Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety
Abstract
1. General Introduction
2. Cheese Starters and Adjunct Cultures
3. Cheese Microbiology
4. The Cheese Microbiota
5. Microbial Interactions in Cheese
5.1. Competition
5.2. Amensalism
5.3. Commensalism
5.4. Mutualism
6. Dynamics of Microbial Communities in Cheese
7. Microbiota-Based Starters
8. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Codex Standard 283-1978. Codex General Standard for Cheese; Adopted in 1973; Revision 1999; Amendments 2006; Codex Alimentarius Commission: Rome, Italy, 2013. [Google Scholar]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Principal families of cheese. In Fundamentals of Cheese Science; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2017; pp. 27–69. [Google Scholar]
- Kosikowski, F.V.; Mistry, V.V. Cheese and Fermented Milk Foods, 3rd ed.; Kosikowski, F.V., Ed.; LLC: Westport, CT, USA, 1997. [Google Scholar]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Microbiology of cheese ripening. In Fundamentals of Cheese Science; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2017; pp. 333–390. [Google Scholar]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Bintis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Carr, F.J.; Chill, D.; Maida, N. The lactic acid bacteria: A literature survey. Crit. Rev. Microbiol. 2002, 28, 281–370. [Google Scholar] [CrossRef] [PubMed]
- Smit, G.; Smit, B.A.; Engels, W.J. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef] [PubMed]
- Fardet, A.; Dupont, D.; Rioux, L.E.; Turgeon, S.L. Influence of food structure on dairy protein, lipid and calcium bioavailability: A narrative review of evidence. Crit. Rev. Food Sci. Nutr. 2019, 59, 1987–2010. [Google Scholar] [CrossRef] [PubMed]
- Metzger, S.A.; Hernandez, L.L.; Suen, G.; Ruegg, P.L. Understanding the milk microbiota. Vet. Clin. North Am. Food Anim. Pract. 2018, 34, 427–438. [Google Scholar] [CrossRef]
- Boor, K.J.; Wiedmann, M.; Murphy, S.; Alcaine, S. A 100-Year Review: Microbiology and safety of milk handling. J. Dairy Sci. 2017, 100, 9933–9951. [Google Scholar] [CrossRef] [PubMed]
- Irlinger, F.; Layec, S.; Hélinck, S.; Dugat-Bony, E. Cheese rind microbial communities: Diversity, composition and origin. FEMS Microbiol. Lett. 2015, 362, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, K.; Touchette, M.; St-Gelais, D.; Labrie, S. Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Sci. Technol. 2012, 92, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Mounier, J.; Goerges, S.; Gelsomino, R.; Vancanneyt, M.; Vandemeulebroecke, K.; Hoste, B.; Brennan, N.M.; Scherer, S.; Swings, J.; Fitzgerald, G.F.; et al. Sources of the adventitious microflora of a smear-ripened cheese. J. Appl. Microbiol. 2006, 101, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Desmasures, N.; Bazin, F.; Guéguen, M. Microbiological composition of raw milk from selected farms in the Camembert region of Normandy. J. Appl. Microbiol. 1997, 83, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Aldrete-Tapia, A.; Escobar-Ramírez, C.M.; Tamplin, M.L.; Hernández-Iturriaga, M. Characterization of bacterial communities in Mexican artisanal raw milk "Bola de Ocosingo" cheese by high-throughput sequencing. Front. Microbiol. 2018, 9, 2598. [Google Scholar] [CrossRef] [PubMed]
- Frétin, M.; Martin, B.; Rifa, E.; Isabelle, V.M.; Pomiès, D.; Ferlay, A.; Montel, M.C.; Delbès, C. Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems. Sci. Rep. 2018, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Haastrup, M.K.; Johansen, P.; Malskær, A.H.; Castro-Mejía, J.L.; Kot, W.; Krych, L.; Arneborg, N.; Jespersen, L. Cheese brines from Danish dairies reveal a complex microbiota comprising several halotolerant bacteria and yeasts. Int. J. Food Microbiol. 2018, 285, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, R.; Cardazzo, B.; Carraro, L.; Negrinotti, M.; Balzan, S.; Novelli, E.; Fasolato, L.; Fasoli, F.; Farina, G. Contribution of natural milk culture to microbiota, safety and hygiene of raw milk cheese produced in alpine malga. Ital. J. Food Saf. 2018, 7, 6967. [Google Scholar] [CrossRef] [PubMed]
- Quijada, N.M.; Mann, E.; Wagner, M.; Rodríguez-Lázaro, D.; Hernández, M.; Schmitz-Esser, S. Autochthonous facility-specific microbiota dominates washed-rind Austrian hard cheese surfaces and its production environment. Int. J. Food Microbiol. 2018, 267, 54–61. [Google Scholar] [CrossRef]
- Santarelli, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Gatti, M. Survey on the community and dynamics of lactic acid bacteria in Grana Padano cheese. Syst. Appl. Microbiol. 2013, 36, 593–600. [Google Scholar] [CrossRef]
- Edalatian, M.R.; Habibi, M.B.; Mortazavi, S.A.; Alegría, A.; Nassiri, M.R.; Bassam, M.R.; Mayo, B. Microbial diversity of the traditional Iranian cheeses Lighvan and Koozeh, as revealed by polyphasic culturing and culture-independent approaches. Dairy Sci. Technol. 2012, 92, 75–90. [Google Scholar] [CrossRef]
- Feutry, F.; Oneca, M.; Berthier, F.; Torre, P. Biodiversity and growth dynamics of lactic acid bacteria in artisanal PDO Ossau-Iraty cheeses made from raw ewe’s milk with different starters. Food Microbiol. 2012, 29, 33–42. [Google Scholar] [CrossRef]
- Alegría, A.; Álvarez-Martín, P.; Sacristán, N.; Fernández, E.; Delgado, S.; Mayo, B. Diversity and evolution of majority microbial populations during manufacturing and ripening of Casín, a Spanish traditional, starter-free cheese made of raw cow’s milk. Int. J. Food Microbiol. 2009, 136, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Callon, C.; Berdagué, J.L.; Dufour, E.; Montel, M.C. The effect of raw milk microbial flora on the sensory characteristics of Salers-type cheeses. J. Dairy Sci. 2005, 88, 3840–3850. [Google Scholar] [CrossRef]
- Morales, P.; Fernández-García, E.; Gaya, P.; Núñez, M. Formation of volatile compounds by wild Lactococcus lactis strains isolated from raw ewes’ milk cheese. Int. Dairy J. 2003, 13, 201–209. [Google Scholar] [CrossRef]
- Baruzzi, F.; Matarante, A.; Morea, M.; Cocconcelli, P.S. Microbial community dynamics during the Scamorza Altamurana cheese natural fermentation. J. Dairy Sci. 2002, 85, 1390–1397. [Google Scholar] [CrossRef]
- Moser, A.; Schafroth, K.; Meile, L.; Egger, L.; Badertscher, R.; Irmler, S. Population dynamics of Lactobacillus helveticus in Swiss Gruyère-type cheese manufactured with natural whey cultures. Front. Microbiol. 2018, 9, 637. [Google Scholar] [CrossRef]
- Castro, R.D.; Oliveira, L.G.; Sant’Anna, F.M.; Luiz, L.M.P.; Sandes, S.H.C.; Silva, C.I.F.; Silva, A.M.; Nunes, A.C.; Penna, C.F.A.M.; Souza, M.R. Lactic acid microbiota identification in water, raw milk, endogenous starter culture, and fresh Minas artisanal cheese from the Campo das Vertentes region of Brazil during the dry and rainy seasons. J. Dairy Sci. 2016, 99, 6086–6096. [Google Scholar] [CrossRef]
- Solieri, L.; Bianchi, A.; Giudici, P. Inventory of non-starter lactic acid bacteria from ripened Parmigiano Reggiano cheese as assessed by a culture dependent multiphasic approach. Syst. Appl. Microbiol. 2012, 35, 270–277. [Google Scholar] [CrossRef]
- Nieto-Arribas, P.; Seseña, S.; Poveda, J.M.; Palop, L.; Cabezas, L. Genotypic and technological characterization of Leuconostoc isolates to be used as adjunct starters in Manchego cheese manufacture. Food Microbiol. 2010, 27, 85–93. [Google Scholar] [CrossRef] [PubMed]
- van Hoorde, K.; van Leuven, I.; Dirinck, P.; Heyndrickx, M.; Coudijzer, K.; Vandamme, P.; Huys, G. Selection, application and monitoring of Lactobacillus paracasei strains as adjunct cultures in the production of Gouda-type cheeses. Int. J. Food Microbiol. 2010, 144, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Henri-Dubernet, S.; Desmasures, N.; Guéguen, M. Diversity and dynamics of lactobacilli populations during ripening of RDO Camembert cheese. Can. J. Microbiol. 2008, 54, 218–228. [Google Scholar] [CrossRef]
- Sánchez, I.; Seseña, S.; Poveda, J.M.; Cabezas, L.; Palop, L. Genetic diversity, dynamics, and activity of Lactobacillus community involved in traditional processing of artisanal Manchego cheese. Int. J. Food Microbiol. 2006, 107, 265–273. [Google Scholar] [CrossRef]
- Dasen, A.; Berthier, F.; Grappin, R.; Williams, A.G.; Banks, J. Genotypic and phenotypic characterization of the dynamics of the lactic acid bacterial population of adjunct-containing Cheddar cheese manufactured from raw and microfiltered pasteurised milk. J. Appl. Microbiol. 2003, 94, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Corroler, D.; Mangin, I.; Desmasures, N.; Gueguen, M. An ecological study of lactococci isolated from raw milk in the camembert cheese registered designation of origin area. Appl. Environ. Microbiol. 1998, 64, 4729–4735. [Google Scholar] [CrossRef] [PubMed]
- Desmasures, N.; Mangin, I.; Corroler, D.; Guéguen, M. Characterization of lactococci isolated from milk produced in the Camembert region of Normandy. J. Appl. Microbiol. 1998, 85, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Starter cultures. In Fundamentals of Cheese Science; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2017; pp. 121–183. [Google Scholar]
- Parente, E.; Cogan, T.M. Starter cultures: General aspects. In Cheese: Chemistry, Physics and Microbiology, 3rd ed.; Fox, P.O., Ed.; Elsevier: Oxford, UK, 2004; pp. 123–147. [Google Scholar]
- Giello, M.; La Storia, A.; Masucci, F.; Di Francia, A.; Ercolini, D.; Villani, F. Dynamics of bacterial communities during manufacture and ripening of traditional Caciocavallo of Castelfranco cheese in relation to cows’ feeding. Food Microbiol. 2017, 63, 170–177. [Google Scholar] [CrossRef]
- Cogan, T.M.; Goerges, S.; Gelsomino, R.; Larpin, S.; Hohenegger, M.; Bora, N.; Jamet, E.; Rea, M.C.; Mounier, J.; Vancanneyt, M.; et al. Biodiversity of the surface microbial consortia from Limburger, Reblochon, Livarot, Tilsit, and Gubbeen cheeses. Microbiol. Spectr. 2014, 2, 219–256. [Google Scholar] [CrossRef]
- Mounier, J.; Monnet, C.; Jacques, N.; Antoinette, A.; Irlinger, F. Assessment of the microbial diversity at the surface of Livarot cheese using culture-dependent and independent approaches. Int. J. Food Microbiol. 2009, 133, 31–37. [Google Scholar] [CrossRef]
