Contribution of Microorganisms to Biogenic Amine Accumulation during Fish Sauce Fermentation and Screening of Novel Starters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.1.1. Fish Sauce Samples Collection
2.1.2. Fermentation Experiments
2.2. Microbiological Analyses
2.2.1. Microbial Community Analysis
2.2.2. Strains Identification
2.3. Biogenic Amines Determination
2.3.1. Determination of BAs
2.3.2. Pretreatment of Samples of Fish Sauce and Other Fermented Foods
2.3.3. Determination of BA Production and Reduction Properties of Strains
2.4. Statistical Analysis
2.5. Nucleotide Sequence Accession Numbers
3. Results
3.1. BA Contents in Fish Sauce Samples at Different Fermentation Stages
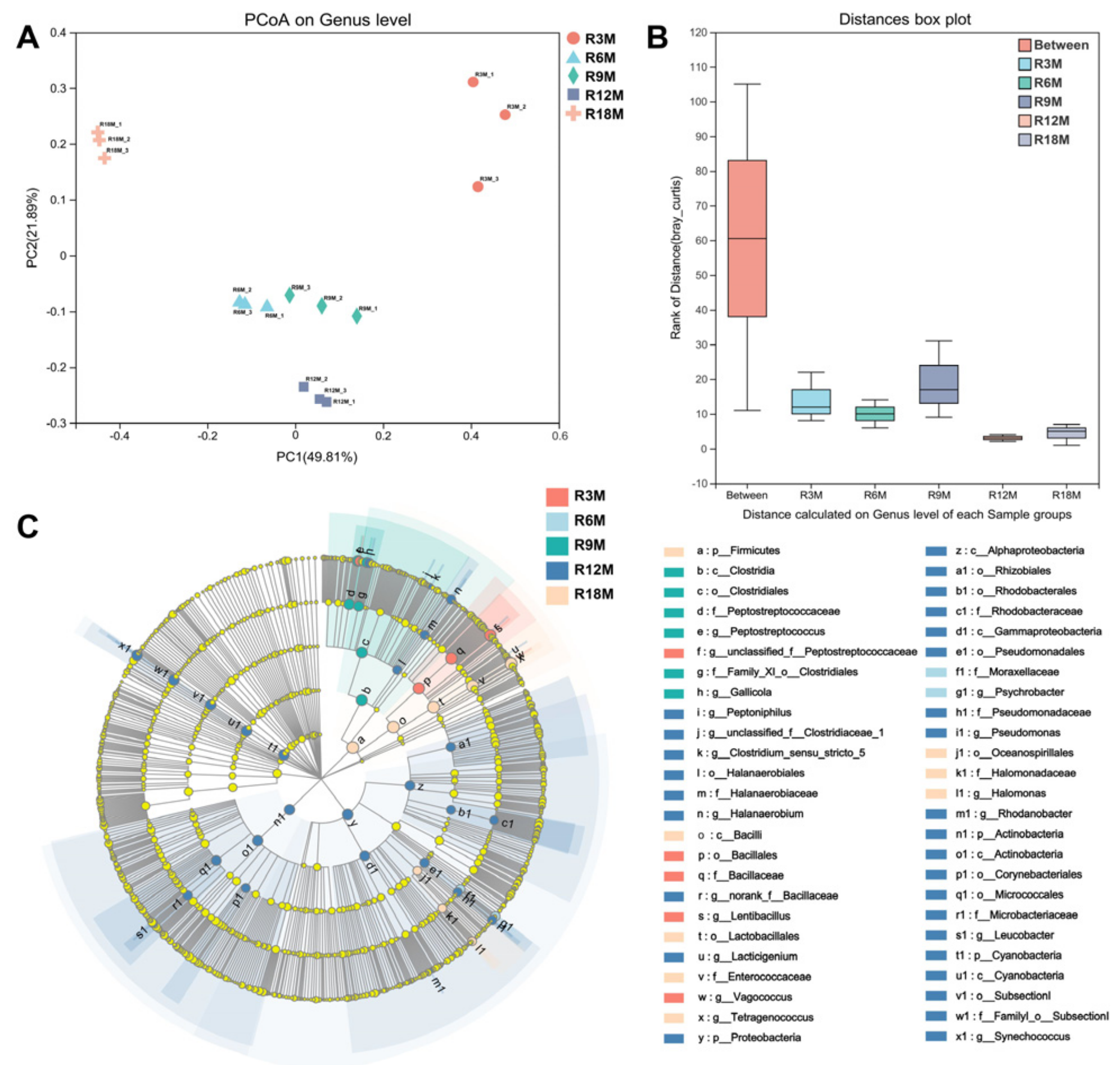
3.2. Dynamic Changes of Microorganisms in Fish Sauce Samples during Fermentation
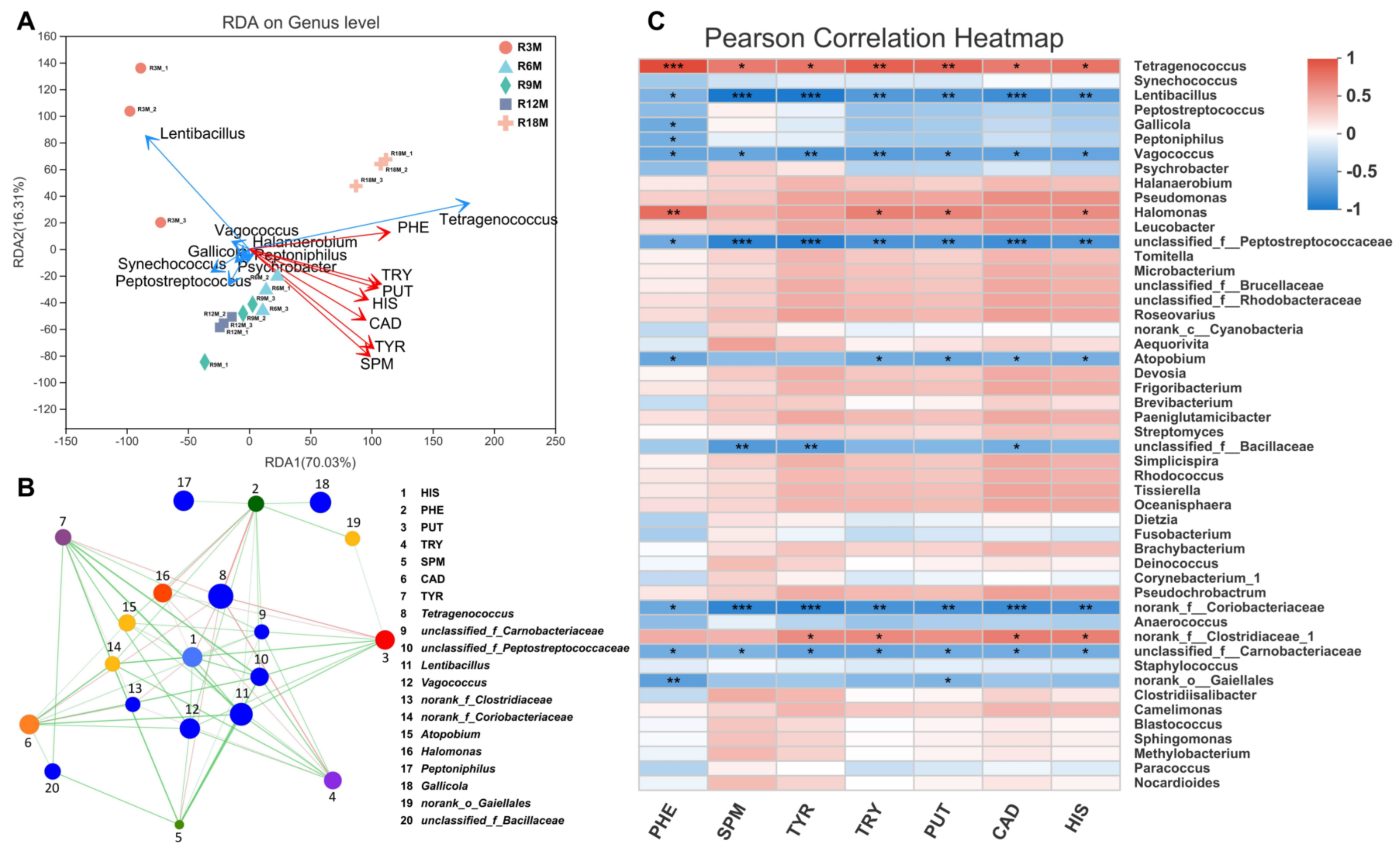
3.3. Microbial Contribution to BA Contents in Fish Sauce
3.4. Screening of Novel Starter Cultures for Reducing Biogenic Amines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fukui, Y.; Yoshida, M.; Shozen, K.-I.; Funatsu, Y.; Takano, T.; Oikawa, H.; Yano, Y.; Satomi, M. Bacterial communities in fish sauce mash using culture-dependent and -independent methods. J. Gen. Appl. Microbiol. 2012, 58, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Jung, J.Y.; Jeon, C.O. Bacterial community dynamics and metabolite changes in myeolchi-aekjeot, a Korean traditional fermented fish sauce, during fermentation. Int. J. Food Microbiol. 2015, 203, 15–22. [Google Scholar] [CrossRef]
- Du, F.; Zhang, X.; Gu, H.; Song, J.; Gao, X. Dynamic Changes in the Bacterial Community During the Fermentation of Traditional Chinese Fish Sauce (TCFS) and Their Correlation with TCFS Quality. Microorganisms 2019, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Xu, Y.; Li, C.; Dong, X.; Wang, D. Biogenic amines in commercially produced Yulu, a Chinese fermented fish sauce. Food Addit. Contam. Part B Surveill. 2014, 7, 25–29. [Google Scholar] [CrossRef]
