Enzyme Production Potential of Penicillium oxalicum M1816 and Its Application in Ferulic Acid Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening and Identification of the Enzyme-Producing Strain
2.2. SSF of Wheat Bran and Enzyme Assays
2.3. Morphology and Molecular Identification
2.4. Whole Genome Sequencing, Assembly, and Annotation
2.5. CAZymes Identification
2.6. Release of FA from DSWB
2.7. Effects of SSF Conditions of Wheat Bran on Enzyme Production
2.8. Application of P. oxalicum M1816 to Huangjiu Fermentation
2.9. Statistical Analyses
3. Results and Discussion
3.1. Isolation, Screening, and Identification of the Enzyme-Producing Strain
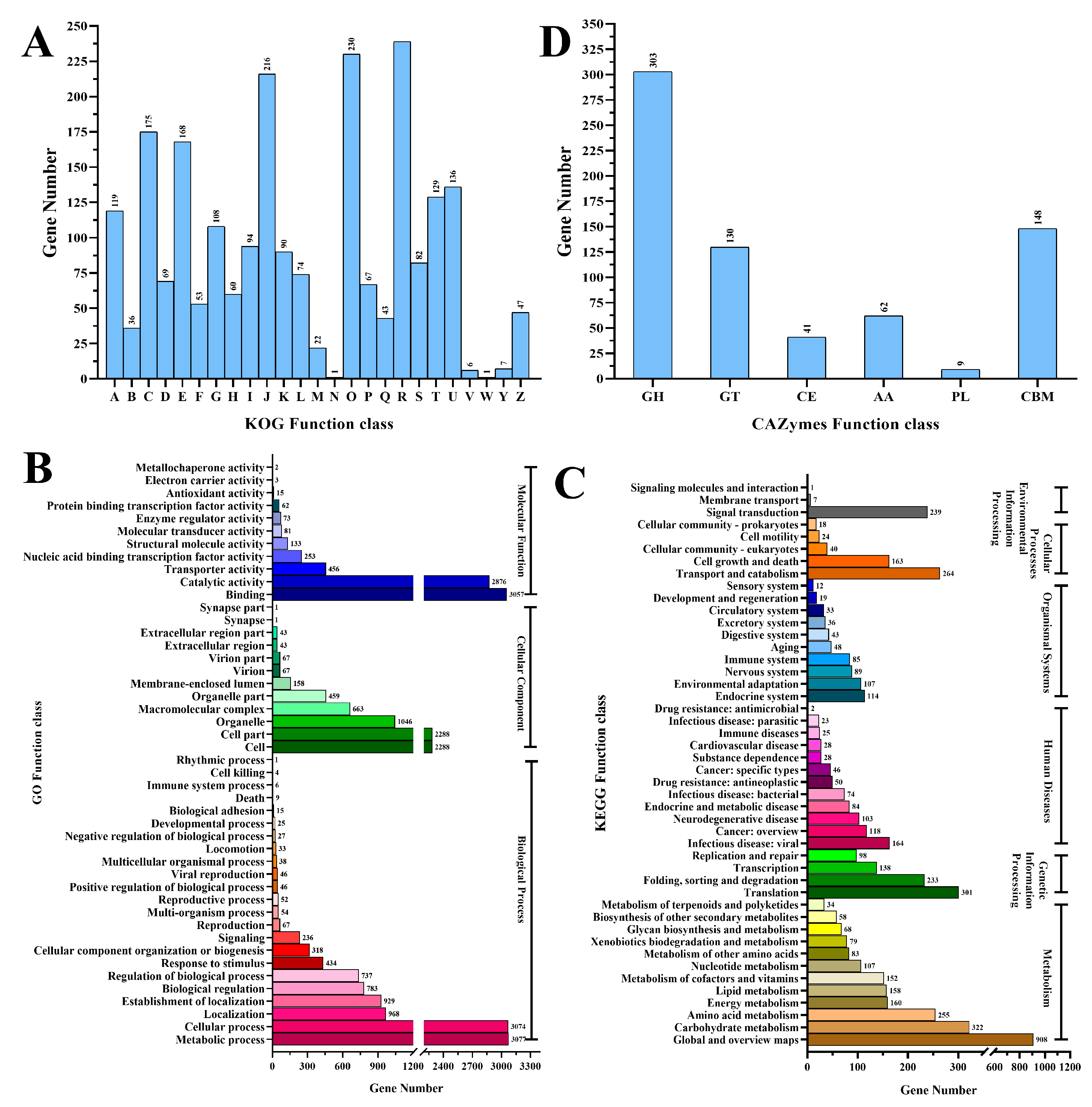
3.2. Genome Features and Gene Function Analysis of Strain M1816
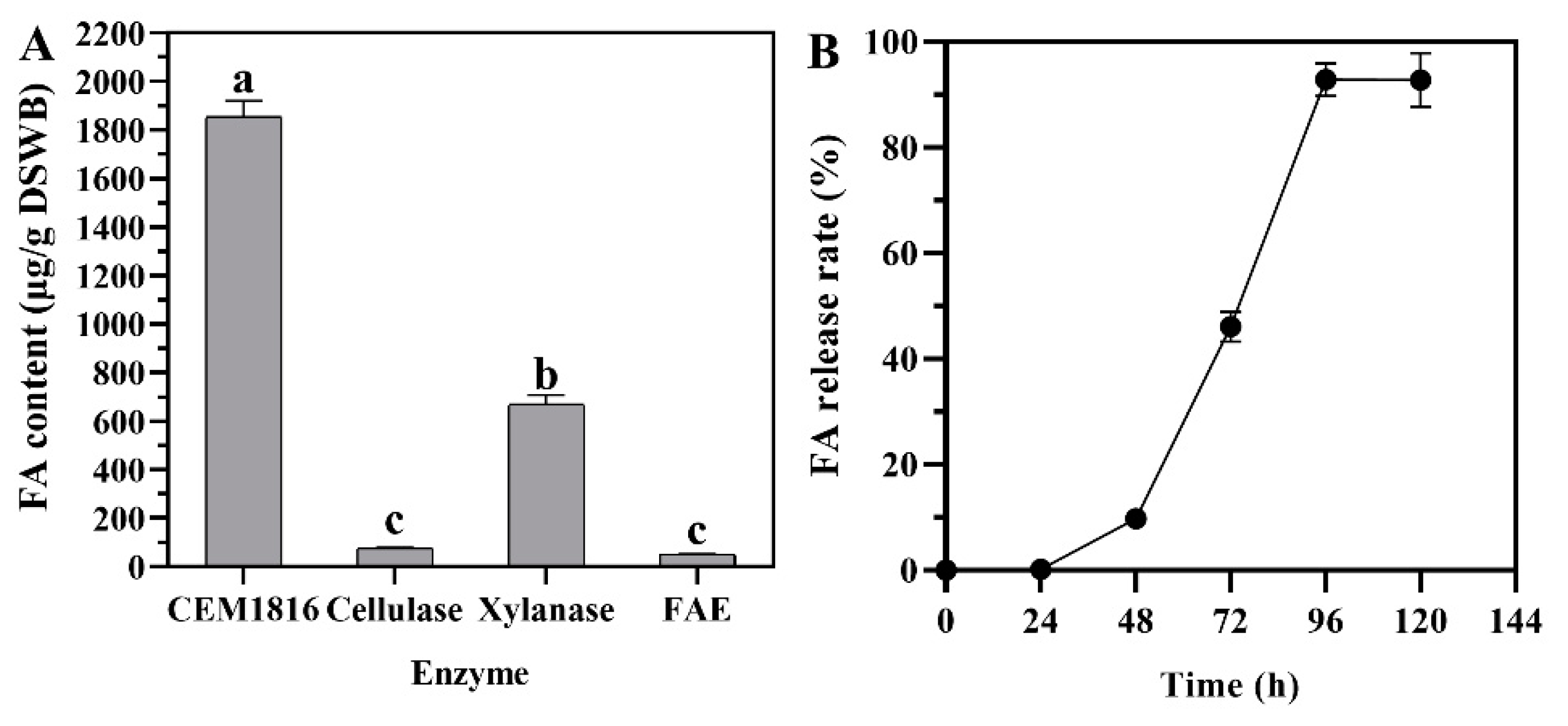
3.3. Potential of P. oxalicum M1816 to Release FA from Wheat Bran
3.4. Release of FA from DSWB
3.5. Optimization of the SSF Conditions by P. oxalicum M1816 for Enzyme Production
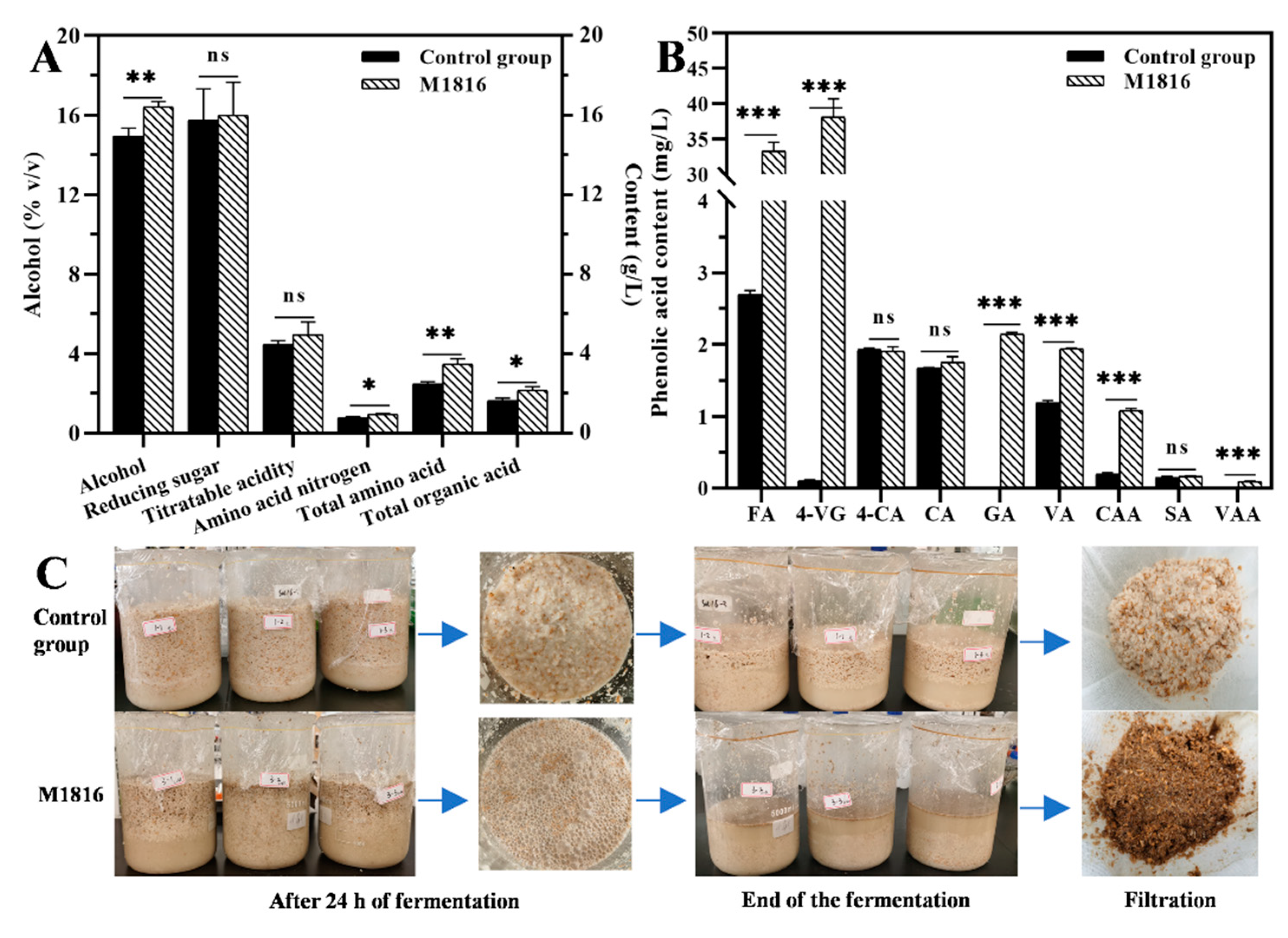
3.6. Production of FA by P. oxalicum M1816 and Its Application in Huangjiu Fermentation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stevenson, L.; Phillips, F.; O’Sullivan, K.; Walton, J. Wheat bran: Its composition and benefits to health, a European perspective. Int. J. Food Sci. Nutr. 2012, 63, 1001–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deroover, L.; Tie, Y.; Verspreet, J.; Courtin, C.M.; Verbeke, K. Modifying wheat bran to improve its health benefits. Crit. Rev. Food Sci. Nutr. 2020, 60, 1104–1122. [Google Scholar] [CrossRef]
- Brouns, F.; Hemery, Y.; Price, R.; Anson, N.M. Wheat Aleurone: Separation, Composition, Health Aspects, and Potential Food Use. Crit. Rev. Food Sci. Nutr. 2012, 52, 553–568. [Google Scholar] [CrossRef] [Green Version]
- Shang, Q.; Liu, S.; Liu, H.; Mahfuz, S.; Piao, X. Impact of sugar beet pulp and wheat bran on serum biochemical profile, inflammatory responses and gut microbiota in sows during late gestation and lactation. J. Anim. Sci. Biotechnol. 2021, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Z.; Ding, L.L.; Pan, Z.H.; Kong, D.H.; Hadiatullah, H.; Fan, Z.C. Proteinase and glycoside hydrolase production is enhanced in solid-state fermentation by manipulating the carbon and nitrogen fluxes in Aspergillus oryzae. Food Chem. 2019, 271, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.L.; Peng, M.M.; Xue, D.N.; Cheng, Y.; Li, C.X.; Wang, D.; Lu, K.; Zheng, Y.; Xia, T.; Song, J.; et al. Development of optimal steam explosion pretreatment and highly effective cell factory for bioconversion of grain vinegar residue to butanol. Biotechnol. Biofuels 2020, 13, 111. [Google Scholar] [CrossRef]
