Investigation of Escherichia coli O157:H7 Survival and Interaction with Meal Components during Gastrointestinal Digestion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Meat Samples
2.2. Strain Culture and Inoculation of Meat
2.3. In Vitro Batch Digestion Model of Meat Bolus and Sampling
2.4. Bacterial Enumeration
2.5. Biochemical Analyses
2.5.1. Protein Digestion Measurement
2.5.2. Oxidation Measurement of Proteins and Lipids
2.5.3. Determination of Free Iron Content and Its Oxidation State
2.5.4. Ascorbate Content and Its State of Oxidation
2.5.5. Nitrite and Nitrate Content
2.5.6. Nitrosothiols and Nonvolatile Nitrosamine Contents
2.5.7. Determination of Heminic Iron and Nitrosylation
2.6. Statistical Analysis
3. Results
3.1. E. coli O157:H7 CM454 Survival and Its Interaction with Meat Components throughout Digestion
3.1.1. E. coli O157:H7 CM454 Survived throughout Meat Digestion
3.1.2. E. coli O157:H7 CM454 had No Impact on Meat Protein Digestion, Contrary to Its Oxidation
3.1.3. E. coli O157:H7 CM454 did Not Impact Nitrosylation
3.1.4. E. coli O157:H7 CM454 Increased Free Iron Release and Modified Its Oxidation State
3.2. E. coli O157:H7 CM454 Survival and Its Interaction with Meal Components (Meat, Nitrite, and Ascorbate) Throughout Digestion
3.2.1. E. coli O157:H7 CM454 Survival Was Decreased by Nitrite/Ascorbate
3.2.2. E. coli O157:H7 CM454 Impacted Nitrite and Ascorbate Chemistry
3.2.3. E. coli O157:H7 CM454 Impacted Free Iron Release and Its Oxidation State
3.2.4. E. coli O157:H7 CM454 did Not Impact Protein Digestion or Lipid and Protein Oxidation
3.2.5. E. coli O157:H7 Impacted Nitrosylation
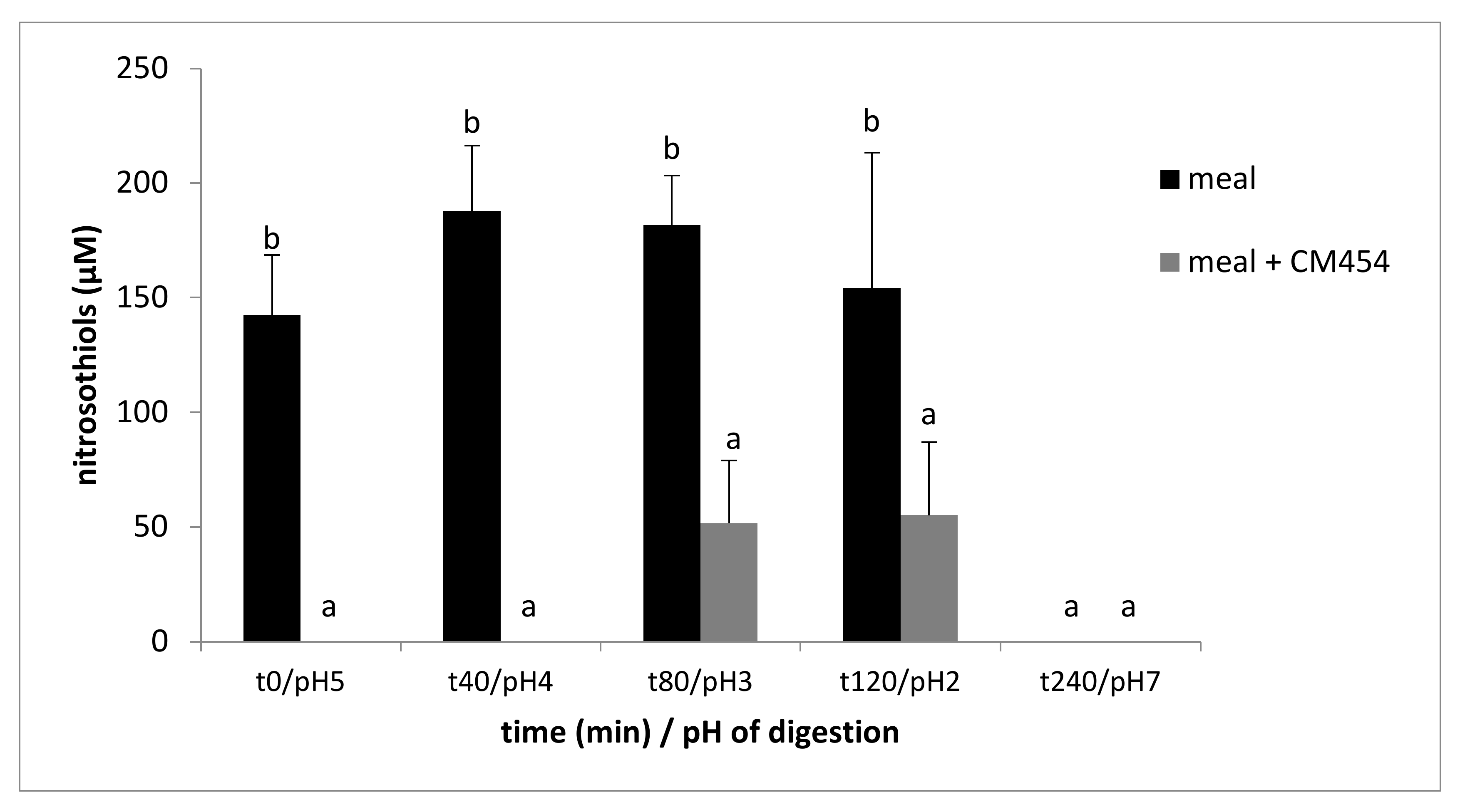
3.2.6. E. coli O157:H7 CM454 did Not Impact N-Nitrosation but Impacted S-Nitrosation
