. Appl. Pharmacol. 2015, 289, 397–408. [Google Scholar] [CrossRef]
- Li, X.; Brejnrod, A.D.; Ernst, M.; Rykær, M.; Herschend, J.; Olsen, N.M.C.; Dorrestein, P.C.; Rensing, C.; Sørensen, S.J. Heavy metal exposure causes changes in the metabolic health-associated gut microbiome and metabolites. Environ. Int. 2019, 126, 454–467. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Qi, Z.; Hou, H.; Qian, L.; Gao, J.; Zhang, X.-X. Structural and functional alterations of gut microbiome in mice induced by chronic cadmium exposure. Chemosphere 2020, 246, 125747. [Google Scholar] [CrossRef]
- Zhai, Q.; Qu, D.; Feng, S.; Yu, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Oral supplementation of lead-intolerant intestinal microbes protects against lead (Pb) toxicity in mice. Front. Microbiol. 2020, 10, 3161. [Google Scholar] [CrossRef]
- Gao, B.; Chi, L.; Mahbub, R.; Bian, X.; Tu, P.; Ru, H.; Lu, K. Multi-omics reveals that lead exposure disturbs gut microbiome development, key metabolites, and metabolic pathways. Chem. Res. Toxicol. 2017, 30, 996–1005. [Google Scholar] [CrossRef]
- Bridges, K.N.; Zhang, Y.; Curran, T.E.; Magnuson, J.T.; Venables, B.J.; Durrer, K.E.; Allen, M.S.; Roberts, A.P. Alterations to the intestinal microbiome and metabolome of Pimephales promelas and Mus musculus following exposure to dietary methylmercury. Environ. Sci. Technol. 2018, 52, 8774–8784. [Google Scholar]
- Shen, G.; Wu, J.; Ye, B.C.; Qi, N. Gut Microbiota-Derived Metabolites in the Development of Diseases. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 6658674. [Google Scholar] [CrossRef]
- Tang, R.; Li, L. Modulation of Short-Chain Fatty Acids as Potential Therapy Method for Type 2 Diabetes Mellitus. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 6632266. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.-Q.; Tseng, K.-Y.; Tsai, Y.-H. Candida gut commensalism and inflammatory disease. Med. Microecol. 2020, 3, 100008. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Y.; Yang, H.; Rao, Y.; Miao, J.; Lu, X. Intestinal Microbiota as an Alternative Therapeutic Target for Epilepsy. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 9032809. [Google Scholar] [CrossRef]
- Zhu, X.; Li, B.; Lou, P.; Dai, T.; Chen, Y.; Zhuge, A.; Yuan, Y.; Li, L. The relationship between the gut microbiome and neurodegenerative diseases. Neurosci. Bull. 2021, 37, 1510–1522. [Google Scholar] [CrossRef]
- Rowland, I.; Robinson, R.; Doherty, R. Effects of diet on mercury metabolism and excretion in mice given methylmercury: Role of gut flora. Arch. Environ. Health Int. J. 1984, 39, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, F.; Halttunen, T.; Tahvonen, R.; Salminen, S. Probiotic bacteria as potential detoxification tools: Assessing their heavy metal binding isotherms. Can. J. Microbiol. 2006, 52, 877–885. [Google Scholar] [CrossRef]
- Medircio, S.N.; Leao, V.A.; Teixeira, M.C. Specific growth rate of sulfate reducing bacteria in the presence of manganese and cadmium. J. Hazard. Mater. 2007, 143, 593–596. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Vítězová, M.; Rittmann, S.K.-M. Environmental Impact of Sulfate-Reducing Bacteria, Their Role in Intestinal Bowel Diseases, and Possible Control by Bacteriophages. Appl. Sci. 2021, 11, 735. [Google Scholar] [CrossRef]
- Stewart, C.S.; Duncan, S.H.; Cave, D.R. Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol. Lett. 2004, 230, 1–7. [Google Scholar] [CrossRef]