- Lin, S.; Jin, X.; Gao, J.; Qiu, Z.; Ying, J.; Wang, Y.; Dong, Z.; Zhou, W. Impact of wheat bran micronization on dough properties and bread quality: Part I—Bran functionality and dough properties. Food Chem. 2021, 353, 129407. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.B.; Luo, H.B.; Ren, Z.Q.; Ahmad, M.S.; Liu, C.G.; Tawab, A.; Al-Ghafari, A.B.; Omar, U.; Gull, M.; Mehmood, M.A. Evaluating the bioenergy potential of Chinese Liquor-industry waste through pyrolysis, thermogravimetric, kinetics and evolved gas analyses. Energy Convers. Manag. 2018, 163, 13–21. [Google Scholar] [CrossRef]
- Kim, M.; Seo, J.A. Fermentation profiling of rice wine produced by Aspergillus oryzae KSS2 and Rhizopus oryzae KJJ39 newly isolated from Korean fermentation starter. Appl. Biol. Chem. 2021, 64, 25. [Google Scholar] [CrossRef]
- Kandil, A.; Li, J.; Vasanthan, T.; Bressler, D.C. Phenolic acids in some cereal grains and their inhibitory effect on starch liquefaction and saccharification. J. Agric. Food. Chem. 2012, 60, 8444–8449. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Happel, A.; Zanaroli, G.; Bertolini, M.; Chiesa, S.; Commisso, M.; Guzzo, F.; Tassoni, A. Advances in combined enzymatic extraction of ferulic acid from wheat bran. New Biotechnol. 2020, 56, 38–45. [Google Scholar] [CrossRef]
- Kim, M.J.; Yoon, W.J.; Kim, S.S. Phytochemical Compositions of Immature Wheat Bran, and Its Antioxidant Capacity, Cell Growth Inhibition, and Apoptosis Induction through Tumor Suppressor Gene. Molecules 2016, 21, 1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.S.; Sheng, J.P.; Cai, T.; Jin, J.; Liu, W.Z.; Lin, Y.M.; Du, Y.X.; Zhang, M.Q.; Shen, L. A Protease-Insensitive Feruloyl Esterase from China Holstein Cow Rumen Metagenomic Library: Expression, Characterization, and Utilization in Ferulic Acid Release from Wheat Straw. J. Agric. Food. Chem. 2012, 60, 2546–2553. [Google Scholar] [CrossRef]