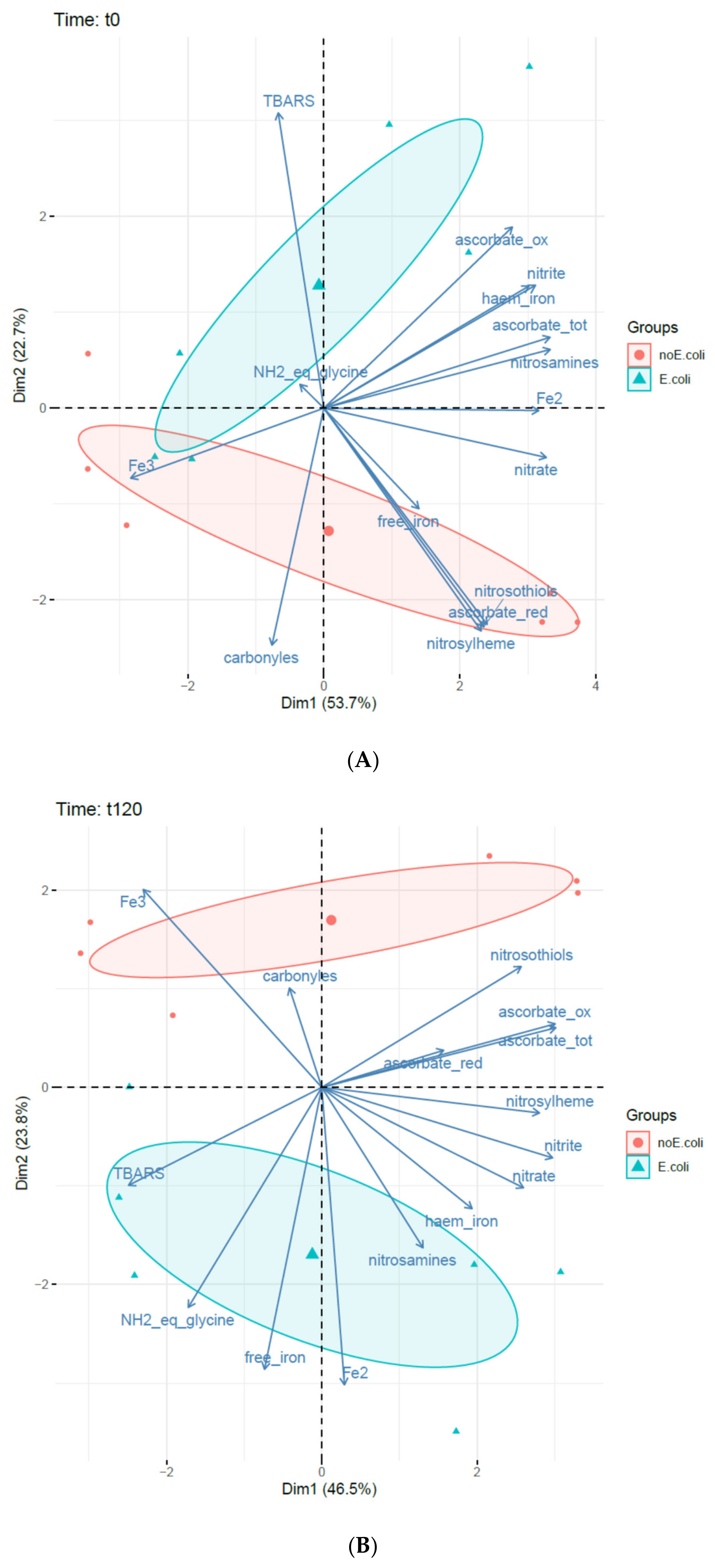
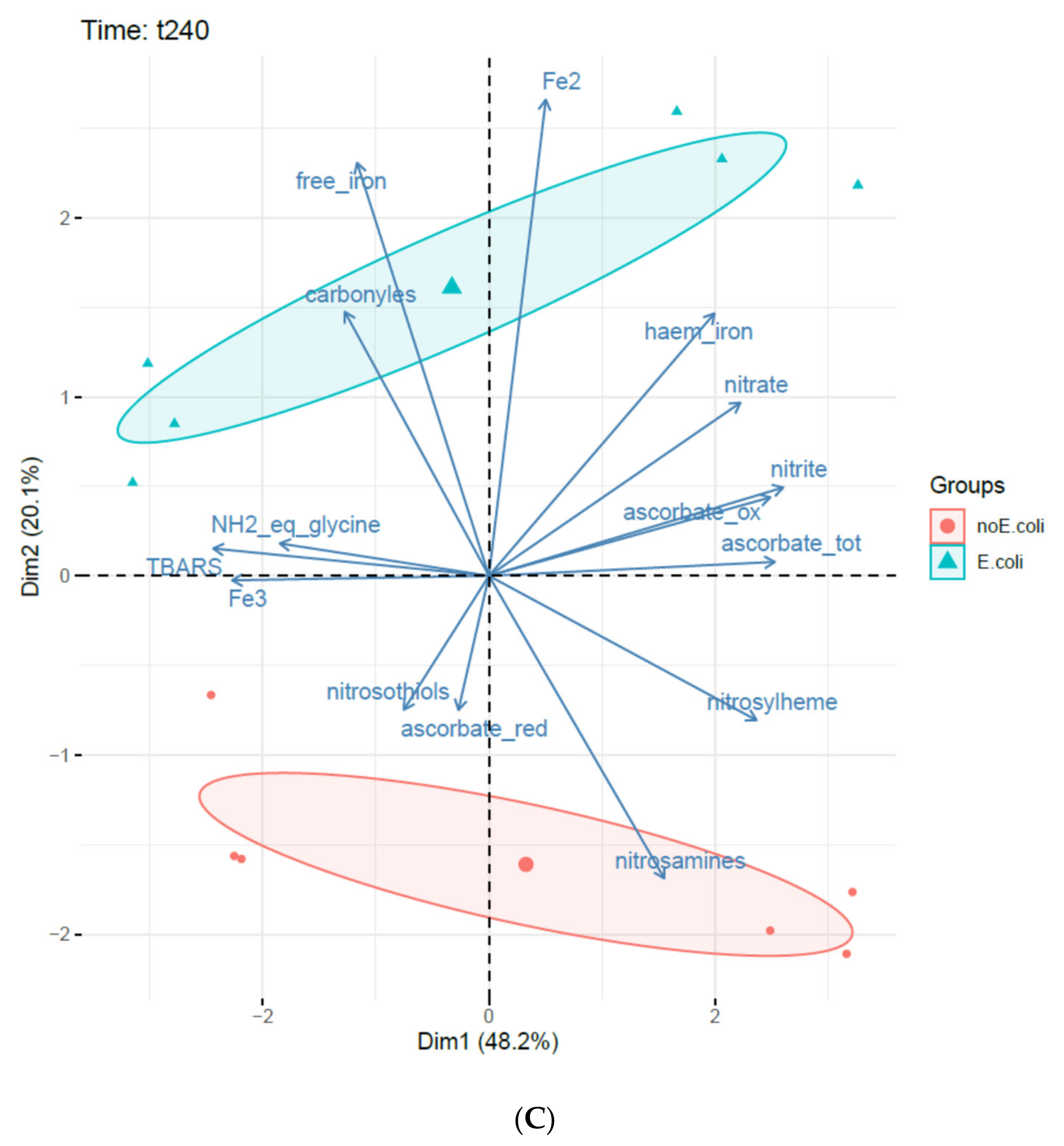
3.3. Principal Component Analysis (PCA)
4. Discussion
4.1. Static Digestive Model of Meat
4.2. Survival of E. coli O157:H7 during Digestion of Meat and Meal
4.3. E. coli O157:H7 CM454 Increased the Release of Iron
4.4. E. coli O157:H7 CM454 Impacted the Formation of Nitroso Compounds
4.5. Oxidative Processes during Digestion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Panel, E.F.S.A.B.; Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Pathogenicity assessment of Shiga toxin-producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020, 18, e05967. [Google Scholar] [CrossRef]
- Launders, N.; Byrne, L.; Jenkins, C.; Harker, K.; Charlett, A.; Adak, G.K. Disease severity of Shiga toxin-producing E. coli O157 and factors influencing the development of typical haemolytic uraemic syndrome: A retrospective cohort study, 2009–2012. BMJ Open 2016, 6, e009933. [Google Scholar] [CrossRef] [Green Version]
- Schmid-Hempel, P.; Frank, S.A. Pathogenesis, Virulence, and Infective Dose. PLoS Pathog. 2007, 3, e147. [Google Scholar] [CrossRef] [Green Version]
- Bibbal, D.; Loukiadis, E.; Kérourédan, M.; Ferré, F.; Dilasser, F.; Peytavin de Garam, C.; Cartier, P.; Oswald, E.; Gay, E.; Auvray, F.; et al. Prevalence of carriage of Shiga toxin-producing Escherichia coli serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 among slaughtered adult cattle in France. Appl. Environ. Microbiol. 2015, 81, 1397–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapountzis, P.; Segura, A.; Desvaux, M.; Forano, E. An Overview of the Elusive Passenger in the Gastrointestinal Tract of Cattle: The Shiga Toxin Producing Escherichia coli. Microorganisms 2020, 8, 877. [Google Scholar] [CrossRef] [PubMed]