- Zaman, M.Z.; Abu Bakar, F.; Jinap, S.; Bakar, J. Novel starter cultures to inhibit biogenic amines accumulation during fish sauce fermentation. Int. J. Food Microbiol. 2011, 145, 84–91. [Google Scholar] [CrossRef]
- Biji, K.B.; Ravishankar, C.N.; Venkateswarlu, R.; Mohan, C.O.; Gopal, T.K. Biogenic amines in seafood: A review. J. Food Sci. Technol. 2016, 53, 2210–2218. [Google Scholar] [CrossRef]
- Onal, A.; Tekkeli, S.E.; Onal, C. A review of the liquid chromatographic methods for the determination of biogenic amines in foods. Food Chem. 2013, 138, 509–515. [Google Scholar] [CrossRef]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [Green Version]
- Yongsawatdigul, J.; Choi, Y.J.; Udomporn, S. Biogenic Amines Formation in Fish Sauce Prepared from Fresh and Temperatureabused Indian Anchovy (Stolephorus indicus). JFS Food Chem. Toxicol. 2004, 69, 312–319. [Google Scholar] [CrossRef]
- Mooraki, N.; Sedaghati, M. Reduction of biogenic amines in fermented fish sauces by using Lactic acid bacteria. J. Surv. Fish. Sci. 2019, 5, 99–110. [Google Scholar] [CrossRef]
- Kuda, T.; Miyawaki, M. Reduction of histamine in fish sauces by rice bran nuka. Food Control 2010, 21, 1322–1326. [Google Scholar] [CrossRef]
- Naila, A.; Flint, S.; Fletcher, G.C.; Bremer, P.J.; Meerdink, G. Chemistry and microbiology of traditional Rihaakuru (fish paste) from the Maldives. Int. J. Food Sci. Nutr. 2011, 62, 139–147. [Google Scholar] [CrossRef]
- Tsai, Y.; Lin, C.; Chien, L.; Lee, T.; Wei, C.; Hwang, D. Histamine contents of fermented fish products in Taiwan and isolation of histamine-forming bacteria. Food Chem. 2006, 98, 64–70. [Google Scholar] [CrossRef]
- FDA. Scombrotoxin (Histamine) Formation; Department of Health and Human Services, Public Health Service, Food and Drug Administration, Center for Food Safety and Applied Nutrition, Office of Seafood: Washington, DC, USA, 2001. [Google Scholar]