- Alam, M.A. Anti-hypertensive Effect of Cereal Antioxidant Ferulic Acid and Its Mechanism of Action. Front. Nutr. 2019, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.L.; Michaelson, L.V.; Shewry, P.R.; Lovegrove, A.; Spencer, J.P.E. Increased bioavailability of phenolic acids and enhanced vascular function following intake of feruloyl esterase-processed high fibre bread: A randomized, controlled, single blind, crossover human intervention trial. Clin. Nutr. 2021, 40, 788–795. [Google Scholar] [CrossRef]
- McCarty, M.; Iloki Assanga, S.B.; Luján, L.; O’Keefe, J.; Dinicolantonio, J. Nutraceutical Strategies for Suppressing NLRP3 Inflammasome Activation: Pertinence to the Management of COVID-19 and Beyond. Nutrients 2020, 13, 47. [Google Scholar] [CrossRef]
- Adom, K.K.; Sorrells, M.E.; Liu, R.H. Phytochemicals and antioxidant activity of milled fractions of different wheat varieties. J. Agric. Food Chem. 2005, 53, 2297–2306. [Google Scholar] [CrossRef]
- Mateo Anson, N.; van den Berg, R.; Havenaar, R.; Bast, A.; Haenen, G.R. Ferulic acid from aleurone determines the antioxidant potency of wheat grain (Triticum aestivum L.). J. Agric. Food Chem. 2008, 56, 5589–5594. [Google Scholar] [CrossRef]
- Mancuso, C.; Santangelo, R. Ferulic acid: Pharmacological and toxicological aspects. Food Chem Toxicol. 2014, 65, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Rondini, L.; Peyrat-Maillard, M.N.; Marsset-Baglieri, A.; Fromentin, G.; Durand, P.; Tome, D.; Prost, M.; Berset, C. Bound ferulic acid from bran is more bioavailable than the free compound in rat. J. Agric. Food. Chem. 2004, 52, 4338–4343. [Google Scholar] [CrossRef]
- Costa, R.D.; de Almeida, S.S.; Cavalcanti, E.D.C.; Freire, D.M.G.; Moura-Nunes, N.; Monteiro, M.; Perrone, D. Enzymes produced by solid state fermentation of agro-industrial by-products release ferulic acid in bioprocessed whole-wheat breads. Food Res. Int. 2021, 140, 109843. [Google Scholar] [CrossRef]
- Wataniyakul, P.; Pavasant, P.; Goto, M.; Shotipruk, A. Microwave pretreatment of defatted rice bran for enhanced recovery of total phenolic compounds extracted by subcritical water. Bioresour. Technol. 2012, 124, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Cengiz, M.F.; Babacan, U.; Akinci, E.; Kesci, S.T.; Kaba, A. Extraction of phenolic acids from ancient wheat bran samples by ultrasound application. J. Chem. Technol. Biotechnol. 2021, 96, 134–141. [Google Scholar] [CrossRef]