- Dolci, P.; Alessandria, V.; Rantsiou, K.; Rolle, L.; Zeppa, G.; Cocolin, L. Microbial dynamics of Castelmagno PDO, a traditional Italian cheese, with a focus on lactic acid bacteria ecology. Int. J. Food Microbiol. 2008, 122, 302–311. [Google Scholar] [CrossRef]
- Martín-Platero, A.M.; Valdivia, E.; Maqueda, M.; Martín-Sánchez, I.; Martínez-Bueno, M. Polyphasic approach to bacterial dynamics during the ripening of Spanish farmhouse cheese, using culture-dependent and -independent methods. Appl. Environ. Microbiol. 2008, 74, 5662–5673. [Google Scholar] [CrossRef]
- Delbès, C.; Ali-Mandjee, L.; Montel, M.C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 2007, 73, 1882–1891. [Google Scholar] [CrossRef]
- Rea, M.C.; Görges, S.; Gelsomino, R.; Brennan, N.M.; Mounier, J.; Vancanneyt, M.; Scherer, S.; Swings, J.; Cogan, T.M. Stability of the biodiversity of the surface consortia of Gubbeen, a red-smear cheese. J. Dairy Sci. 2007, 90, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Brennan, N.M.; Ward, A.C.; Beresford, T.P.; Fox, P.F.; Goodfellow, M.; Cogan, T.M. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 2002, 68, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Callon, C.; Millet, L.; Montel, M.C. Diversity of lactic acid bacteria isolated from AOC Salers cheese. J. Dairy Res. 2004, 71, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D. High-throughput sequencing and metagenomics: Moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 2011, 150, 81–94. [Google Scholar] [CrossRef]
- Jany, J.L.; Barbier, G. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 2008, 25, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J.; Abt, B.; Sikorski, J. Present and future of culturing bacteria. Ann. Rev. Microbiol. 2017, 71, 711–730. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.S.; Ghosh, A.R. Assessment of bacterial viability: A comprehensive review on recent advances and challenges. Microbiology 2019, 165, 593–610. [Google Scholar] [CrossRef]
- Yunita, D.; Dodd, C.E.R. Microbial community dynamics of a blue-veined raw milk cheese from the United Kingdom. J. Dairy Sci. 2018, 101, 4923–4935. [Google Scholar] [CrossRef] [PubMed]
- Larpin-Laborde, S.; Imran, M.; Bonaïti, C.; Bora, N.; Gelsomino, R.; Goerges, S.; Irlinger, F.; Goodfellow, M.; Ward, A.C.; Vancanneyt, M.; et al. Surface microbial consortia from Livarot, a French smear-ripened cheese. Can. J. Microbiol. 2011, 57, 651–660. [Google Scholar] [CrossRef]
- Feligini, M.; Panelli, S.; Buffoni, J.N.; Bonacina, C.; Andrighetto, C.; Lombardi, A. Identification of microbiota present on the surface of Taleggio cheese using PCR-DGGE and RAPD-PCR. J. Food Sci. 2012, 77, M609–M615. [Google Scholar] [CrossRef]
- Ercolini, D.; Moschetti, G.; Blaiotta, G.; Coppola, S. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo Mozzarella cheese production: Bias of culture-dependent and culture-independent analyzes. Syst. Appl. Microbiol. 2001, 24, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Dolci, P.; Zenato, S.; Pramotton, R.; Barmaz, A.; Alessandria, V.; Rantsiou, K.; Cocolin, L. Cheese surface microbiota complexity: RT-PCR-DGGE, a tool for a detailed picture? Int. J. Food Microbiol. 2013, 162, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Flórez, A.B.; Mayo, B. Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int. J. Food Microbiol. 2006, 110, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, C.L.; Torriani, S.; Akkermans, A.D.; de Vos, W.M.; Vaughan, E.E. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 2002, 68, 1882–1892. [Google Scholar] [CrossRef]
- Roth, E.; Miescher Schwenninger, S.; Hasler, M.; Eugster-Meier, E.; Lacroix, C. Population dynamics of two antilisterial cheese surface consortia revealed by temporal temperature gradient gel electrophoresis. BMC Microbiol. 2010, 10, 74. [Google Scholar] [CrossRef]
- Bertani, G.; Levante, A.; Lazzi, C.; Bottari, B.; Gatti, M.; Neviani, E. Dynamics of a natural bacterial community under technological and environmental pressures: The case of natural whey starter for Parmigiano Reggiano cheese. Food Res. Int. 2020, 129, 108860. [Google Scholar] [CrossRef]
- Agrimonti, C.; Bottari, B.; Sardaro, M.L.S.; Marmiroli, N. Application of real-time PCR (qPCR) for characterization of microbial populations and type of milk in dairy food products. Crit. Rev. Food Sci. Nutr. 2019, 59, 423–442. [Google Scholar] [CrossRef] [PubMed]
- Hermet, A.; Mounier, J.; Keravec, M.; Vasseur, V.; Barbier, G.; Jany, J.L. Application of capillary electrophoresis single-stranded conformation polymorphism (CE-SSCP) analysis for identification of fungal communities in cheese. Food Microbiol. 2014, 41, 82–90. [Google Scholar] [CrossRef]
- Duthoit, F.; Godon, J.J.; Montel, M.C. Bacterial community dynamics during production of registered designation of origin Salers cheese as evaluated by 16S rRNA gene single-strand conformation polymorphism analysis. Appl. Environ. Microbiol. 2003, 69, 3840–3848. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.I.; Rossetti, L.; Martínez, B.; Rodríguez, A.; Giraffa, G. Application of reverse transcriptase PCR-based T-RFLP to perform semi-quantitative analysis of metabolically active bacteria in dairy fermentations. J. Microbiol. Methods 2006, 65, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Alegría, A.; Szczesny, P.; Mayo, B.; Bardowski, J.; Kowalczyk, M. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and -independent approaches. Appl. Environ. Microbiol. 2012, 78, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A review on the applications of Next Generation Sequencing technologies as applied to food-related microbiome studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef] [PubMed]
- Kergourlay, G.; Taminiau, B.; Daube, G.; Champomier Vergès, M.C. Metagenomic insights into the dynamics of microbial communities in food. Int. J. Food Microbiol. 2015, 213, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Mayo, B.; Rachid, C.T.C.C.; Alegría, A.; Leite, A.M.O.; Peixoto, R.S.; Delgado, S. Impact of next generation sequencing techniques in Food Microbiology. Curr. Genom. 2014, 15, 293–309. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 2012, 45, 377–389. [Google Scholar] [CrossRef]
- Ercolini, D.; De Filippis, F.; La Storia, A.; Iacono, M. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo Mozzarella cheese. Appl. Environ. Microbiol. 2012, 78, 8142–8145. [Google Scholar] [CrossRef]
- Lusk, T.S.; Ottensen, A.R.; White, J.R.; Allard, M.W.; Brown, E.W.; Kase, J.E. Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BCM Microbiol. 2012, 12, 254. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef]
- Masoud, W.; Takamiya, M.; Vogensen, F.K.; Lillevang, S.; Al-Soud, W.A.; Sørensen, S.J.; Jakobsen, M. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturing gradient gel electrophoresis and pyrosequencing. Int. Dairy J. 2010, 21, 142–148. [Google Scholar] [CrossRef]
- Yeluri Jonnala, B.R.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the cheese microbiome and its relevance to industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef]
- Jin, H.; Mo, L.; Pan, L.; Hou, Q.; Li, C.; Darima, I.; Yu, J. Using PacBio sequencing to investigate the bacterial microbiota of traditional Buryatian cottage cheese and comparison with Italian and Kazakhstan artisanal cheeses. J. Dairy Sci. 2018, 101, 6885–6896. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, Y.; Xu, H.; Xi, X.; Hou, Q.; Feng, S.; Wuri, L.; Bian, Y.; Yu, Z.; Kwok, L.Y.; et al. Bacterial microbiota of Kazakhstan cheese revealed by single molecule real time (SMRT) sequencing and its comparison with Belgian, Kalmykian and Italian artisanal cheeses. BMC Microbiol. 2017, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Ritschard, J.S.; Amato, L.; Kumar, Y.; Müller, B.; Meile, L.; Schuppler, M. The role of the surface smear microbiome in the development of defective smear on surface-ripened red-smear cheese. AIMS Microbiol. 2018, 4, 622–641. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco Baruch, M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Ceugniez, A.; Taminiau, B.; Coucheney, F.; Jacques, P.; Delcenserie, V.; Daube, G.; Drider, D. Use of a metagenetic approach to monitor the bacterial microbiota of “Tomme d’Orchies” cheese during the ripening process. Int. J. Food Microbiol. 2017, 247, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Cleary, J.L.; Kolachina, S.; Wolfe, B.E.; Sanchez, L.M. Coproporphyrin III produced by the bacterium Glutamicibacter arilaitensis binds zinc and is upregulated by fungi in cheese rinds. mSystems 2018, 3, e00036-18. [Google Scholar] [CrossRef] [PubMed]
- Kamelamela, N.; Zalesne, M.; Morimoto, J.; Robbat, A.; Wolfe, B.E. Indigo- and indirubin-producing strains of Proteus and Psychrobacter are associated with purple rind defect in a surface-ripened cheese. Food Microbiol. 2018, 76, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Delgado, D.; Rachid, C.T.C.C.; Fernández, E.; Rychlik, T.; Alegría, A.; Peixoto, R.S.; Mayo, B. Diversity of thermophilic bacteria in raw, pasteurized and selectively-cultured milk, as assessed by culturing, PCR-DGGE and pyrosequencing. Food Microbiol. 2013, 36, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Skizuka, T.; Kishi, N.; Yamahita, A.; Kuroda, M. Conventional culture methods with commercially available media unveil the presence of novel culturable bacteria. Gut Microbes 2018, 10, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, Q.; Nie, Y.; Wu, J.; Xu, Y. Construction of a synthetic microbiota for reproducible flavor compound metabolism in Chinese light-aroma-type liquor produced by solid-state fermentation. Appl. Environ. Microbiol. 2019, 85, e03090-18. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, I.; Di Cagno, R.; Buchin, S.; De Angelis, M.; Gobbetti, M. Spatial distribution of the metabolically active microbiota within Italian PDO ewes’ milk cheeses. PLoS ONE 2016, 11, e0153213. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Hébert, A.; Abraham, A.L.; Rasmussen, S.; Monnet, C.; Pons, N.; Delbès, C.; Loux, V.; Batto, J.M.; Leonard, P.; et al. Construction of a dairy microbial genome catalog opens new perspectives for the metagenomic analysis of dairy fermented products. BMC Genom. 2014, 15, 1101. [Google Scholar] [CrossRef] [PubMed]