- Wang, Y.; Li, C.; Li, L.; Yang, X.; Chen, S.; Wu, Y.; Zhao, Y.; Wang, J.; Wei, Y.; Yang, D. Application of UHPLC-Q/TOF-MS-based metabolomics in the evaluation of metabolites and taste quality of Chinese fish sauce (Yu-lu) during fermentation. Food Chem. 2019, 296, 132–141. [Google Scholar] [CrossRef]
- Bao, X.; Xiang, S.; Chen, J.; Shi, Y.; Chen, Y.; Wang, H.; Zhu, X. Effect of Lactobacillus reuteri on vitamin B12 content and microbiota composition of furu fermentation. LWT Food Sci. Technol. 2019, 100, 138–143. [Google Scholar] [CrossRef]
- Sang, X.; Ma, X.; Hao, H.; Bi, J.; Zhang, G.; Hou, H. Evaluation of biogenic amines and microbial composition in the Chinese traditional fermented food grasshopper sub shrimp paste. LWT Food Sci. Technol. 2020, 134, 109979. [Google Scholar] [CrossRef]
- Li, L.; Wen, X.; Wen, Z.; Chen, S.; Wang, L.; Wei, X. Evaluation of the Biogenic Amines Formation and Degradation Abilities of Lactobacillus curvatus From Chinese Bacon. Front. Microbiol. 2018, 9, 1015. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Zhang, Q.; Zhang, B.; Zhang, W.; Wang, W. Insights into the Biogenic Amine Metabolic Landscape during Industrial Semidry Chinese Rice Wine Fermentation. J. Agric. Food Chem. 2016, 64, 7385–7393. [Google Scholar] [CrossRef]
- Mah, J.-H.; Hwang, H.-J. Inhibition of biogenic amine formation in a salted and fermented anchovy by Staphylococcus xylosus as a protective culture. Food Control 2009, 20, 796–801. [Google Scholar] [CrossRef]
- Sang, X.; Li, K.; Zhu, Y.; Ma, X.; Hao, H.; Bi, J.; Zhang, G.; Hou, H. The Impact of Microbial Diversity on Biogenic Amines Formation in Grasshopper Sub Shrimp Paste During the Fermentation. Front. Microbiol. 2020, 11, 782. [Google Scholar] [CrossRef] [Green Version]
- Jung, W.Y.; Lee, S.H.; Jin, H.M.; Jeon, C.O. Lentibacillus garicola sp. nov., isolated from myeolchi-aekjeot, a Korean fermented anchovy sauce. Antonie Van Leeuwenhoek 2015, 107, 1569–1576. [Google Scholar] [CrossRef]