- Gong, L.X.; Huang, L.L.; Zhang, Y. Effect of Steam Explosion Treatment on Barley Bran Phenolic Compounds and Antioxidant Capacity. J. Agric. Food. Chem. 2012, 60, 7177–7184. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, B.G.; Cao, Y.P.; Tian, Y.A.; Li, X.H. Optimisation of ultrasound-assisted extraction of phenolic compounds from wheat bran. Food Chem. 2008, 106, 804–810. [Google Scholar] [CrossRef]
- Kim, K.-H.; Tsao, R.; Yang, R.; Cui, S.W. Phenolic acid profiles and antioxidant activities of wheat bran extracts and the effect of hydrolysis conditions. Food Chem. 2006, 95, 466–473. [Google Scholar] [CrossRef]
- Arranz, S.; Calixto, F.S. Analysis of polyphenols in cereals may be improved performing acidic hydrolysis: A study in wheat flour and wheat bran and cereals of the diet. J. Cereal Sci. 2010, 51, 313–318. [Google Scholar] [CrossRef]
- Xie, C.Y.; Gu, Z.X.; You, X.J.; Liu, G.X.; Tan, Y.X.; Zhang, H. Screening of edible mushrooms for release of ferulic acid from wheat bran by fermentation. Enzym. Microb. Technol. 2010, 46, 125–128. [Google Scholar] [CrossRef]
- Yin, Z.N.; Wu, W.J.; Sun, C.Z.; Lei, Z.G.; Chen, H.M.; Liu, H.F.; Chen, W.B.; Ma, J.J.; Min, T.; Zhang, M.M.; et al. Comparison of releasing bound phenolic acids from wheat bran by fermentation of three Aspergillus species. Int. J. Food Sci. Technol. 2018, 53, 1120–1130. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, T.; Zhang, S. Extracellular secretion of feruloyl esterase derived from Lactobacillus crispatus in Escherichia coli and its application for ferulic acid production. Bioresour Technol. 2019, 288, 121526. [Google Scholar] [CrossRef]
- Tabka, M.G.; Herpoel-Gimbert, I.; Monod, F.; Asther, M.; Sigoillot, J.C. Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enzym. Microb. Technol. 2006, 39, 897–902. [Google Scholar] [CrossRef]
- Liu, S.P.; Hu, J.; Xu, Y.Z.; Xue, J.B.; Zhou, J.D.; Han, X.; Ji, Z.W.; Mao, J. Combined use of single molecule real-time DNA sequencing technology and culture-dependent methods to analyze the functional microorganisms in inoculated raw wheat Qu. Food Res. Int. 2020, 132, 109062. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, S.; Sun, H.; Jiang, Z.; Xu, Y.; Mao, J.; Qian, B.; Wang, L.; Mao, J. Metagenomics-based insights into the microbial community profiling and flavor development potentiality of Baijiu Daqu and Huangjiu wheat Qu. Food Res. Int. 2021, 110707. [Google Scholar] [CrossRef]