- Bonaïti, C.; Irlinger, F.; Spinnler, H.E.; Engel, E. An iterative sensory procedure to select odor-active associations in complex consortia of microorganisms: Application to the construction of a cheese model. J. Dairy Sci. 2005, 88, 1671–1684. [Google Scholar] [CrossRef]
- Mayo, B.; Ammor, M.S.; Delgado, S.; Alegría, A. Fermented milk products. In Fermented Food and Beverages of the World; Tmang, J.P., Kilasapathy, K., Eds.; Taylor & Francis: Abingdon, UK, 2010; pp. 263–288. [Google Scholar]
- Wouters, J.T.M.; Ayad, E.H.E.; Hugenholtz, J.; Smit, G. Microbes from raw milk for fermented dairy products. Int. Dairy J. 2002, 12, 91–109. [Google Scholar] [CrossRef]
- Sheedan, A.; O’Cuinn, G.; Fitzgerald, R.J.; Wilkinson, M.G. Distribution of microbial flora, intracellular enzymes and compositional indices throughout a 12 kg Cheedar cheese block during ripening. Int. Dairy J. 2009, 19, 321–329. [Google Scholar]
- Alessandria, V.; Ferrocino, I.; De Filippis, F.; Fontana, M.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Microbiota of an Italian Grana-like cheese during manufacture and ripening, unraveled by 16S rRNA-based approaches. Appl. Environ. Microbiol. 2016, 82, 3988–3995. [Google Scholar] [CrossRef]
- Ropars, J.; Cruaud, C.; Lacoste, S.; Dupont, J. A taxonomic and ecological overview of cheese fungi. Int. J. Food Microbiol. 2012, 155, 199–210. [Google Scholar] [CrossRef]
- Oliveira, J.; Mahony, J.; Hanemaaijer, L.; Kouwen, T.R.H.M.; van Sinderen, D. Biodiversity of bacteriophages infecting Lactococcus lactis starter cultures. J. Dairy Sci. 2018, 101, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Alexeeva, S.; Guerra-Martínez, J.A.; Spus, M.; Smid, E.J. Spontaneously induced prophages are abundant in a naturally evolved bacterial starter culture and deliver competitive advantage to the host. BMC Microbiol. 2018, 18, 120. [Google Scholar] [CrossRef]
- Mahony, J.; Moscarelli, A.; Kelleher, P.; Lugli, G.A.; Ventura, M.; Settanni, L.; van Sinderen, D. Phage biodiversity in artisanal cheese wheys reflects the complexity of the fermentation process. Viruses 2017, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, H.R.; Saad, S.M.; Gibson, G.R.; Vulevic, J. Functional petit-suisse cheese: Measure of the prebiotic effect. Anaerobe 2007, 13, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Ferreiro, T.; Martínez, S.; Gayoso, L.; Rodríguez-Otero, J.L. 2016. Evolution of phospholipid contents during the production of quark cheese from buttermilk. J. Dairy Sci. 2016, 99, 4154–4159. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Seo, Y.; Ha, J.; Kim, S.; Choi, Y.; Oh, H.; Lee, Y.; Kim, Y.; Kang, J.; Park, E.; et al. Influence of milk microbiota on Listeria monocytogenes survival during cheese ripening. Food Sci. Nutr. 2020, 8, 5071–5076. [Google Scholar] [CrossRef] [PubMed]
- Delcenserie, V.; Taminiau, B.; Delhalle, L.; Nezer, C.; Doyen, P.; Crevecoeur, S.; Roussey, D.; Korsak, N.; Daube, G. Microbiota characterization of a Belgian protected designation of origin cheese, Herve cheese, using metagenomic analysis. J. Dairy Sci. 2014, 97, 6046–6056. [Google Scholar] [CrossRef]
- Fuka, M.M.; Wallisch, S.; Engel, M.; Welzl, G.; Havranek, J.; Schloter, M. Dynamics of bacterial communities during the ripening process of different Croatian cheese types derived from raw ewe’s milk cheeses. PLoS ONE 2013, 8, e80734. [Google Scholar] [CrossRef]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Biochemistry of cheese ripening. In Fundamentals of Cheese Science; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H., Eds.; Springer: Boston, MA, USA, 2017; pp. 391–442. [Google Scholar]
- Laurencík, M.; Sulo, P.; Sláviková, E.; Piecková, E.; Seman, M.; Ebringer, L. The diversity of eukaryotic microbiota in the traditional Slovak sheep cheese-bryndza. Int. J. Food Microbiol. 2008, 127, 176–179. [Google Scholar] [CrossRef]
- Flórez, A.B.; Álvarez-Martín, P.; López-Díaz, T.M.; Mayo, B. Microbiological characterisation of the traditional Spanish blue-veined Cabrales cheese: Identification of dominant lactic acid bacteria. Eur. Food Res. Technol. 2006, 223, 503–508. [Google Scholar] [CrossRef]
- Pangallo, D.; Saková, N.; Koreňová, J.; Puškárová, A.; Kraková, L.; Valík, L.; Kuchta, T. Microbial diversity and dynamics during the production of May bryndza cheese. Int. J. Food Microbiol. 2014, 170, 38–43. [Google Scholar] [CrossRef]
- De Pasquale, I.; Calasso, M.; Mancini, L.; Ercolini, D.; La Storia, A.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Causal relationship between microbial ecology dynamics and proteolysis during manufacture and ripening of protected designation of origin (PDO) cheese Canestrato Pugliese. Appl. Environ. Microbiol. 2014, 80, 4085–4094. [Google Scholar] [CrossRef] [PubMed]
- Afshari, R.; Pillidge, C.J.; Read, E.; Rochfort, S.; Dias, D.A.; Osborn, A.M.; Gill, H. New insights into cheddar cheese microbiota-metabolome relationships revealed by integrative analysis of multi-omics data. Sci. Rep. 2020, 10, 3164. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.K.; Carstens, C.K.; Ramachandran, P.; Shazer, A.G.; Narula, S.S.; Reed, E.; Ottesen, A.; Schill, K.M. Metagenomics of pasteurized and unpasteurized Gouda cheese using targeted 16S rDNA sequencing. BMC Microbiol. 2018, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Ceugniez, A.; Taminiau, B.; Coucheney, F.; Jacques, P.; Delcenserie, V.; Daube, G.; Drider, D. Fungal diversity of “Tomme d’Orchies” cheese during the ripening process as revealed by a metagenomic study. Int. J. Food Microbiol. 2017, 258, 89–93. [Google Scholar] [CrossRef]
- Ceugniez, A.; Drider, D.; Jacques, P.; Coucheney, F. Yeast diversity in a traditional French cheese “Tomme d’orchies” reveals infrequent and frequent species with associated benefits. Food Microbiol. 2015, 52, 177–184. [Google Scholar] [CrossRef]
- Bodinaku, I.; Shaffer, J.; Connors, A.B.; Steenwyk, J.L.; Biango-Daniels, M.N.; Kastman, E.K.; Rokas, A.; Robbat, A.; Wolfe, B.E. Rapid phenotypic and metabolomic domestication of wild Penicillium molds on cheese. mBio 2019, 10, e02445-19. [Google Scholar] [CrossRef] [PubMed]
- Laroute, V.; Tormo, H.; Couderc, C.; Mercier-Bonin, M.; Le Bourgeois, P.; Cocaign-Bousquet, M.; Daveran-Mingot, M.L. From genome to phenotype: An integrative approach to evaluate the biodiversity of Lactococcus lactis. Microorganisms 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D.; Fitzgerald, G.F.; McAuliffe, O. From field to fermentation: The origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiol. 2015, 47, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.L.; Klaenhammer, T.R. Genomic evolution of domesticated microorganisms. Annu. Rev. Food Sci. Technol. 2010, 1, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liu, F.; Shi, X.; Wang, B.; Li, K.; Li, B.; Zhuge, B. Dynamic correlations between microbiota succession and flavor development involved in the ripening of Kazak artisanal cheese. Food Res. Int. 2018, 105, 733–742. [Google Scholar] [CrossRef]
- Flórez, A.B.; Álvarez-Martín, P.; López-Díaz, T.M.; Mayo, B. Morphotypic and molecular identification of filamentous fungi from Spanish blue-veined Cabrales cheese and technological characterisation of Penicillium roqueforti and Geotrichum candidum strains. Int. Dairy J. 2007, 17, 350–357. [Google Scholar] [CrossRef]
- Guzzon, R.; Carafa, I.; Tuohy, K.; Cervantes, G.; Vernetti, L.; Barmaz, A.; Larcher, R.; Franciosi, E. Exploring the microbiota of the red-brown defect in smear-ripened cheese by 454-pyrosequencing and its prevention using different cleaning systems. Food Microbiol. 2017, 62, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Bassi, D.; Puglisi, E.; Cocconcelli, P.S. Understanding the bacterial communities of hard cheese with blowing defect. Food Microbiol. 2015, 52, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Sharma, K.; Swanson, B.G.; Yüksel, G.U.; Clark, S. Nonstarter lactic acid bacteria biofilms and calcium lactate crystals in Cheddar cheese. J. Dairy Sci. 2006, 89, 1452–1466. [Google Scholar] [CrossRef]
- Rosengren, A.; Fabricius, A.; Guss, B.; Sylvén, S.; Lindqvist, R. Occurrence of foodborne pathogens and characterization of Staphylococcus aureus in cheese produced on farm-dairies. Int. J. Food Microbiol. 2010, 144, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Linares, D.M.; Del Río, B.; Ladero, V.; Martínez, N.; Fernández, M.; Martín, M.C.; Alvarez, M.A. Factors influencing biogenic amines accumulation in dairy products. Front. Microbiol. 2012, 3, 180. [Google Scholar] [CrossRef] [PubMed]
- Thommes, M.; Wang, T.; Zhao, Q.; Paschalidis, I.C.; Segrè, D. Designing metabolic division of labor in microbial communities. mSystems 2019, 4, e00263-18. [Google Scholar] [CrossRef] [PubMed]
- Embree, M.; Liu, J.K.; Al-Bassam, M.M.; Zengler, K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc. Natl. Acad. Sci. USA 2015, 112, 15450–15455. [Google Scholar] [CrossRef]
- O’Connor, P.M.; Kuniyoshi, T.M.; Oliveira, R.P.; Hill, C.; Ross, R.P.; Cotter, P.D. Antimicrobials for food and feed; a bacteriocin perspective. Curr. Opin. Biotechnol. 2020, 61, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Leyva Salas, M.; Mounier, J.; Maillard, M.B.; Valence, F.; Coton, E.; Thierry, A. Identification and quantification of natural compounds produced by antifungal bioprotective cultures in dairy products. Food Chem. 2019, 301, 125260. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Baran, R.; Brodie, E.L.; Mayberry-Lewis, J.; Hummel, E.; Da Rocha, U.N.; Chakraborty, R.; Bowen, B.P.; Karaoz, U.; Cadillo-Quiroz, H.; Garcia-Pichel, F. Exometabolite niche partitioning among sympatric soil bacteria. Nat. Commun. 2015, 6, 8289. [Google Scholar] [CrossRef] [PubMed]