- Stringer, S.C.; George, S.M.; Peck, M.W. Thermal inactivation of Escherichia coli O157:H7. J. Appl. Microbiol. 2000, 88, 79S–89S. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, G.; Desvaux, M.; Schüller, S.; Etienne-Mesmin, L.; Muniesa, M.; Blanquet-Diot, S. Modulation of Enterohaemorrhagic Escherichia coli Survival and Virulence in the Human Gastrointestinal Tract. Microorganisms 2018, 6, 115. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.W. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat. Rev. Microbiol. 2004, 2, 898–907. [Google Scholar] [CrossRef]
- King, T.; Lucchini, S.; Hinton, J.C.D.; Gobius, K. Transcriptomic analysis of Escherichia coli O157:H7 and K-12 cultures exposed to inorganic and organic acids in stationary phase reveals acidulant- and strain-specific acid tolerance responses. Appl. Environ. Microbiol. 2010, 76, 6514–6528. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.-H.; Breidt, F.; Fratamico, P.; Oh, D.-H. Acid Resistance and Molecular Characterization of Escherichia coli O157:H7 and Different Non-O157 Shiga Toxin-Producing E. coli Serogroups. J. Food Sci. 2015, 80, M2257–M2264. [Google Scholar] [CrossRef]
- Etienne-Mesmin, L.; Livrelli, V.; Privat, M.; Denis, S.; Cardot, J.-M.; Alric, M.; Blanquet-Diot, S. Effect of a new probiotic Saccharomyces cerevisiae strain on survival of Escherichia coli O157:H7 in a dynamic gastrointestinal model. Appl. Environ. Microbiol. 2011, 77, 1127–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo-López, F.N.; Blanquet-Diot, S.; Denis, S.; Thévenot, J.; Chalancon, S.; Alric, M.; Rodríguez-Gómez, F.; Romero-Gil, V.; Jiménez-Díaz, R.; Garrido-Fernández, A. Survival of pathogenic and lactobacilli species of fermented olives during simulated human digestion. Front. Microbiol. 2014, 5, 540. [Google Scholar] [CrossRef] [PubMed]