- Xie, G.F.; Li, W.J.; Lu, J.; Cao, Y.; Fang, H.; Zou, H.J.; Hu, Z.M. Isolation and identification of representative fungi from Shaoxing rice wine wheat Qu using a polyphasic approach of culture-based and molecular-based methods. J. Inst. Brew. 2007, 113, 272–279. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, L.; Wei, X.; Zou, G.; Qin, Y.; Ma, L.; Li, J.; Zheng, H.; Wang, S.; Wang, C.; et al. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 2013, 8, e55185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Yan, Y.S.; He, Q.P.; Yang, L.; Yin, X.; Li, C.X.; Mao, L.C.; Liao, L.S.; Huang, J.Q.; Xie, S.B.; et al. Comparative genomic, transcriptomic and secretomic profiling of Penicillium oxalicum HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106, and identification of two novel regulatory genes of cellulase and xylanase gene expression. Biotechnol. Biofuels 2016, 9, 203. [Google Scholar] [CrossRef] [Green Version]
- Donaghy, J.; Kelly, P.F.; McKay, A.M. Detection of ferulic acid esterase production by Bacillus spp. and lactobacilli. Appl. Microbiol. Biotechnol. 1998, 50, 257–260. [Google Scholar] [CrossRef]
- Pennacchio, A.; Ventorino, V.; Cimini, D.; Pepe, O.; Schiraldi, C.; Inverso, M.; Faraco, V. Isolation of new cellulase and xylanase producing strains and application to lignocellulosic biomasses hydrolysis and succinic acid production. Bioresour. Technol. 2018, 259, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Hankin, L.; Anagnostakis, S.L. The Use of Solid Media for Detection of Enzyme Production by Fungi. Mycologia 1975, 67, 597. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 1969, 31, 426–428. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.J.; Lam, M.Q.; Thevarajoo, S.; Abd Manan, F.; Yahya, A.; Chong, C.S. Genome analysis of cellulose and hemicellulose degrading Micromonospora sp. CP22. 3 Biotech 2020, 10, 160. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Singh, R.K.; Mitra, A.; Sen, S.K. Ferulic acid esterase production by Streptomyces sp. Bioresour. Technol. 2007, 98, 211–213. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Jia, P.; Jiang, J.; Bai, Y.; Fan, T.P.; Zheng, X.; Cai, Y. Expression and characterisation of feruloyl esterases from Lactobacillus fermentum JN248 and release of ferulic acid from wheat bran. Int. J. Biol. Macromol. 2019, 138, 272–277. [Google Scholar] [CrossRef]