- Irlinger, F.; Yung, S.A.; Sarthou, A.S.; Delbes-Paus, C.; Montel, M.C.; Coton, E.; Coton, M.; Helinck, S. Ecological and aromatic impact of two Gram-negative bacteria (Psychrobacter celer and Hafnia alvei) inoculated as part of the whole microbial community of an experimental smear soft cheese. Int. J. Food Microbiol. 2012, 153, 332–338. [Google Scholar] [CrossRef]
- Helinck, S.; Perello, M.-C.; Deetae, P.; de Revel, G.; Spinnler, H.-E. Debaryomyces hansenii, Proteus vulgaris, Psychrobacter sp. and Microbacterium foliorum are able to produce biogenic amines. Dairy Sci. Technol. 2013, 93, 191–200. [Google Scholar] [CrossRef]
- Ge, Y.; Zhu, J.; Ye, X.; Yang, Y. Spoilage potential characterization of Shewanella and Pseudomonas isolated from spoiled large yellow croaker (Pseudosciaena crocea). Lett. Appl. Microbiol. 2017, 64, 86–93. [Google Scholar] [CrossRef]
- Amadoro, C.; Rossi, F.; Piccirilli, M.; Colavita, G. Tetragenococcus koreensis is part of the microbiota in a traditional Italian raw fermented sausage. Food Microbiol. 2015, 50, 78–82. [Google Scholar] [CrossRef]
- He, G.; Wu, C.; Huang, J.; Zhou, R. Metabolic response of Tetragenococcus halophilus under salt stress. Biotechnol. Bioprocess Eng. 2017, 22, 366–375. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, S.H.; Chun, B.H.; Jeong, S.E.; Jeon, C.O. Tetragenococcus halophilus MJ4 as a starter culture for repressing biogenic amine (cadaverine) formation during saeu-jeot (salted shrimp) fermentation. Food Microbiol. 2019, 82, 465–473. [Google Scholar] [CrossRef]
- Kimura, B.; Konagaya, Y.; Fujii, T. Histamine formation by Tetragenococcus muriaticus, a halophilic lactic acid bacterium isolated from fish sauce. Int. J. Food Microbiol. 2001, 70, 71–77. [Google Scholar] [CrossRef]
- Li, L.; Zou, D.; Ruan, L.; Wen, Z.; Chen, S.; Xu, L.; Wei, X. Evaluation of the Biogenic Amines and Microbial Contribution in Traditional Chinese Sausages. Front. Microbiol. 2019, 10, 872. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Ospina, J.; Acquaticci, L.; Molina-Hernandez, J.B.; Rantsiou, K.; Martuscelli, M.; Kamgang-Nzekoue, A.F.; Vittori, S.; Paparella, A.; Chaves-Lopez, C. Exploring the Capability of Yeasts Isolated from Colombian Fermented Cocoa Beans to Form and Degrade Biogenic Amines in a Lab-Scale Model System for Cocoa Fermentation. Microorganisms 2020, 9, 28. [Google Scholar] [CrossRef]