- McDermott, T.R.; Stolz, J.F.; Oremland, R.S. Arsenic and the gastrointestinal tract microbiome. Environ. Microbiol. Rep. 2020, 12, 136–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qin, N.; Guo, J.; Qian, G.; Fang, D.; Shi, D.; Xu, M.; Yang, F.; He, Z.; Van Nostrand, J.D. Functional gene arrays-based analysis of fecal microbiomes in patients with liver cirrhosis. BMC Genom. 2014, 15, 753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, C.; Lu, Z. Health promoting activities of probiotics. J. Food Biochem. 2019, 43, e12944. [Google Scholar] [CrossRef]
- Mathew, B.B.; Tiwari, A.; Jatawa, S.K. Free radicals and antioxidants: A review. J. Pharm. Res. 2011, 4, 4340–4343. [Google Scholar]
- Flora, S.; Mittal, M.; Mehta, A. Heavy metal induced oxidative stress & its possible reversal by chelation therapy. Indian J. Med. Res. 2008, 128, 501. [Google Scholar]
- Majlesi, M.; Shekarforoush, S.S.; Ghaisari, H.R.; Nazifi, S.; Sajedianfard, J.; Eskandari, M.H. Effect of probiotic Bacillus coagulans and Lactobacillus plantarum on alleviation of mercury toxicity in rat. Probiotics Antimicrob. Proteins 2017, 9, 300–309. [Google Scholar] [CrossRef]
- Yu, L.; Zhai, Q.; Tian, F.; Liu, X.; Wang, G.; Zhao, J.; Zhang, H.; Narbad, A.; Chen, W. Potential of Lactobacillus plantarum CCFM639 in protecting against aluminum toxicity mediated by intestinal barrier function and oxidative stress. Nutrients 2016, 8, 783. [Google Scholar] [CrossRef]
- Jiang, X.; Gu, S.; Liu, D.; Zhao, L.; Xia, S.; He, X.; Chen, H.; Ge, J. Lactobacillus brevis 23017 relieves mercury toxicity in the colon by modulation of oxidative stress and inflammation through the interplay of MAPK and NF-κB signaling cascades. Front. Microbiol. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Bennur, T.; Kumar, A.R.; Zinjarde, S.; Javdekar, V. Nocardiopsis species: Incidence, ecological roles and adaptations. Microbiol. Res. 2015, 174, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Dubey, V.; Mishra, A.K.; Ghosh, A.R.; Mandal, B.K. Probiotic Pediococcus pentosaceus GS 4 shields brush border membrane and alleviates liver toxicity imposed by chronic cadmium exposure in Swiss albino mice. J. Appl. Microbiol. 2019, 126, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Enos, M.K.; Mwanga, J.R.; Changalucha, J.; Burton, J.P.; Gloor, G.B.; Reid, G. Randomized open-label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. MBio 2014, 5, e01580-14. [Google Scholar] [CrossRef] [PubMed]
- Anadón, A.; Martínez-Larrañaga, M.R.; Martínez, M.A. Probiotics for animal nutrition in the European Union. Regulation and safety assessment. Regul. Toxicol. Pharmacol. 2006, 45, 91–95. [Google Scholar] [CrossRef]
- Musa, H.H.; Wu, S.; Zhu, C.; Seri, H.; Zhu, G. The potential benefits of probiotics in animal production and health. J. Anim. Vet. Adv. 2009, 8, 313–321. [Google Scholar]
- Karimi, O.; Peña, A.S. Indications and challenges of probiotics, prebiotics, and synbiotics in the management of arthralgias and spondyloarthropathies in inflammatory bowel disease. J. Clin. Gastroenterol. 2008, 42, S136–S141. [Google Scholar] [CrossRef]
- Peng, M.; Tabashsum, Z.; Anderson, M.; Truong, A.; Houser, A.K.; Padilla, J.; Akmel, A.; Bhatti, J.; Rahaman, S.O.; Biswas, D. Effectiveness of probiotics, prebiotics, and prebiotic-like components in common functional foods. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1908–1933. [Google Scholar] [CrossRef]