- Miszczycha, S.D.; Thévenot, J.; Denis, S.; Callon, C.; Livrelli, V.; Alric, M.; Montel, M.-C.; Blanquet-Diot, S.; Thevenot-Sergentet, D. Survival of Escherichia coli O26:H11 exceeds that of Escherichia coli O157:H7 as assessed by simulated human digestion of contaminated raw milk cheeses. Int. J. Food Microbiol. 2014, 172, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Rysman, T.; Van Hecke, T.; Van Poucke, C.; De Smet, S.; Van Royen, G. Protein oxidation and proteolysis during storage and in vitro digestion of pork and beef patties. Food Chem. 2016, 209, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Papuc, C.; Goran, G.V.; Predescu, C.N.; Nicorescu, V. Mechanisms of Oxidative Processes in Meat and Toxicity Induced by Postprandial Degradation Products: A Review. Compr. Rev. Food Sci. Food Saf. 2017, 16, 96–123. [Google Scholar] [CrossRef] [PubMed]
- de La Pomélie, D.; Santé-Lhoutellier, V.; Sayd, T.; Gatellier, P. Oxidation and nitrosation of meat proteins under gastro-intestinal conditions: Consequences in terms of nutritional and health values of meat. Food Chem. 2018, 243, 295–304. [Google Scholar] [CrossRef]
- de La Pomélie, D.; Santé-Lhoutellier, V.; Sayd, T.; Théron, L.; Gatellier, P. Using a dynamic artificial digestive system to investigate heme iron nitrosylation during gastro-intestinal transit. Food Chem. 2019, 281, 231–235. [Google Scholar] [CrossRef]
- Sheridan, W.G.; Lowndes, R.H.; Young, H.L. Intraoperative tissue oximetry in the human gastrointestinal tract. Am. J. Surg. 1990, 159, 314–319. [Google Scholar] [CrossRef]