- Xia, X.; Luo, Y.; Zhang, Q.; Huang, Y.; Zhang, B. Mixed Starter Culture Regulate Biogenic Amines Formation via Decarboxylation and Transamination during Chinese Rice Wine Fermentation. J. Agric. Food Chem. 2018, 66, 1–32. [Google Scholar] [CrossRef]
- Zaman, M.Z.; Bakar, F.A.; Selamat, J.; Bakar, J.; Ang, S.S.; Chong, C.Y. Degradation of histamine by the halotolerant Staphylococcus carnosus FS19 isolate obtained from fish sauce. Food Control 2014, 40, 58–63. [Google Scholar] [CrossRef]
- Namwong, S.; Tanasupawat, S.; Smitinont, T.; Visessanguan, W.; Kudo, T.; Itoh, T. Isolation of Lentibacillus salicampi strains and Lentibacillus juripiscarius sp. nov. from fish sauce in Thailand. Int. J. Syst. Evol. Microbiol. 2005, 55, 315–320. [Google Scholar] [CrossRef] [Green Version]



| BAs (mg/kg) | R3M | R6M | R9M | R12M | R18M |
|---|---|---|---|---|---|
| TRY | 9.59 ± 0.40 e | 10.78 ± 0.24 d | 12.77 ± 0.34 c | 17.76 ± 0.26 b | 20.41 ± 0.42 a |
| PHE | 7.63 ± 0.25 e | 9.14 ± 0.12 d | 13.56 ± 0.62 c | 21.24 ± 0.25 b | 27.26 ± 0.06 a |
| PUT | 14.71 ± 0.92 e | 19.77 ± 0.10 d | 28.15 ± 1.25 c | 47.67 ± 0.42 b | 60.41 ± 0.27 a |
| CAD | 23.44 ± 1.21 e | 29.75 ± 0.20 d | 42.08 ± 1.90 c | 71.03 ± 0.55 b | 90.61 ± 0.28 a |
| HIS | 18.39 ± 0.91 e | 22.99 ± 0.17 d | 32.75 ± 1.48 c | 55.59 ± 0.45 b | 74.5 ± 0.17 a |
| TYR | 13.23 ± 0.50 e | 15.18 ± 0.28 d | 17.86 ± 0.59 c | 19.07 ± 0.16 b | 20.88 ± 0.06 a |
| SPD | ND | ND | 2.41 ± 0.01 c | 2.7 ± 0.01 b | 2.75 ± 0.002 a |
| SPM | 2.39 ± 0.04 a | 2.41 ± 0.01 a | 1.98 ± 0.04 b | ND | ND |
| Total | 89.38 ± 3.97 e | 110.01 ± 1.01 d | 151.55 ± 7.24 c | 235.05 ± 1.79 b | 296.82 ± 0.71 a |
| Genus | Number of Total Strains | The Percent of BA-Production Strains |
|---|---|---|