- Koropatkin, N.M.; Cameron, E.A.; Martens, E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012, 10, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; McDonnell, B.; Casey, E.; van Sinderen, D. Phage-host interactions of cheese-making lactic acid bacteria. Annu. Rev. Food Sci. Technol. 2016, 7, 267–285. [Google Scholar] [CrossRef]
- Erkus, O.; de Jager, V.C.L.; Spus, M.; van Alen-Boerritger, I.J.; van Rijswijck, I.M.H.; Hazelwood, L.; Janssen, P.W.M.; van Hijum, S.A.F.T.; Kleerebezem, M.; Smid, E.J. Multifactorial diversity sustains microbial community stability. ISME J. 2013, 7, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Valera, F.; Martín-Cuadrado, A.B.; Rodríguez-Brito, B.; Pasic, L.; Thingstad, T.F.; Rohwer, F.; Mira, A. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 2009, 7, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Braga, R.M.; Dourado, M.N.; Araújo, W.L. Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 2016, 47, 86–98. [Google Scholar] [CrossRef]
- Pacheco, A.R.; Segrè, D. A multidimensional perspective on microbial interactions. FEMS Microbiol. Lett. 2019, 366, fnz125. [Google Scholar] [CrossRef] [PubMed]
- Smid, E.J.; Lacroix, C. Microbe-microbe interactions in mixed culture food fermentations. Curr. Opin. Biotechnol. 2013, 24, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Irlinger, F.; Mounier, J. Microbial interactions in cheese: Implications for cheese quality and safety. Curr. Opin. Biotechnol. 2009, 20, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Mounier, J.; Monnet, C.; Vallaeys, T.; Arditi, R.; Sarthou, A.S.; Hélias, A.; Irlinger, F. Microbial interactions within a cheese microbial community. Appl. Environ. Microbiol. 2008, 74, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, S.; de Bok, F.A.; Hugenholtz, J.; van Hylckama Vlieg, J.E. Unraveling microbial interactions in food fermentations: From classical to genomics approaches. Appl. Environ. Microbiol. 2008, 74, 4997–5007. [Google Scholar] [CrossRef]
- Iskandar, C.F.; Cailliez-Grimal, C.; Borges, F.; Revol-Junelles, A.M. Review of lactose and galactose metabolism in Lactic Acid Bacteria dedicated to expert genomic annotation. Trends Food Sci. Technol. 2019, 88, 121–132. [Google Scholar] [CrossRef]
- Upadhyay, V.K.; McSweeney, P.L.H.; Magboul, A.A.A.; Fox, P.F. Proteolysis in cheese during ripening. In Cheese Chemistry, Physics and Microbiology; Fox, P.F., McSweeney, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Elsevier Academic Press: London, UK, 2004; Volume 1, pp. 391–434. [Google Scholar]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef]
- Noordman, W.H.; Reissbrodt, R.; Bongers, R.S.; Rademaker, J.L.W.; Bockelmann, W.; Smit, G. Growth stimulation of Brevibacterium sp. by siderophores. J. Appl. Microbiol. 2006, 101, 637–646. [Google Scholar] [CrossRef]
- Özcan, E.; Seven, M.; Şirin, B.; Çakır, T.; Nikerel, E.; Teusink, B.; Toksoy Öner, E. Dynamic co-culture metabolic models reveal the fermentation dynamics, metabolic capacities and interplays of cheese starter cultures. Biotechnol. Bioeng. 2020, 1–15. [Google Scholar] [CrossRef]
- Monnet, C.; Back, A.; Irlinger, F. Growth of aerobic ripening bacteria at the cheese surface is limited by the availability of iron. Appl. Environ. Microbiol. 2012, 78, 3185–3192. [Google Scholar] [CrossRef]
- Schnürer, J.; Magnusson, J. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 2005, 16, 70–78. [Google Scholar] [CrossRef]
- Arqués, J.L.; Rodríguez, E.; Langa, S.; Landete, J.M.; Medina, M. Antimicrobial activity of lactic acid bacteria in dairy products and gut: Effect on pathogens. Biomed. Res. Int. 2015, 2015, 584183. [Google Scholar] [CrossRef] [PubMed]
- Garnier, L.; Mounier, J.; Lê, S.; Pawtowski, A.; Pinon, N.; Camier, B.; Chatel, M.; Garric, G.; Thierry, A.; Coton, E.; et al. Development of antifungal ingredients for dairy products: From in vitro screening to pilot scale application. Food Microbiol. 2019, 81, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.C.; Staliano, C.D.; Vieira, A.D.; Villarreal, M.L.; Todorov, S.D.; Saad, S.M.; Franco, B.D. Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. sakei 2a in a potentially synbiotic cheese spread. Food Microbiol. 2015, 48, 143–152. [Google Scholar] [CrossRef]
- Imran, M.; Bré, J.M.; Vernoux, J.P.; Desmasures, N. Reduced growth of Listeria monocytogenes is not associated with individual microbial strains. Food Microbiol. 2013, 33, 30–39. [Google Scholar] [CrossRef]
- Loessner, M.; Guenther, S.; Steffan, S.; Scherer, S. A pediocin-producing Lactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Appl. Environ. Microbiol. 2003, 69, 1854–1857. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, O.; Hill, C.; Ross, R.P. Inhibition of Listeria monocytogenes in cottage cheese manufactured with a lacticin 3147-producing starter culture. J. Appl. Microbiol. 1999, 86, 251–256. [Google Scholar] [CrossRef]
- Ennahar, S.; Assobhel, O.; Hasselmann, C. Inhibition of Listeria monocytogenes in a smear-surface soft cheese by Lactobacillus plantarum WHE 92, a pediocin AcH producer. J. Food Prot. 1998, 61, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Núñez, M.; Rodríguez, J.L.; García, E.; Gaya, P.; Medina, M. Inhibition of Listeria monocytogenes by enterocin 4 during the manufacture and ripening of Manchego cheese. J. Appl. Microbiol. 1997, 83, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Harmark, K.; Davidson, B.E.; Hillier, A.J.; Gordon, J.B.; Wilcock, A.; Hickey, M.W.; Coventry, M.J. Inhibition of Listeria monocytogenes by piscicolin 126 in milk and Camembert cheese manufactured with a thermophilic starter. J. Appl. Microbiol. 1997, 82, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Delbès-Paus, C.; Dorchies, G.; Chaabna, Z.; Callon, C.; Montel, M.C. Contribution of hydrogen peroxide to the inhibition of Staphylococcus aureus by Lactococcus garvieae in interaction with raw milk microbial community. Food Microbiol. 2010, 27, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Rilla, N.; Martínez, B.; Rodríguez, A. Inhibition of a methicillin-resistant Staphylococcus aureus strain in Afuega’l Pitu cheese by the nisin Z-producing strain Lactococcus lactis subsp. lactis IPLA 729. J. Food Prot. 2004, 67, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Ferrari Ida, S.; de Souza, J.V.; Ramos, C.L.; da Costa, M.M.; Schwan, R.F.; Dias, F.S. Selection of autochthonous lactic acid bacteria from goat dairies and their addition to evaluate the inhibition of Salmonella typhi in artisanal cheese. Food Microbiol. 2016, 60, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Garde, S.; Ávila, M.; Arias, R.; Gaya, P.; Núñez, M. Outgrowth inhibition of Clostridium beijerinckii spores by a bacteriocin-producing lactic culture in ovine milk cheese. Int. J. Food Microbiol. 2011, 150, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Mathot, A.G.; Beliard, E.; Thuault, D. Streptococcus thermophilus 580 produces a bacteriocin potentially suitable for inhibition of Clostridium tyrobutyricum in hard cheese. J. Dairy Sci. 2003, 86, 3068–3074. [Google Scholar] [CrossRef]
- Rilla, N.; Martínez, B.; Delgado, T.; Rodríguez, A. Inhibition of Clostridium tyrobutyricum in Vidiago cheese by Lactococcus lactis ssp. lactis IPLA 729, a nisin Z producer. Int. J. Food Microbiol. 2003, 85, 23–33. [Google Scholar] [CrossRef]
- Bassi, D.; Gazzola, S.; Sattin, E.; Dal Bello, F.; Simionati, B.; Cocconcelli, P.S. Lactic acid bacteria adjunct cultures exert a mitigation effect against spoilage microbiota in fresh cheese. Microorganisms 2020, 8, 1199. [Google Scholar] [CrossRef]
- Murado, M.A.; Vazquez, J.A. Biphasic toxicodynamic features of some antimicrobial agents on microbial growth: A dynamic mathematical model and its implications on hormesis. BMC Microbiol. 2010, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Davies, J. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 2006, 33, 496–499. [Google Scholar] [CrossRef] [PubMed]
- Herve-Jimenez, L.; Guillouard, I.; Guedon, E.; Boudebbouze, S.; Hols, P.; Monnet, V.; Maguin, E.; Rul, F. Postgenomic analysis of Streptococcus thermophilus cocultivated in milk with Lactobacillus delbrueckii subsp. bulgaricus: Involvement of nitrogen, purine, and iron metabolism. Appl. Environ. Microbiol. 2009, 75, 2062–2073. [Google Scholar] [CrossRef]
- Callon, C.; Saubusse, M.; Didienne, R.; Buchin, S.; Montel, M.C. Simplification of a complex microbial antilisterial consortium to evaluate the contribution of its flora in uncooked pressed cheese. Int. J. Food Microbiol. 2011, 145, 379–389. [Google Scholar] [CrossRef]
- Bleicher, A.; Stark, T.; Hofmann, T.; Bogovic Matijasić, B.; Rogelj, I.; Scherer, S.; Neuhaus, K. Potent antilisterial cell-free supernatants produced by complex red-smear cheese microbial consortia. J. Dairy Sci. 2010, 93, 4497–4505. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Desmasures, N.; Vernoux, J.P. From undefined red smear cheese consortia to minimal model communities both exhibiting similar anti-listerial activity on a cheese-like matrix. Food Microbiol. 2010, 27, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Monnet, C.; Bleicher, A.; Neuhaus, K.; Sarthou, A.S.; Leclercq-Perlat, M.N.; Irlinger, F. Assessment of the anti-listerial activity of microfloras from the surface of smear-ripened cheeses. Food Microbiol. 2010, 27, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Maoz, A.; Mayr, R.; Scherer, S. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl. Environ. Microbiol. 2003, 69, 4012–4018. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Tsao, M. Inhibition of spoilage yeasts in cheese by killer yeast Williopsis saturnus var. saturnus. Int. J. Food Microbiol. 2009, 131, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Malek, R.; Bonnarme, P.; Irlinger, F.; Frey-Klett, P.; Onésime, D.; Aubert, J.; Loux, V.; Beckerich, J.M. Transcriptomic response of Debaryomyces hansenii during mixed culture in a liquid model cheese medium with Yarrowia lipolytica. Int. J. Food Microbiol. 2018, 264, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Senaka Ranadheera, C.; Evans, C.A.; Adams, M.C.; Baines. S.K. Probiotic viability and physico-chemical and sensory properties of plain and stirred fruit yogurts made from goat’s milk. Food Chem. 2012, 135, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Juillard, V.; Richard, J. Indirect interaction in milk between proteolytic and isogenic nonproteolytic strains of Lactococcus lactis. II. Effect of pre-culturing by a proteolytic strain. Lait 1991, 71, 55–64. [Google Scholar] [CrossRef]