- Alemayehu, D.; Ross, R.P.; O’Sullivan, O.; Coffey, A.; Stanton, C.; Fitzgerald, G.F.; McAuliffe, O. Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 2009, 448, 29–39. [Google Scholar] [CrossRef]
- Guglielmotti, D.M.; Marcó, M.B.; Golowczyc, M.; Reinheimer, J.A.; Quiberoni, A.d.L. Probiotic potential of Lactobacillus delbrueckii strains and their phage resistant mutants. Int. Dairy J. 2007, 17, 916–925. [Google Scholar] [CrossRef]
- Sonnenborn, U.; Schulze, J. The non-pathogenic Escherichia coli strain Nissle 1917–features of a versatile probiotic. Microb. Ecol. Health Dis. 2009, 21, 122–158. [Google Scholar]
- Schultz, M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Wehkamp, J.; Harder, J.R.; Wehkamp, K.; Meissner, B.W.-V.; Schlee, M.; Enders, C.; Sonnenborn, U.; Nuding, S.; Bengmark, S.; Fellermann, K. NF-κB-and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: A novel effect of a probiotic bacterium. Infect. Immun. 2004, 72, 5750–5758. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.-P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.-P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.-J.; Weibel, S.; Wehkamp, J.; Oelschlaeger, T.A. Construction of recombinant E. coli Nissle 1917 (EcN) strains for the expression and secretion of defensins. Int. J. Med. Microbiol. 2012, 302, 276–287. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, H.; Lu, J.; Ma, C.; Chen, T. Antidiabetic effect of an engineered bacterium Lactobacillus plantarum-pMG36e-GLP-1 in monkey model. Synth. Syst. Biotechnol. 2021, 6, 272–282. [Google Scholar] [CrossRef]
- Zou, Y.; Chen, T. Engineered Akkermansia muciniphila: A promising agent against diseases. Exp. Ther. Med. 2020, 20, 285. [Google Scholar] [CrossRef]
- Fang, X.; Zhou, X.; Miao, Y.; Han, Y.; Wei, J.; Chen, T. Therapeutic effect of GLP-1 engineered strain on mice model of Alzheimer’s disease and Parkinson’s disease. Amb Express 2020, 10, 80. [Google Scholar] [CrossRef]
- Amalaradjou, M.A.R.; Bhunia, A.K. Bioengineered probiotics, a strategic approach to control enteric infections. Bioengineered 2013, 4, 379–387. [Google Scholar] [CrossRef]
- Rasheed, T.; Bilal, M.; Nabeel, F.; Adeel, M.; Iqbal, H.M. Environmentally-related contaminants of high concern: Potential sources and analytical modalities for detection, quantification, and treatment. Environ. Int. 2019, 122, 52–66. [Google Scholar] [CrossRef]
- Samuelson, P.; Gunneriusson, E.; Nygren, P.-Å.; Ståhl, S. Display of proteins on bacteria. J. Biotechnol. 2002, 96, 129–154. [Google Scholar] [CrossRef]
- Wernérus, H.; Ståhl, S. Biotechnological applications for surface-engineered bacteria. Biotechnol. Appl. Biochem. 2004, 40, 209–228. [Google Scholar] [PubMed]
- Valls, M.; de Lorenzo, V.C.; Gonzàlez-Duarte, R.; Atrian, S.L. Engineering outer-membrane proteins in Pseudomonas putida for enhanced heavy-metal bioadsorption. J. Inorg. Biochem. 2000, 79, 219–223. [Google Scholar] [CrossRef]
- Saffar, B.; Yakhchali, B.; Arbabi, M. Development of a bacterial surface display of hexahistidine peptide using CS3 pili for bioaccumulation of heavy metals. Curr. Microbiol. 2007, 55, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Schneewind, O.; Fowler, A.; Faull, K.F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 1995, 268, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Wernérus, H.; Lehtiö, J.; Teeri, T.; Nygren, P.-Å.; Ståhl, S. Generation of metal-binding staphylococci through surface display of combinatorially engineered cellulose-binding domains. Appl. Environ. Microbiol. 2001, 67, 4678–4684. [Google Scholar] [CrossRef]