- GBT13662-2018 Huangjiu; China Standardization Administration: Beijing, China, 2018.
- Liu, S.P.; Chen, Q.L.; Zou, H.; Yu, Y.; Zhou, Z.L.; Mao, J.; Zhang, S. A metagenomic analysis of the relationship between microorganisms and flavor development in Shaoxing mechanized huangjiu fermentation mashes. Int. J. Food Microbiol. 2019, 303, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Luz, B.; Sarrouh, B.; Bicas, J.L.; Lofrano, R.C.Z. Lipase production by microorganisms isolated from the Serra de Ouro Branco State Park. Anais da Academia Brasileira de Ciências 2021, 93, e20190672. [Google Scholar] [CrossRef]
- Ward, O.P. Production of recombinant proteins by filamentous fungi. Biotechnol. Adv. 2012, 30, 1119–1139. [Google Scholar] [CrossRef]
- Sakamoto, T.; Fujita, T.; Kawasaki, H. Transglycosylation catalyzed by a Penicillium chrysogenum exo-1,5-alpha-L-arabinanase. Biochim. Biophys. Acta 2004, 1674, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, G.L.; de Azevedo-Martins, A.C.; Albano, R.M.; de Souza, W.; Frases, S. Comprehensive analysis of the cellulolytic system reveals its potential for deconstruction of lignocellulosic biomass in a novel Streptomyces sp. Appl. Microbiol. Biotechnol. 2017, 101, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Eibinger, M.; Ganner, T.; Bubner, P.; Rosker, S.; Kracher, D.; Haltrich, D.; Ludwig, R.; Plank, H.; Nidetzky, B. Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J. Biol. Chem. 2014, 289, 35929–35938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheawchanlertfa, P.; Sutheeworapong, S.; Jenjaroenpun, P.; Wongsurawat, T.; Nookaew, I.; Cheevadhanarak, S.; Kosugi, A.; Pason, P.; Waeonukul, R.; Ratanakhanokchai, K.; et al. Clostridium manihotivorum sp. nov., a novel mesophilic anaerobic bacterium that produces cassava pulp-degrading enzymes. PeerJ 2020, 8, e10343. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, G.; Kumar, J.; Sangwan, R.S.; Singh, S.P. Metagenomic analysis of geothermal water reservoir sites exploring carbohydrate-related thermozymes. Int. J. Biol. Macromol. 2018, 119, 882–895. [Google Scholar] [CrossRef]
- Crepin, V.F.; Faulds, C.B.; Connerton, I.F. Identification of a type-D feruloyl esterase from Neurospora crassa. Appl. Microbiol. Biotechnol. 2004, 63, 567–570. [Google Scholar] [CrossRef]