- Baer, A.; Ryba, I. Influence of casein proteolysis by starter bacteria, rennet and plasmin on the growth of propionibacteria in Swiss-type cheese. Lait 1995, 75, 391–400. [Google Scholar] [CrossRef]
- Sudun, W.; Arakawa, K.; Miyamoto, M.; Miyamoto, T. Interaction between lactic acid bacteria and yeasts in airag, an alcoholic fermented milk. Anim. Sci. J. 2013, 84, 66–74. [Google Scholar] [CrossRef]
- Álvarez-Martín, P.; Flórez, A.B.; Hernández-Barranco, A.; Mayo, B. Interaction between dairy yeasts and lactic acid bacteria strains during milk fermentation. Food Control 2008, 1, 62–70. [Google Scholar] [CrossRef]
- Ruiz-Barba, J.L.; Jiménez-Díaz, R. Availability of essential B-group vitamins to Lactobacillus plantarum in green olive fermentation brines. Appl. Environ. Microbiol. 1995, 61, 1294–1297. [Google Scholar] [CrossRef]
- De Freitas, I.; Pinon, N.; Maubois, J.L.; Lortal, S.; Thierry, A. The addition of a cocktail of yeasts species to Cantalet cheese changes bacterial survival and enhances aroma compound formation. Int. J. Food Microbiol. 2009, 129, 37–42. [Google Scholar] [CrossRef]
- Leclercq-Perlat, M.N.; Corrieu, G.; Spinnler, H.E. The color of Brevibacterium linens depends on the yeast used for cheese deacidification. J. Dairy Sci. 2004, 87, 1536–1544. [Google Scholar] [CrossRef]
- Sieuwerts, S.; Molenaar, D.; van Hijum, S.A.F.T.; Beerthuyzen, M.; Stevens, M.J.A.; Janssen, P.W.M.; Ingham, C.J.; de Bok, F.A.M.; de Vos, W.M.; van Hylckama Vlieg, J.E.T. Mixed-culture transcriptome analysis reveals the molecular basis of mixed-culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Appl. Environ. Microbiol. 2010, 76, 7775–7784. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, R.; Maguin, E.; Horiuchi, H.; Hosokawa, M.; Sasaki, Y. The critical role of urease in yogurt fermentation with various combinations of Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 2019, 102, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Desfossés-Foucault, E.; LaPointe, G.; Roy, D. Transcription profiling of interactions between Lactococcus lactis subsp. cremoris SK11 and Lactobacillus paracasei ATCC 334 during Cheddar cheese simulation. Int. J. Food Microbiol. 2014, 178, 76–86. [Google Scholar] [CrossRef]
- Viljoen, B.C. The interaction between yeasts and bacteria in dairy environments. Int. J. Food Microbiol. 2001, 69, 37–44. [Google Scholar] [CrossRef]
- van den Tempel, T.; Nielsen, M.S. Effects of atmospheric conditions, NaCl and pH on growth and interactions between moulds and yeasts related to blue cheese production. Int. J. Food Microbiol. 2000, 57, 193–199. [Google Scholar] [CrossRef]
- Ponomarova, O.; Gabrielli, N.; Sévin, D.C.; Mülleder, M.; Zirngibl, K.; Bulyha, K.; Andrejev, S.; Kafkia, E.; Typas, A.; Sauer, U.; et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017, 5, 345–357. [Google Scholar] [CrossRef]
- Castellote, J.; Fraud, S.; Irlinger, F.; Swennen, D.; Fer, F.; Bonnarme, P.; Monnet, C. Investigation of Geotrichum candidum gene expression during the ripening of Reblochon-type cheese by reverse transcription-quantitative PCR. Int. J. Food Microbiol. 2015, 194, 54–61. [Google Scholar] [CrossRef]
- Dugat-Bony, E.; Straub, C.; Teissandier, A.; Onésime, D.; Loux, V.; Monnet, C.; Irlinger, F.; Landaud, S.; Leclercq-Perlat, M.N.; Bento, P.; et al. Overview of a surface-ripened cheese community functioning by meta-omics analyses. PLoS ONE 2015, 10, e0124360. [Google Scholar] [CrossRef] [PubMed]
- Pham, N.P.; Landaud, S.; Lieben, P.; Bonnarme, P.; Monnet, C. Transcription profiling reveals cooperative metabolic interactions in a microbial cheese-ripening community composed of Debaryomyces hansenii, Brevibacterium aurantiacum, and Hafnia alvei. Front. Microbiol. 2019, 10, 1901. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.; Bailly, J.; Landaud, S.; Monnet, C.; Sarthou, A.S.; Cocaign-Bousquet, M.; Leroy, S.; Irlinger, F.; Bonnarme, P. Investigation of associations of Yarrowia lipolytica, Staphylococcus xylosus, and Lactococcus lactis in culture as a first step in microbial interaction analysis. Appl. Environ. Microbiol. 2009, 75, 6422–6430. [Google Scholar] [CrossRef] [PubMed]
- Blaya, J.; Barzideh, Z.; LaPointe, G. Symposium review: Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 2018, 101, 3611–3629. [Google Scholar] [CrossRef]
- De Pasquale, I.; Di Cagno, R.; Buchin, S.; De Angelis, M.; Gobbetti, M. Microbial ecology dynamics reveal a succession in the core microbiota involved in the ripening of pasta filata Caciocavallo Pugliese cheese. Appl. Environ. Microbiol. 2014, 80, 6243–6255. [Google Scholar] [CrossRef]
- Settanni, L.; Moschetti, G. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 2010, 27, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Marzotto, M.; Cremonese, S.; Rizzotti, L.; Torriani, S. Diversity of Streptococcus thermophilus in bacteriocin production; inhibitory spectrum and occurrence of thermophilin genes. Food Microbiol. 2013, 35, 27–33. [Google Scholar] [CrossRef]
- Simova, E.D.; Beshkova, D.M.; Angelov, M.P.; Dimitrov, Z.P. Bacteriocin production by strain Lactobacillus delbrueckii ssp. bulgaricus BB18 during continuous prefermentation of yogurt starter culture and subsequent batch coagulation of milk. J. Ind. Microbiol. Biotechnol. 2008, 35, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Medlock, G.L.; Carey, M.A.; McDuffie, D.G.; Mundy, M.B.; Giallourou, N.; Swann, J.R.; Kolling, G.L.; Papin, J.A. Inferring metabolic mechanisms of interaction within a defined gut microbiota. Cell Syst. 2018, 7, 245–257. [Google Scholar] [CrossRef]
- De Filippis, F.; Genovese, A.; Ferranti, P.; Gilbert, J.A.; Ercolini, D. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci. Rep. 2016, 6, 21871. [Google Scholar] [CrossRef]
- Fischer, C.N.; Trautman, E.P.; Crawford, J.M.; Stabb, E.V.; Handelsman, J.; Broderick, N.A. Metabolite exchange between microbiome members produces compounds that influence Drosophila behabiour. eLife 2017, 6, e18855. [Google Scholar] [CrossRef]
- Acinas, S.G.; Klepac-Ceraj, V.; Hunt, D.E.; Pharino, C.; Ceraj, I.; Distel, D.L.; Polz, M.F. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 2004, 430, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Cosetta, C.M.; Wolfe, B.E. Causes and consequences of biotic interactions within microbiomes. Curr. Opin. Microbiol. 2019, 50, 35–41. [Google Scholar] [CrossRef]
- O’Donnell, S.T.; Ross, R.P.; Stanton, C. The progress of multi-omics technologies: Determining function in lactic acid bacteria using a systems level approach. Front. Microbiol. 2020, 10, 3084. [Google Scholar] [CrossRef]
- Castellanos-Rozo, J.; Pérez Pulido, R.; Grande, M.J.; Lucas, R.; Gálvez, A. Analysis of the bacterial diversity of Paipa cheese (a traditional raw cow’s milk cheese from Colombia) by high-throughput sequencing. Microorganisms 2020, 8, 218. [Google Scholar] [CrossRef]
- Gonçalves, M.T.P.; Benito, M.J.; Córdoba, M.G.; Egas, C.; Merchán, A.V.; Galván, A.I.; Ruiz-Moyano, S. Bacterial communities in Serpa cheese by culture dependent techniques, 16S rRNA gene sequencing and high-throughput sequencing analysis. J. Food Sci. 2018, 83, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Monnet, C.; Dugat-Bony, E.; Swennen, D.; Beckerich, J.M.; Irlinger, F.; Fraud, S.; Bonnarme, P. Investigation of the activity of the microorganisms in a Reblochon-style cheese by metatranscriptomic analysis. Front. Microbiol. 2016, 7, 536. [Google Scholar] [CrossRef] [PubMed]
- Valente, N.I.P.; Rudnitskaya, A.; Oliveira, J.A.B.P.; Gaspar, E.M.M.; Gomes, M.T.S.R. Cheeses made from raw and pasteurized cow’s milk analysed by an electronic nose and an electronic tongue. Sensors 2018, 18, 2415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kastman, E.K.; Guasto, J.S.; Wolfe, B.E. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat. Commun. 2018, 9, 336. [Google Scholar] [CrossRef]
- Kastman, E.K.; Kamelamela, N.; Norville, J.W.; Cosetta, C.M.; Dutton, R.J.; Wolfe, B.E. Biotic interactions shape the ecological distributions of Staphylococcus species. mBio 2016, 7, e01157-16. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.E. Using cultivated microbial communities to dissect microbiome assembly: Challenges, limitations, and the path ahead. mSystems 2018, 3, e00161-17. [Google Scholar] [CrossRef]
- Park, W.; Yoo, J.; Oh, S.; Ham, J.S.; Jeong, S.G.; Kim, Y. Microbiological characteristics of Gouda cheese manufactured with pasteurized and raw milk during ripening using Next Generation Sequencing. Food Sci. Anim. Resour. 2019, 39, 585–600. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Dutton, R.J. Towards an ecosystem approach to cheese microbiology. Microbiol. Spectr. 2013, 1, CM0012-2012. [Google Scholar] [CrossRef]
- Niccum, B.A.; Kastman, E.K.; Kfoury, N.; Robbat, A., Jr.; Wolfe, B.E. Strain-level diversity impacts cheese rind microbiome assembly and function. mSystems 2020, 5, e00149-20. [Google Scholar] [CrossRef]
- Cosetta, C.M.; Kfoury, N.; Robbat, A.; Wolfe, B.E. Fungal volatiles mediate cheese rind microbiome assembly. Environ. Microbiol. 2020, 22, 4745–4760. [Google Scholar] [CrossRef] [PubMed]
- Cosetta, C.M.; Wolfe, B.E. Deconstructing and reconstructing cheese rind microbiomes for experiments in Microbial Ecology and Evolution. Curr. Protoc. Microbiol. 2020, 56, e95. [Google Scholar] [CrossRef]
- Tveit, A.T.; Urich, T.; Frenzel, P.; Svenning, M.M. Metabolic and trophic interactions modulate methane production by Arctic peat microbiota in response to warming. Proc. Natl. Acad. Sci. USA 2015, 112, E2507–E2516. [Google Scholar] [CrossRef] [PubMed]
- Tormo, J.; Barral, J. Accidents de Fromagerie. 2004. Available online: http://www.accident-fromagerie.fr/spip.php (accessed on 20 November 2020).