- Zou, C.; Chen, Y.; Li, H.; Li, W.; Wei, J.; Li, Z.; Wang, X.; Chen, T.; Huang, H. Engineered bacteria EcN-MT alleviate liver injury in cadmium-exposed mice via its probiotics characteristics and expressing of metallothionein. Front. Pharmacol. 2022, 13, 506. [Google Scholar] [CrossRef]
- Sousa, C.; Kotrba, P.; Ruml, T.; Cebolla, A.; De Lorenzo, V.C. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J. Bacteriol. 1998, 180, 2280–2284. [Google Scholar] [CrossRef]
- Schmoger, M.E.; Oven, M.; Grill, E. Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 2000, 122, 793–802. [Google Scholar] [CrossRef]
- Valls, M.; Atrian, S.; de Lorenzo, V.; Fernández, L.A. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 2000, 18, 661–665. [Google Scholar] [CrossRef]
- Nadarajan, S.P.; Ganesh, I.; Ravikumar, S.; Yun, H.; Yoo, I.-K.; Hong, S.H. Construction of a high efficiency copper adsorption bacterial system via peptide display and its application on copper dye polluted wastewater. Bioprocess Biosyst. Eng. 2015, 38, 2077–2084. [Google Scholar]
- Kim, W.; Kim, D.; Back, S.; Lee, Y.-S.; Abari, A.H.; Kim, J. Removal of Ni2+ and Cd2+ by surface display of polyhistidine on bacillus subtilis spore using CotE anchor protein. Biotechnol. Bioprocess Eng. 2019, 24, 375–381. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, B.; Yu, Q. Genetic engineering-facilitated coassembly of synthetic bacterial cells and magnetic nanoparticles for efficient heavy metal removal. ACS Appl. Mater. Interfaces 2020, 12, 22948–22957. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Li, Y.; Xu, T.; Wu, K. Display of lead-binding proteins on Escherichia coli surface for lead bioremediation. Biotechnol. Bioeng. 2020, 117, 3820–3834. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr. Transport protein evolution deduced from analysis of sequence, topology and structure. Curr. Opin. Struct. Biol. 2016, 38, 9–17. [Google Scholar] [CrossRef]
- Diep, P.; Mahadevan, R.; Yakunin, A.F. Heavy Metal Removal by Bioaccumulation Using Genetically Engineered Microorganisms. Front. Bioeng. Biotechnol. 2018, 6, 157. [Google Scholar] [CrossRef]
- Verma, P.K.; Verma, S.; Meher, A.K.; Tripathi, R.D.; Chakrabarty, D.; Pandey, N. Bioremediation of Heavy Metals using the Interaction between Plants and Genetically Engineered Microbes. Int. J. Plant Environ. 2020, 6, 241–252. [Google Scholar] [CrossRef]
- Ueno, D.; Milner, M.J.; Yamaji, N.; Yokosho, K.; Koyama, E.; Clemencia Zambrano, M.; Kaskie, M.; Ebbs, S.; Kochian, L.V.; Ma, J.F. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011, 66, 852–862. [Google Scholar] [CrossRef]
- Shen, M.W.; Shah, D.; Chen, W.; Da Silva, N. Enhanced arsenate uptake in Saccharomyces cerevisiae overexpressing the Pho84 phosphate transporter. Biotechnol. Prog. 2012, 28, 654–661. [Google Scholar] [CrossRef]
- Shah, D.; Shen, M.W.; Chen, W.; Da Silva, N.A. Enhanced arsenic accumulation in Saccharomyces cerevisiae overexpressing transporters Fps1p or Hxt7p. J. Biotechnol. 2010, 150, 101–107. [Google Scholar] [CrossRef]