- Brandon, A.G.; Scheller, H.V. Engineering of Bioenergy Crops: Dominant Genetic Approaches to Improve Polysaccharide Properties and Composition in Biomass. Front. Plant. Sci. 2020, 11, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, L.C.; Chen, R.; Xie, W.; Liu, Y.H. Enhancing the Thermostability of Feruloyl Esterase EstF27 by Directed Evolution and the Underlying Structural Basis. J. Agric. Food Chem. 2015, 63, 8225–8233. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, Z.; Du, F.; Wang, H.; Ng, T.B. Feruloyl esterase from the edible mushroom Panus giganteus: A potential dietary supplement. J. Agric. Food Chem. 2014, 62, 7822–7827. [Google Scholar] [CrossRef]
- Nieter, A.; Kelle, S.; Takenberg, M.; Linke, D.; Bunzel, M.; Popper, L.; Berger, R.G. Heterologous production and characterization of a chlorogenic acid esterase from Ustilago maydis with a potential use in baking. Food Chem. 2016, 209, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Radhai, R.; Sundaram, K.S.; Rajalakshmi, V. Utilization of cellulosic biomass as a substrate for the production of bioethanol. Int. J. Environ. Sci. 2015, 5, 743–753. [Google Scholar] [CrossRef]
- Mallek-Fakhfakh, H.; Fakhfakh, J.; Walha, K.; Hassairi, H.; Gargouri, A.; Belghith, H. Enzymatic hydrolysis of pretreated Alfa fibers (Stipa tenacissima) using β-d-glucosidase and xylanase of Talaromyces thermophilus from solid-state fermentation. Int. J. Biol. Macromol. 2017, 103, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Venkata Mohan, S.; Sirisha, K.; Sreenivasa Rao, R.; Sarma, P.N. Bioslurry phase remediation of chlorpyrifos contaminated soil: Process evaluation and optimization by Taguchi design of experimental (DOE) methodology. Ecotoxicol. Environ. Saf. 2007, 68, 252–262. [Google Scholar] [CrossRef]
- Krauss, G.J.; Sole, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Barlocher, F. Fungi in freshwaters: Ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 2011, 35, 620–651. [Google Scholar] [CrossRef] [PubMed]
- Antoine, A.A.; Jacqueline, D.; Thonart, P. Xylanase production by Penicillium canescens on soya oil cake in solid-state fermentation. Appl Biochem Biotechnol. 2010, 160, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Amadi, O.C.; Egong, E.J.; Nwagu, T.N.; Okpala, G.; Onwosi, C.O.; Chukwu, G.C.; Okolo, B.N.; Agu, R.C.; Moneke, A.N. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon 2020, 6, e04566. [Google Scholar] [CrossRef]