- Le Bars-Bailly, S.; Bailly, J.D.; Brugere, H. Mold-realted failings in cheesemaking. Rev. Med. Vet. 1999, 150, 413–430. [Google Scholar]
- Salazar, J.K.; Gonsalves, L.J.; Natarajan, V.; Shazer, A.; Reineke, K.; Mhetras, T.; Sule, C.; Carstens, C.K.; Schill, K.M.; Tortorello, M.L. Population dynamics of Listeria monocytogenes, Escherichia coli O157:H7, and native microflora during manufacture and aging of Gouda cheese made with unpasteurized milk. J. Food Prot. 2020, 21, 266–276. [Google Scholar] [CrossRef]
- Daly, D.F.M.; McSweeney, P.L.H.; Sheehan, J.J. Split defect and secondary fermentation in Swiss-type cheeses—A review. Dairy Sci. Technol. 2010, 90, 3–26. [Google Scholar] [CrossRef]
- Sattin, E.; Andreani, N.A.; Carraro, L.; Fasolato, L.; Balzan, S.; Novelli, E.; Squartini, A.; Telatin, A.; Simionati, B.; Cardazzo, B. Microbial dynamics during shelf-life of industrial Ricotta cheese and identification of a Bacillus strain as a cause of a pink discolouration. Food Microbiol. 2016, 57, 8–15. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, O.; Kilcawley, K.; Stefanovic, E. Symposium review: Genomic investigations of flavor formation by dairy microbiota. J. Dairy Sci. 2019, 102, 909–922. [Google Scholar] [CrossRef]
- Alonso, R.; Picón, A.; Gaya, P.; Núñez, M. Proteolysis, lipolysis, volatile compounds and sensory characteristics of Hispanico cheeses made using frozen curd from raw and pasteurized ewe milk. J. Dairy Res. 2013, 80, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, L.; Kojic, M.; Fira, D.; Golic, N.; Strahinic, I.; Lozo, J. Potential of lactic acid bacteria isolated from specific natural niches in food production and preservation. Int. J. Food Microbiol. 2006, 112, 230–235. [Google Scholar] [CrossRef]
- Minty, J.J.; Singer, M.E.; Scholz, S.A.; Bae, C.H.; Ahn, J.H.; Foster, C.E.; Liao, J.C.; Lin, X.N. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc. Natl. Acad. Sci. USA 2013, 110, 14592–14597. [Google Scholar] [CrossRef]
- Ben-Harb, S.; Saint-Eve, A.; Panouillé, M.; Souchon, I.; Bonnarme, P.; Dugat-Bony, E.; Irlinger, F. Design of microbial consortia for the fermentation of pea-protein-enriched emulsions. Int. J. Food Microbiol. 2019, 293, 124–136. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Friman, V.P.; Gu, S.H.; Wang, X.F.; Eisenhauer, N.; Yang, T.J.; Ma, J.; Shen, Q.R.; Xu, Y.C.; et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 2016, 7pii, e01790-16. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Castellanos, J.F.; Biclot, A.; Vrancken, G.; Huys, G.R.; Raes, J. Design of synthetic microbial consortia for gut microbiota modulation. Curr. Opin. Pharmacol. 2019, 49, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Carriço, J.A.; Rossi, M.; Moran-Gilad, J.; van Domselaar, G.; Ramirez, M. A primer on microbial bioinformatics for nonbioinformaticians. Clin. Microbiol. Infect. 2018, 24, 342–349. [Google Scholar] [CrossRef]
- Mataragas, M.; Alessandria, V.; Ferrocino, I.; Rantsiou, K.; Cocolin, L. A bioinformatics pipeline integrating predictive metagenomics profiling for the analysis of 16S rDNA/rRNA sequencing data originated from foods. Food Microbiol. 2018, 76, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Parente, E.; Cocolin, L.; De Filippis, F.; Zotta, T.; Ferrocino, I.; O’Sullivan, O.; Neviani, E.; De Angelis, M.; Cotter, P.D.; Ercolini, D. FoodMicrobionet: A database for the visualisation and exploration of food bacterial communities based on network analysis. Int. J. Food Microbiol. 2016, 219, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Bockelmann, W. Secondary cheese starter cultures. In Technology of Cheesemaking; Law, B.A., Tamime, A.Y., Eds.; Wiley-Blackwell: Oxford, UK, 2010; pp. 193–230. [Google Scholar] [CrossRef]
- Irlinger, F.; Yung, S.A.; Sarthou, A.S.; Delbès-Paus, C.; Montel, M.C.; Coton, E.; Coton, M.; Helinck, S. Ecological and aromatic impact of two Gram-negative bacteria (Psychrobacter celer and Hafnia alvei) inoculated as part of the whole microbial community of an experimental smear soft cheese. Int. J. Food Microbiol. 2012, 153, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Deetae, P.; Mounier, J.; Bonnarme, P.; Spinnler, H.E.; Irlinger, F.; Helinck, S. Effects of Proteus vulgaris growth on the establishment of a cheese microbial community and on the production of volatile aroma compounds in a model cheese. J. Appl. Microbiol. 2009, 107, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Goerges, S.; Mounier, J.; Rea, M.C.; Gelsomino, R.; Heise, V.; Beduhn, R.; Cogan, T.M.; Vancanneyt, M.; Scherer, S. Commercial ripening starter microorganisms inoculated into cheese milk do not successfully establish themselves in the resident microbial ripening consortia of a South German red smear cheese. Appl. Environ. Microbiol. 2008, 74, 2210–2217. [Google Scholar] [CrossRef] [PubMed]
- Feurer, C.; Vallaeys, T.; Corrieu, G.; Irlinger, F. Does smearing inoculum reflect the bacterial composition of the smear at the end of the ripening of a French soft, red-smear cheese? J. Dairy Sci. 2004, 87, 3189–3197. [Google Scholar] [CrossRef]