- Singh, S.; Kang, S.H.; Lee, W.; Mulchandani, A.; Chen, W. Systematic engineering of phytochelatin synthesis and arsenic transport for enhanced arsenic accumulation in E. coli. Biotechnol. Bioeng. 2010, 105, 780–785. [Google Scholar] [CrossRef]
- Villadangos, A.F.; Ordóñez, E.; Pedre, B.; Messens, J.; Gil, J.A.; Mateos, L.M. Engineered coryneform bacteria as a bio-tool for arsenic remediation. Appl. Microbiol. Biotechnol. 2014, 98, 10143–10152. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Jia, P. Construction and characterization of a photosynthetic bacterium genetically engineered for Hg2+ uptake. Bioresour. Technol. 2011, 102, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Saylor, Z.; Maier, R. Helicobacter pylori nickel storage proteins: Recognition and modulation of diverse metabolic targets. Microbiology 2018, 164, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Jiang, Y.-L.; Wang, S.; Jin, H.; Zhang, R.-G.; Virolle, M.-J.; Chen, Y.; Zhou, C.-Z. Streptomyces coelicolor SCO4226 is a nickel binding protein. PLoS ONE 2014, 9, e109660. [Google Scholar] [CrossRef]
- Priyadarshanee, M.; Chatterjee, S.; Rath, S.; Dash, H.R.; Das, S. Cellular and genetic mechanism of bacterial mercury resistance and their role in biogeochemistry and bioremediation. J. Hazard. Mater. 2022, 423, 126985. [Google Scholar] [CrossRef]
- Sasaki, Y.; Minakawa, T.; Miyazaki, A.; Silver, S.; Kusano, T. Functional dissection of a mercuric ion transporter, MerC, from Acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 2005, 69, 1394–1402. [Google Scholar] [CrossRef]
- Deng, X.; Wilson, D. Bioaccumulation of mercury from wastewater by genetically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2001, 56, 276–279. [Google Scholar] [CrossRef]
- Chen, J.; Sun, G.-X.; Wang, X.-X.; Lorenzo, V.C.D.; Rosen, B.P.; Zhu, Y.-G. Volatilization of arsenic from polluted soil by Pseudomonas putida engineered for expression of the arsM arsenic (III) S-adenosine methyltransferase gene. Environ. Sci. Technol. 2014, 48, 10337–10344. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, F.; Chen, J.; Sun, G. Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J. Environ. Sci. 2011, 23, 1544–1550. [Google Scholar] [CrossRef]
- Hui, C.; Guo, Y.; Zhang, W.; Gao, C.; Yang, X.; Chen, Y.; Li, L.; Huang, X. Surface display of PbrR on Escherichia coli and evaluation of the bioavailability of lead associated with engineered cells in mice. Sci. Rep. 2018, 8, 5685. [Google Scholar] [CrossRef]
- Liu, M.; Kakade, A.; Liu, P.; Wang, P.; Tang, Y.; Li, X. Hg(2+)-binding peptide decreases mercury ion accumulation in fish through a cell surface display system. Sci. Total Environ. 2019, 659, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lu, X.; Khan, A.; Ling, Z.; Wang, P.; Tang, Y.; Liu, P.; Li, X. Reducing methylmercury accumulation in fish using Escherichia coli with surface-displayed methylmercury-binding peptides. J. Hazard. Mater. 2019, 367, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Marinov, G.K. The bioenergetic costs of a gene. Proc. Natl. Acad. Sci. USA 2015, 112, 15690–15695. [Google Scholar] [CrossRef]
- Khan, M.A.; Ghouri, A.M. Environmental pollution: Its effects on life and its remedies. Res. World J. Arts Sci. Commer. 2011, 2, 276–825. [Google Scholar]
- Lin, Y.; Krogh-Andersen, K.; Pelletier, J.; Marcotte, H.; Östenson, C.-G.; Hammarström, L. Oral delivery of pentameric glucagon-like peptide-1 by recombinant Lactobacillus in diabetic rats. PLoS ONE 2016, 11, e0162733. [Google Scholar] [CrossRef] [PubMed]