| Tetragenococcus | 47 (16) | 65.96% |
| Staphylococcus | 48 (26) | 45.83% |
| Lentibacillus | 3 (2) | 33.33% |
| Psychrobacter | 1 (0) | 100.00% |
| Pseudomonas | 1 (0) | 100.00% |
| Strains | PUT (%) | CAD (%) | HIS (%) |
|---|---|---|---|
| Staphylococcus nepalensis 5-5 | 17.64 ± 0.44 a | 19.80 ± 0.93 a | 16.77 ± 1.04 a |
| Staphylococcus xylosus JCM 2418 | 11.96 ± 0.71 b | 6.74 ± 0.31 c | 5.04 ± 0.27 d |
| Staphylococcus hominis ICC_10-1_SCI_contig_1 | 9.32 ± 0.34 c | 8.68 ± 0.69 b | 7.81 ± 0.73 b |
| Staphylococcus capitis +Y36 | 9.78 ± 0.37 c | 7.26 ± 0.51 c | 5.64 ± 0.79 cd |
| Lentibacillus salicamp SF-20 | 8.62 ± 0.12 d | 8.21 ± 0.62 bc | 6.87 ± 0.47 c |
| Lentibacillus amyloliquefaciens LAM0015 | 8.78 ± 0.89 cd | 5.26 ± 0.68 d | 4.54 ± 0.65 d |
| Tetragenococcus halophilus NBRC 12172 | 5.34 ± 0.64 e | 3.50 ± 0.61 e | 2.59 ± 0.63 e |
| Tetragenococcus muriaticus LMG 18498 | 4.68 ± 0.78 e | 3.91 ± 0.24 e | 2.00 ± 0.15 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Bi, J.; Li, X.; Zhang, G.; Hao, H.; Hou, H. Contribution of Microorganisms to Biogenic Amine Accumulation during Fish Sauce Fermentation and Screening of Novel Starters. Foods 2021, 10, 2572. https://doi.org/10.3390/foods10112572
Ma X, Bi J, Li X, Zhang G, Hao H, Hou H. Contribution of Microorganisms to Biogenic Amine Accumulation during Fish Sauce Fermentation and Screening of Novel Starters. Foods. 2021; 10(11):2572. https://doi.org/10.3390/foods10112572
Chicago/Turabian StyleMa, Xinxiu, Jingran Bi, Xinyu Li, Gongliang Zhang, Hongshun Hao, and Hongman Hou. 2021. "Contribution of Microorganisms to Biogenic Amine Accumulation during Fish Sauce Fermentation and Screening of Novel Starters" Foods 10, no. 11: 2572. https://doi.org/10.3390/foods10112572
APA StyleMa, X., Bi, J., Li, X., Zhang, G., Hao, H., & Hou, H. (2021). Contribution of Microorganisms to Biogenic Amine Accumulation during Fish Sauce Fermentation and Screening of Novel Starters. Foods, 10(11), 2572. https://doi.org/10.3390/foods10112572