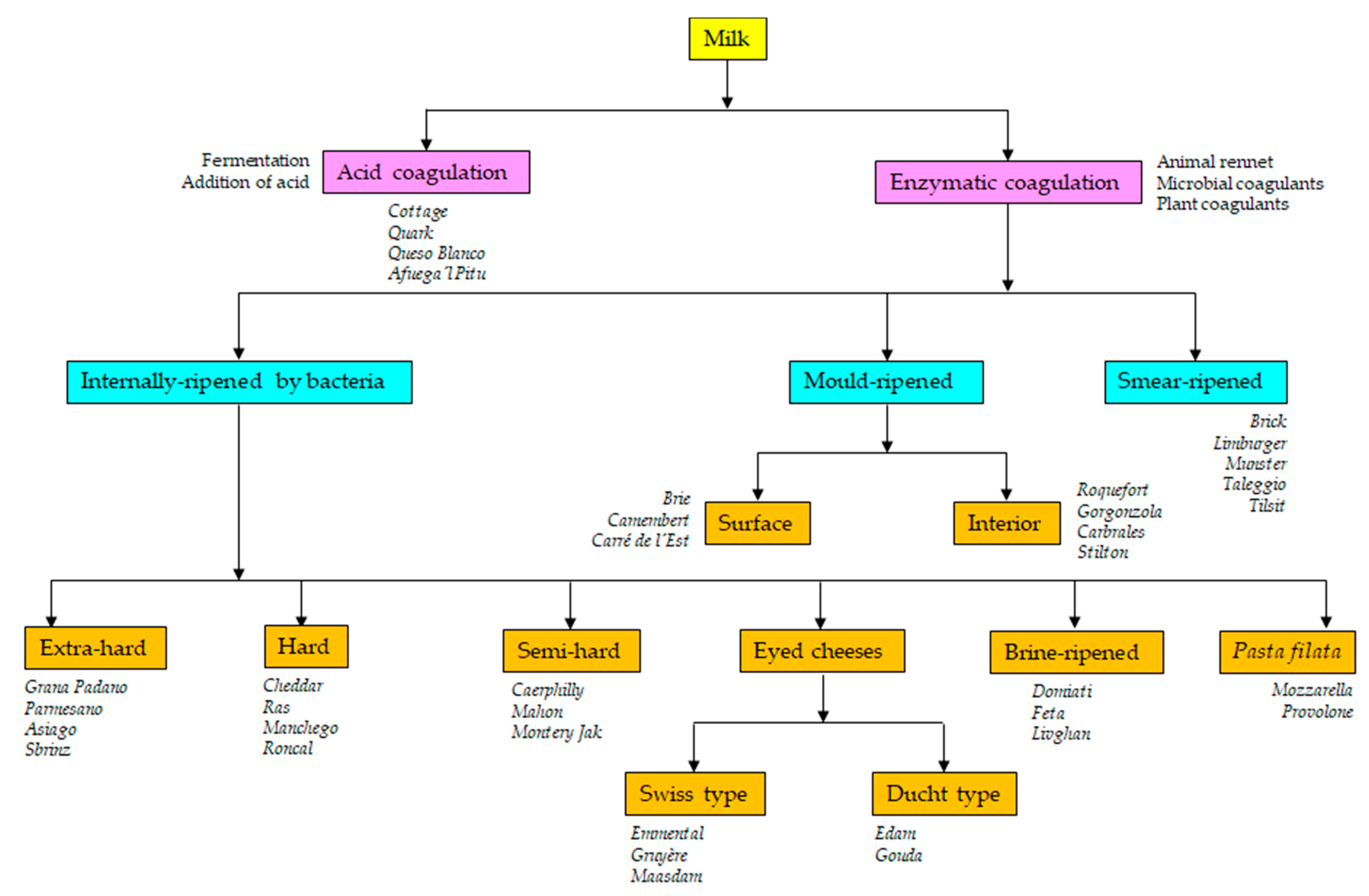
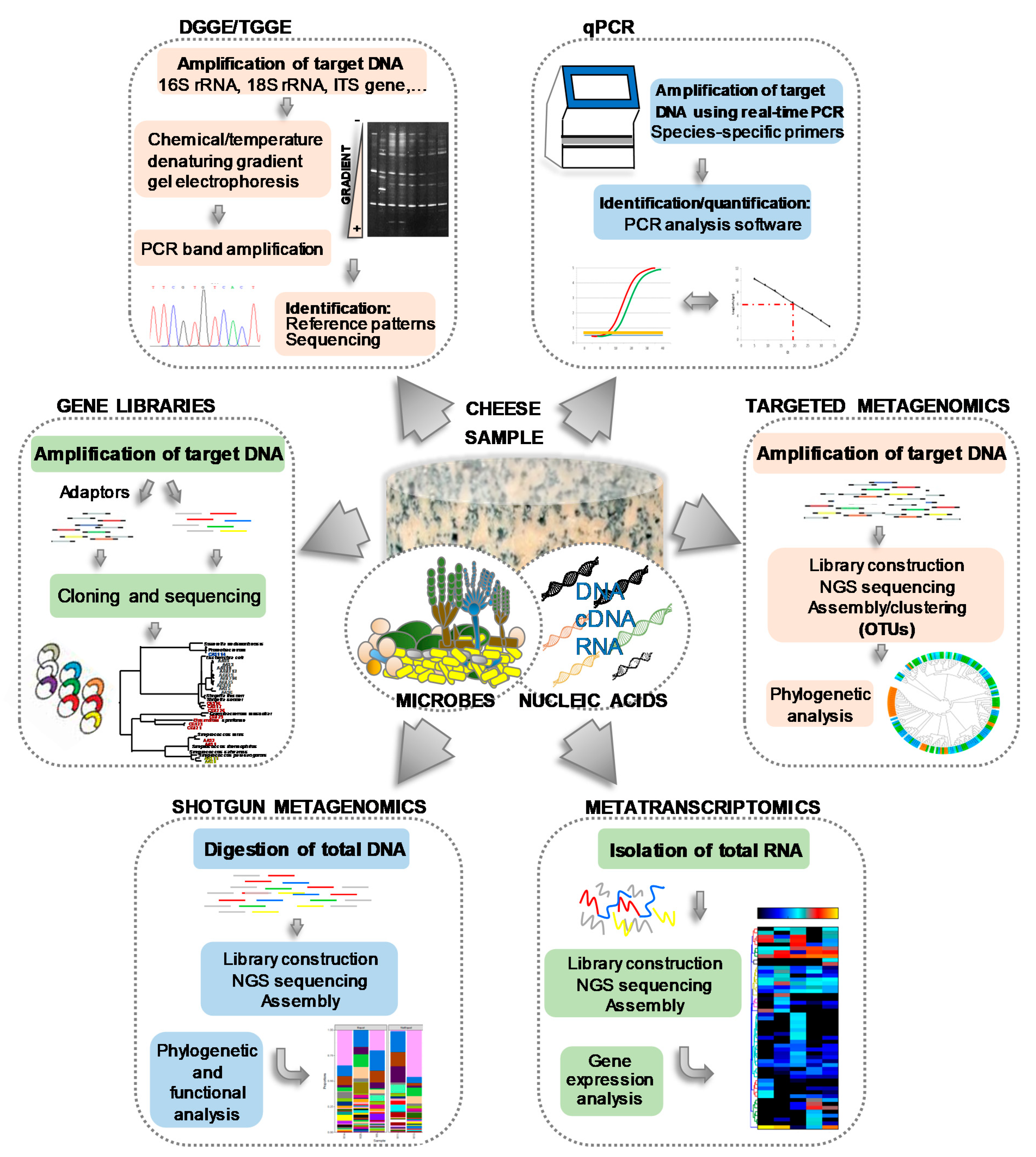
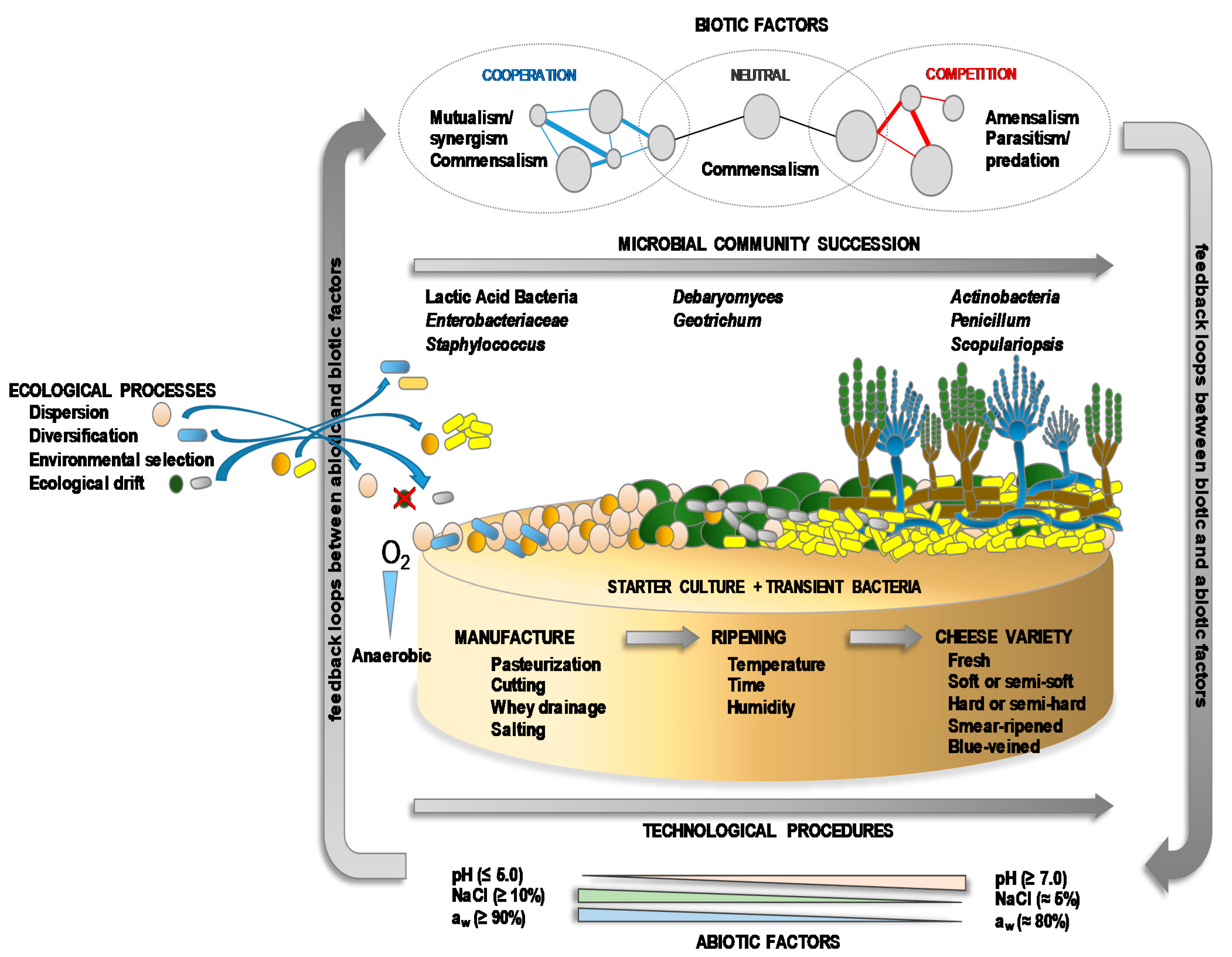
| Microbial Group/Species | Cheese | Type of Starter | Main Role/s |
|---|---|---|---|
| Lactic acid bacteria | |||
| Lc. lactis subsp. lactis Lc. lactis subsp. cremoris | Most cheeses | Primary | Acidification, flavor development |
| S. thermophilus Lb. delbrueckii subsp. lactis | Italian and Swiss types | Primary | Acidification, flavor development |
| Leuc. mesenteroides subsp. cremoris Leuc. lactis | Soft and semi-hard | Secondary/adjunct | Flavor development, CO2 production |
| Lb. helveticus | Semi-hard, hard | Secondary/adjunct | Flavor development, health benefits |
| Lb. casei/Lb. paracasei | Artisanal | Secondary/adjunct | Flavor development |
| Lb. plantarum | Artisanal | Secondary/adjunct | Flavor development |
| Propionibacteria | |||
| Propionibacterium freudenreichii | Swiss-type | Secondary/ripening | Hole formation, flavor development |
| Other bacteria | |||
| Brevibacterium linens | Smear-ripened | Secondary/ripening | Color, flavor development |
| Corynebacterium casei | Smear-ripened | Secondary/ripening | Flavor development |
| Fungi | |||
| P. camemberti | White moldy | Secondary/ripening | Aspect, texture, and flavor development |
| P. roqueforti | Blue-veined | Secondary/ripening | |
| G. candidum | Smear-ripened | Secondary/ripening |
| Cheese/Type, Country (Milk Type) | Technique | Microbial Target | No. of Specimens | Main Families/Genera/Species (Relative Abundance); Sampling Point | Reference |
|---|---|---|---|---|---|
| Culturing | |||||
| Bryndza/soft Feta-type, Slovakia (Sheep) | Culturing | Fungi | 5 species | Geotrichum candidum > Kluyveromyces marxianus > Pichia fermentans > Candida inconspicua > Trichosporon cutaneum | Laurencík et al. [106] |
| Cabrales/blue-veined, Spain (Cow, sheep, and goat) | Culturing | LAB | 15 species | Lc. lactis subsp. lactis > Lb. plantarum > Leuc. mesenteroides > Leuc. citreum > Enterococcus > Lb. paracasei | Flórez et al. [107] |
| Gubbeen/smear-ripened, Ireland (Cow) | Culturing | Corynebacteria | 39 species | Corynebacterium casei (50.2%) > Corynebacterium mooreparkense (26%) > Microbacterium gubbeenense (12.8%); cheese rind | Brennan et al. [48] |
| May bryndza/soft, Slovakia (Sheep) | Culturing | Bacteria Fungi | 5 species 17 species | Lc. lactis subsp. cremoris > Lc. lactis subsp. lactis > Mannheimia glucosida G. candidum > Penicillium > Beauveria brongniartii > Alternaria alternata | Pangallo et al. [108] |
| Rinds of 33 cheeses/smear-ripened, various countries (Cow, sheep, or goat) | Culturing, sequencing | Microbes | 104 bacterial genera, 39 fungal genera | Staphylococcus (78%) > Brevibacterium (75%) > Corynebacterium (75%) > Arthrobacter (66%) > Lactococcus (50%) > Enterococcus (41%) > Brachybacterium (38%) > Microbacterium (38%) > Psychrobacter (33%) > Halomonas (31%) > Lactobacillus (25%) > Streptococcus (22%) > Marinilactibacillus (22%) > Pseudoalteromonas (22%) > Agrococcus (19%) > Micrococcus (19%) > Vibrio (19%) > Vagococcus (16%) > Facklamia (16%) Debaryomyces (86%) > Yarrowia (57%) > Candida (54%) > Geotrichum (49%) > Kluyveromyces (32%) > Pichia (22%) > Penicillium (19%) > Scopulariopsis (8%) > Fusarium (8%) | Irlinger et al. [12] |
| Scamorza Altamurana/pasta filata, Italy (Cow) | Culturing | LAB | 10 species | Lb. delbrueckii > Streptococcus macedonicus > S. thermophilus > Enterococcus durans > Lb. fermentum > Lb. paracasei | Baruzzi et al. [28] |
| Culturing and molecular methods | |||||
| Casín/kneaded, Spain (Cow) | Culturing DGGE | Bacteria | 14 species | Lc. lactis subsp. lactis > Lactococcus garvieae > Staphylococcus saprophyticus > Klebsiella > Lb. plantarum | Alegría et al. [25] |
| Bacteria (V1-V2 16S rDNA) | 14 OTUs | Lc. lactis, Streptococcus parauberis, S. thermophilus, Lc. garvieae, Lb. plantarum, Enterobacter, Corynebacterium variabile, Lb. paracasei, Macrococcus caseolyticus | |||
| Castelmagno/semi-hard, Italy (Cow) | Culturing PCR-DGGE | LAB Bacteria (V1 16S rDNA) | 11 species 7 OTUs | Lc. lactis subsp. lactis > Lb. plantarum > Lc. paracasei >Enterococcus faecium >E. durans Lb. plantarum, Lb. kefiranofaciens, Lactobacillus, Lc. lactis, Streptococcus agalactiae, M. caseolyticus | Dolci et al. [44] |
| Cueva de la Magahá/hard, Spain (Goat) | Culturing PCR-TTGE | Bacteria Bacteria (V3 16S rDNA) | 10 species 8 species | Lb. paracasei > Lb. plantarum > Lb. brevis > Lactobacillus > Enterococcus Lb. plantarum, Lb. brevis, Lc. Lactis, S. thermophilus, Staphylococcus equorum, Lb. curvatus, Lb. paracasei | Martín-Platero et al. [45] |
| Grana Padano/hard, Italy (Cow) | LH-PCR | LAB | 6 species | Lb. rhamnosus > Lb. paracasei > Lb. delbrueckii > Pediococcus acidilactici | Santarelli et al. [22] |
| Livarot/smear-ripened, France (Cow) | Culturing Cloning | Bacteria/yeasts Bacteria (V4 16S rDNA) | 8 bacteria, 5 yeasts species 8 species | M. gubbeenense > Leucobacter komagatae > Halomonas; cheese rind Candida catenulata > Candida intermedia > G. candidum > Geotrichum > Yarrowia lipolytica; cheese rindHalomonas > L. komagatae > M. gubbeenense; cheese rind | Mounier et al. [43] |