- Danino, T.; Lo, J.; Prindle, A.; Hasty, J.; Bhatia, S.N. In vivo gene expression dynamics of tumor-targeted bacteria. ACS Synth. Biol. 2012, 1, 465–470. [Google Scholar] [CrossRef]
- Ceroni, F.; Boo, A.; Furini, S.; Gorochowski, T.E.; Borkowski, O.; Ladak, Y.N.; Awan, A.R.; Gilbert, C.; Stan, G.-B.; Ellis, T. Burden-driven feedback control of gene expression. Nat. Methods 2018, 15, 387–393. [Google Scholar] [CrossRef]
- De Groote, M.A.; Frank, D.N.; Dowell, E.; Glode, M.P.; Pace, N.R. Lactobacillus rhamnosus GG bacteremia associated with probiotic use in a child with short gut syndrome. Pediatr. Infect. Dis. J. 2005, 24, 278–280. [Google Scholar] [CrossRef]
- Salminen, M.K.; Rautelin, H.; Tynkkynen, S.; Poussa, T.; Saxelin, M.; Valtonen, V.; Järvinen, A. Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. rhamnosus GG. Clin. Infect. Dis. 2004, 38, 62–69. [Google Scholar] [CrossRef]
- Marteau, P.; Pochart, P.; Flourie, B.; Pellier, P.; Santos, L.; Desjeux, J.; Rambaud, J. Effect of chronic ingestion of a fermented dairy product containing Lactobacillus acidophilus and Bifidobacterium bifidum on metabolic activities of the colonic flora in humans. Am. J. Clin. Nutr. 1990, 52, 685–688. [Google Scholar] [CrossRef]
- Bongaerts, G.; Bakkeren, J.; Severijnen, R.; Sperl, W.; Willems, H.; Naber, T.; Wevers, R.; van Meurs, A.; Tolboom, J. Lactobacilli and acidosis in children with short small bowel. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Danielsen, M.; Huys, G.; Swings, J. Molecular characterization of tet (M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 2003, 69, 1270–1275. [Google Scholar] [CrossRef] [PubMed]
- Snydman, D.R. The safety of probiotics. Clin. Infect. Dis. 2008, 46, S104–S111. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.A.; Schoeni, J.L.; Vegge, C.; Pane, M.; Stahl, B.; Bradley, M.; Goldman, V.S.; Burguière, P.; Atwater, J.B.; Sanders, M.E. Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 2019, 10, 739. [Google Scholar] [CrossRef]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Yissachar, N.; Zhou, Y.; Ung, L.; Lai, N.Y.; Mohan, J.F.; Ehrlicher, A.; Weitz, D.A.; Kasper, D.L.; Chiu, I.M.; Mathis, D. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 2017, 168, 1135–1148.e1112. [Google Scholar] [CrossRef]
- De Boever, P.; Deplancke, B.; Verstraete, W. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J. Nutr. 2000, 130, 2599–2606. [Google Scholar] [CrossRef]
| Heavy Metal | The Effects on Gut Microbiota Composition | The Effects on Metabolic Profiles | Reference |
|---|---|---|---|
| As | Erysipelotrichaceae↑ Clostridiaceae↓, Catabacteriaceae↓ Cyanobacteria↓ | The secretion of bile acids, amino acid, lipids, fatty acids, glucuronide, isoflavones and indole derivatives were altered | [18] |
| Clostridium sulfatireducens↑ L. johnsonii↑ Butyricicoccus↑ Parasporobacterium↑ Intestinimonas↓ | The fecal concentration of pro-/anti-inflammatory cytokine and chemokines was increased | [89] | |
| Bacteroides↑, Porphyromonadaceae↑ Lactobacillus↑ Lachnospiraceae↓ Ruminococcaceae↓ | The metabolism of nitrogen and amino acid was enhanced | [90] | |
| Cd | Clostridium_XlVb↓ Syntrophococcus↓ Cellulosilyticum↓ Prevotella↑ | The amino acid and bile acid secretions were altered | [23,82,91] |