- Fan, G.; Zhu, Y.; Fu, Z.; Sun, B.; Teng, C.; Yang, R.; Li, X. Optimization of fermentation conditions for the production of recombinant feruloyl esterase from Burkholderia pyrrocinia B1213. 3 Biotech 2020, 10, 216. [Google Scholar] [CrossRef]
- Mo, X.L.; Xu, Y. Ferulic acid release and 4-vinylguaiacol formation during Chinese rice wine brewing and fermentation. J. Inst. Brew. 2010, 116, 304–311. [Google Scholar] [CrossRef]
- Mathew, S.; Abraham, T.E. Bioconversions of ferulic acid, an hydroxycinnamic acid. Crit. Rev. Microbiol. 2006, 32, 115–125. [Google Scholar] [CrossRef]
- Boratynski, F.; Szczepanska, E.; Grudniewska, A.; Gnilka, R.; Olejniczak, T. Improving of hydrolases biosythesis by solid-state fermentation of Penicillium camemberti on rapeseed cake. Sci. Rep. 2018, 8, 10157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
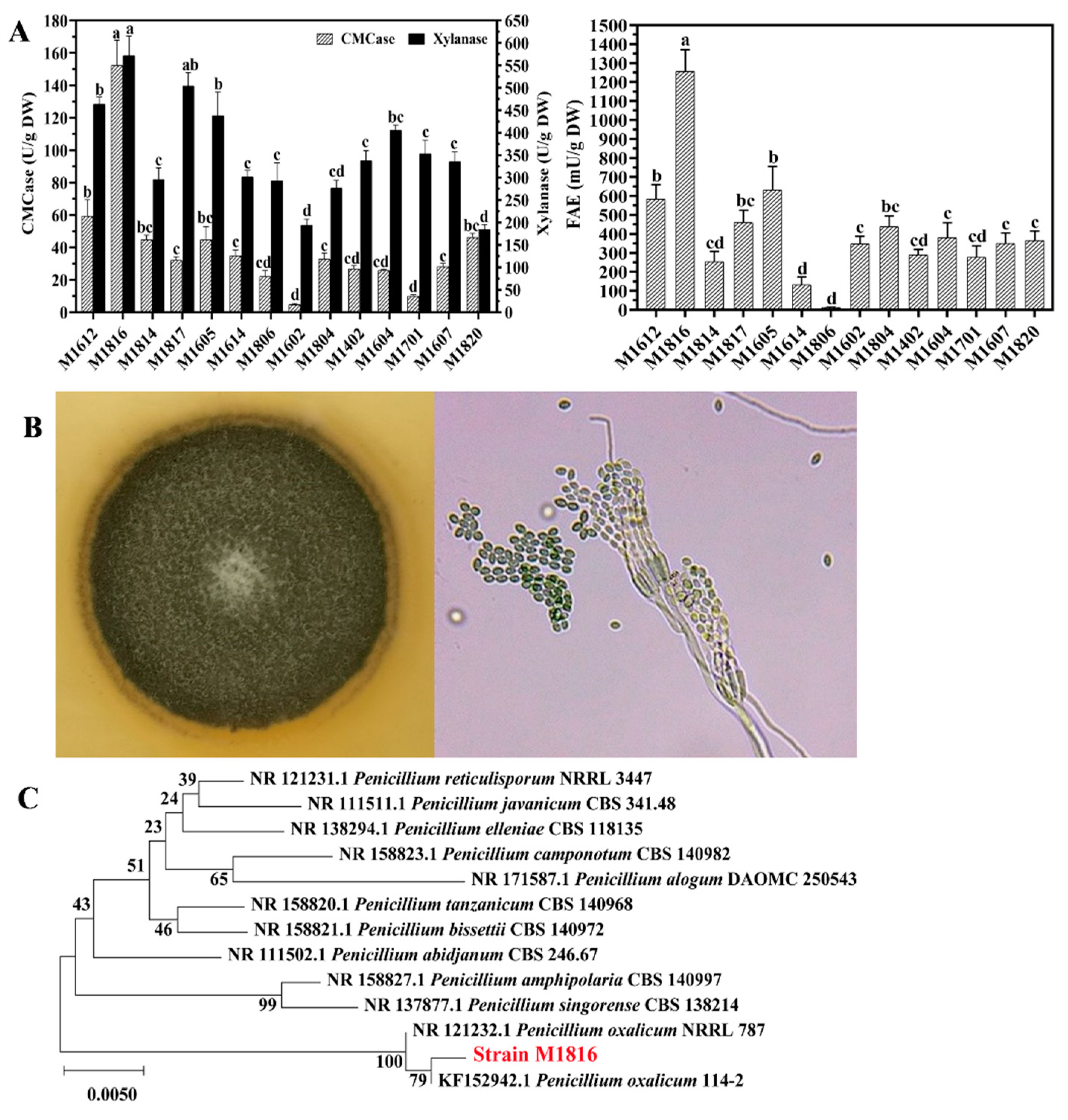

| Strains | FAE EI | CMCase EI | Xylanase EI |
|---|---|---|---|
| M1612 | + | + | +++ |
| M1816 | ++ | + | +++ |
| M1814 | − | + | +++ |
| M1817 | − | + | ++ |
| M1605 | ++ | + | ++ |
| M1614 | − | + | ++ |
| M1806 | + | + | ++ |
| M1602 | − | + | ++ |
| M1804 | + | + | + |
| M1402 | + | + | + |
| M1604 | ++ | + | + |
| M1701 | + | + | + |
| M1607 | +++ | + | + |
| M1820 | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, S.; Sun, H.; Jiang, Z.; Zhou, Z.; Han, X.; Zhou, Y.; Sun, H.; Zhou, W.; Mao, J. Enzyme Production Potential of Penicillium oxalicum M1816 and Its Application in Ferulic Acid Production. Foods 2021, 10, 2577. https://doi.org/10.3390/foods10112577
Zhang J, Liu S, Sun H, Jiang Z, Zhou Z, Han X, Zhou Y, Sun H, Zhou W, Mao J. Enzyme Production Potential of Penicillium oxalicum M1816 and Its Application in Ferulic Acid Production. Foods. 2021; 10(11):2577. https://doi.org/10.3390/foods10112577
Chicago/Turabian StyleZhang, Jing, Shuangping Liu, Hailong Sun, Zhengfei Jiang, Zhilei Zhou, Xiao Han, Yongxiang Zhou, Honggen Sun, Weibiao Zhou, and Jian Mao. 2021. "Enzyme Production Potential of Penicillium oxalicum M1816 and Its Application in Ferulic Acid Production" Foods 10, no. 11: 2577. https://doi.org/10.3390/foods10112577
APA StyleZhang, J., Liu, S., Sun, H., Jiang, Z., Zhou, Z., Han, X., Zhou, Y., Sun, H., Zhou, W., & Mao, J. (2021). Enzyme Production Potential of Penicillium oxalicum M1816 and Its Application in Ferulic Acid Production. Foods, 10(11), 2577. https://doi.org/10.3390/foods10112577