| Nottinghamshire/blue-veined, UK (Cow) | Culturing PCR-DGGE | Bacteria Bacteria (V3, V4-V5, V6-V8 16S rDNA) | 12 species 11 OTUs | Lc. lactis subsp. lactis > E. faecalis > Kokuria > Lactobacillus Lc. lactis, Lb. plantarum, Staph. equorum | Yunita and Dodd [55] |
| Ragusano/pasta filata, Italy (Cow) | PCR-DGGE | Bacteria (V6-V8, V1-V3 16S rDNA or rRNA) | 12 species | S. thermophilus, Lb. fermentum, Lb. delbrueckii, Lc. lactis, Leuc. mesenteroides, Lb. casei, Enterococcus hirae > E. faecalis | Randazzo et al. [61] |
| Salers/semi-hard, France (Cow) | PCR-SSCP | Bacteria (V2 16S rDNA) | 9 OTUs | E. faecium, Leuconostoc, Enterobacteriaceae, Bacillus thuringiensis, S. thermophilus, Leuc. pseudomesenteroides, Lb. pentosus, Corynebacterium variabilis, Brachybacterium nesterenkovii | Duthoit et al. [66] |
| Saint Nectaire/smear-ripened, France (Cow) | Culturing SSCP-PCR | Bacteria Bacteria | 21 species 12 OTUs | Lc. lactis > Staphylococcus fleurettii > E. faecalis > S. thermophilus > Marinilactibacillus psychrotolerans > Chryseobacterium > Klebsiella Lc. lactis, S. thermophilus, Clostridium confusum, Nocardioides dubius, Arthrobacter psychrolactophilus, Enterobacter agglomerans | Delbès et al. [46] |
| Molecular methods/high throughput sequencing | |||||
| Artisan cheeses/various, Ireland (Cow, goat, or sheep) | Pyrosequencing | Bacteria (V4 16S rDNA) | 5 phyla 21 genera | Lactococcus (50–90%) > Lactobacillus > Leuconostoc > Pseudomonas > Psychrobacter > Staphylococcus > Arthrobacter > Faecalibacterium; common to 62 cheeses | Quigley et al. [76] |
| Buryatian/soft, Kazakhstan (Cow) | PacBio sequencing | Microbes | 7 phyla, 82 genera, 145 species | Lactococcus (51.46%) > Streptococcus (17.81%) > Pseudomonas (5.48%) > Acetobacter (4.83%) > Klebsiella (3.36%) > Lactobacillus (2.36%) > Acinetobacter (1.84%) > Raoultella (1.63%) | Jin et al. [79] |
| Canestrato Pugliese/hard, Italy (Sheep) | Pyrosequencing | Bacteria (V1-V3 16S rDNA) | 28 genera | Lactococcus (87.2%) > Lactobacillus (4.8%; mainly Lb. plantarum and Lb. sakei) > Leuconostoc (3.9%) | De Pasquale et al. [109] |
| Cheddar/semi-hard, UK (Cow) | Illumina sequencing | Bacteria (V4 16S rDNA) | 159 OTUs | Streptococcus > Lactococcus > Lactobacillus > Staphylococcus (70%); interior | Afshari et al. [110] |
| Gouda-like cheese/semi-hard, USA (Cow) | Illumina sequencing | Bacteria (V4 16S rDNA) | 36 genera | Bacillaceae > Lactococcus > Lactobacillus > Streptococcus > Staphylococcus | Salazar et al. [111] |
| Cotija/hard, Mexico (Cow) | Illumina sequencing | Microbes | 31 phyla, 574 genera | Lb. plantarum > Leuc. mesenteroides > Weissella paramesenteroides (>80%) Aerococcus > Enterococcus > Lactococcus > Staphylococcus (<10%) | Escobar-Zepeda et al. [82] |
| Grana/hard, Italy (Cow) | RT-PCR-DGGE Pyrosequencing | Bacteria (V1 16S rDNA) Bacteria (V1-V3 16S rDNA) | 16 OTUs 25 genera | Lb. helveticus, Lb. delbrueckii, S. thermophilus, Lb. acidophilus, Lb. rhamnosus, Acetobacter baumanii, Propionibacterium Lb. helveticus > Propionibacterium > Lb. delbrueckiii > Lb. casei > Lb. rhamnosus > S. thermophilus > Staphylococcus > Lb. brevis | Alessandria et al. [95] |
| Artisanal cheeses/soft, Kazakhstan (Cow) | PacBio sequencing | Microbes | 14 phyla, 140 genera, 238 species | Lc. lactis (28.93%) > Lb. helveticus (26.43%) > S. thermophilus (12.18%) > Lb. delbrueckii (12.15%) | Li et al. [80] |
| Ocosingo/semi-hard, Mexico (Cow) | Pyrosequencing | Bacteria (V1 16S rDNA) | 162 OTUs | S. thermophilus > Lc. lactis > Lb. helveticus > Lb. delbrueckii > Lb. plantarum (70%); interior | Aldrete-Tapia et al. [17] |
| Tomme d’Orchies/semi-hard, France (Cow) | Illumina sequencing | Bacteria (V1-V3 16S rDNA) | 10 species core, 21 species surface | Lactococcuss > Streptococcus (66%); interior Lactobacillus > Lactococcus > Corynebacterium > Micrococcales > Psychrobacter (80%); surface | Ceugniez et al. [83] |
| Tomme d’Orchies/semi-hard, France (Cow) | Illumina sequencing | Fungi 5.8S-ITS2 | 30 OTUs | Y. lipolytica > G. candidum/Galactomyces geotrichum (99%); interior Y. lipolytica > G. candidum/Galactomyces geotrichum (98%); surface | Ceugniez et al. [112] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayo, B.; Rodríguez, J.; Vázquez, L.; Flórez, A.B. Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety. Foods 2021, 10, 602. https://doi.org/10.3390/foods10030602
Mayo B, Rodríguez J, Vázquez L, Flórez AB. Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety. Foods. 2021; 10(3):602. https://doi.org/10.3390/foods10030602
Chicago/Turabian StyleMayo, Baltasar, Javier Rodríguez, Lucía Vázquez, and Ana Belén Flórez. 2021. "Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety" Foods 10, no. 3: 602. https://doi.org/10.3390/foods10030602
APA StyleMayo, B., Rodríguez, J., Vázquez, L., & Flórez, A. B. (2021). Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety. Foods, 10(3), 602. https://doi.org/10.3390/foods10030602