| Bacteroides↑ Shewanella↑ Anaerorhabdus↑ Alistipes↑ Chryseobacterium↑ Hafnia↓, Buttiauxella↓ Arcobacter↓ | The metabolism of carbohydrate, amino acid and nucleotide were promoted | [92] | |
| Pb | Ruminococcaceae↓ Lachnospiraceae↓ Oscillibacter↓ Anaerotruncus↓ Lachnoclostridium↓ | - | [93] |
| Desulfovibrionaceae↑ Enterorhabdus↓ Pseudomonas↓ Desulfovibrio↓ | - | [81] | |
| Ruminococcus↓ Coprococcus↓ Oscillospira↓ Blautia↓ | The production of vitamin E and bile acids was reduced and the nitrogen and energy metabolism was altered, also induction of oxidative stress | [94] | |
| Hg | Sutterellaceae↓ Desulfovibrionaceae↑ Helicobacteraceae↑ Rhodospirillaceae↑ | Amino acid, carbohydrate, and lipid were disrupted | [80] |
| Xanthomonadaceae↑ Acinetobacter↑ Nocardia↓ Aeromonas↑ Comamonadaceae families↑ Pseudomonas↑ | Lipid metabolism and secretion of neurotransmission was altered | [95] |
| Probiotics | Heavy Metals | Mechanism | Reference |
|---|---|---|---|
| Xanthomonadaceae, Comamonadaceae, Pirellula, Cloacibacterium, Deltaproteobacteria FAC87 | Hg | Convert methylated Hg to Hg0 that reduces its absorption | [79,89] |
| Lactobacillus plantarum (L. plantarum) TW1-1 | Cd | Convert Cd into a less absorbable form and reduce its intra-intestinal absorption | [23] |
| sulfate-reducing bacteria (SRB), Fe-reducing bacteria methanogens, Desulfovibrio spp. | As, Cd, Fe | Chemical modification of HMs by methylation | [63] |
| L. plantarum CCFM8610, CCFM 8611, and Bacillus cereus | Hg | Increase the HM excretion accompanied by bile acid production | [23,24] |
| L. plantarum LC-705 and Propionibacterium freudenreichii | Pb, Cd | Decrease the intestinal PH | [90] |
| Pseudomonas, Oxalobacter formigens (O. formigens) | Pb, Cd, Hg, Cr, As | Form insoluble complex with HMs via siderophores and hydrogen sulfide | [92,93] |
| Faecalibacterium prausnitzii (F. prausnitzii), Bacteroides and Faecalibacterium | As | Synthesize As-detoxifying enzymes | [27,94,95] |
| L. plantarum CCFM639, Bacillus Coagulans (B. coagulans) | As, Cd, Pb | Promote the expression of antioxidant-related genes to synthesis antioxidative enzymes | [99] |
| L. plantarum CCFM639 [100], CCFM8610 [23], L. brevis 23017, Nocardia and Bacteroidales | Hg | Release the HM-induced inflammatory responses by reducing the levels of proinflammatory cytokines | [23,101,102] |
| Pediococcus pentosaceus GS4, Akkermansia muciniphila (A. muciniphila) and Lactobacillus rhamnosus (L. rhamnosus) GR-1 | Cd, Pb | Re-establishing the structural balance by reverse the HM-induced compositional changes in gut microbiota | [19,23,102,104] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Tu, H.; Chen, T. Potential Application of Living Microorganisms in the Detoxification of Heavy Metals. Foods 2022, 11, 1905. https://doi.org/10.3390/foods11131905
Chen R, Tu H, Chen T. Potential Application of Living Microorganisms in the Detoxification of Heavy Metals. Foods. 2022; 11(13):1905. https://doi.org/10.3390/foods11131905
Chicago/Turabian StyleChen, Runqiu, Huaijun Tu, and Tingtao Chen. 2022. "Potential Application of Living Microorganisms in the Detoxification of Heavy Metals" Foods 11, no. 13: 1905. https://doi.org/10.3390/foods11131905
APA StyleChen, R., Tu, H., & Chen, T. (2022). Potential Application of Living Microorganisms in the Detoxification of Heavy Metals. Foods, 11(13), 1905. https://doi.org/10.3390/foods11131905