- Locato, V.; Cimini, S.; Gara, L.D. Strategies to increase vitamin C in plants: From plant defense perspective to food biofortification. Front. Plant Sci. 2013, 4, 152. [Google Scholar] [CrossRef] [Green Version]
- Rosier, B.T.; Moya-Gonzalvez, E.M.; Corell-Escuin, P.; Mira, A. Isolation and Characterization of Nitrate-Reducing Bacteria as Potential Probiotics for Oral and Systemic Health. Front. Microbiol. 2020, 11, 555465. [Google Scholar] [CrossRef]
- Thomson, B. Nitrates and nitrites dietary exposure and risk assessment. In ESR Client Report FW0392; New Zealand Food Safety Authority: Wellington, New Zealand, 2004. [Google Scholar]
- Kuhnle, G.G.C.; Story, G.W.; Reda, T.; Mani, A.R.; Moore, K.P.; Lunn, J.C.; Bingham, S.A. Diet-induced endogenous formation of nitroso compounds in the GI tract. Free Radic. Biol. Med. 2007, 43, 1040–1047. [Google Scholar] [CrossRef]
- Bechaux, J.; de La Pomélie, D.; Théron, L.; Santé-Lhoutellier, V.; Gatellier, P. Iron-catalysed chemistry in the gastrointestinal tract: Mechanisms, kinetics and consequences. A review. Food Chem. 2018, 268, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Hoebler, M.-F.; Devaux, A.; Karinthi, C.; Belleville, J.-L.; Barry, C. Particle size of solid food after human mastication and in vitro simulation of oral breakdown. Int. J. Food Sci. Nutr. 2000, 51, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Dumas, J.B.A. Procedes de l’analyse organique. Ann. Chim. Phys. 1831, 2, 198. [Google Scholar]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Gobert, A.P.; Vareille, M.; Glasser, A.-L.; Hindré, T.; de Sablet, T.; Martin, C. Shiga Toxin Produced by Enterohemorrhagic Escherichia coli Inhibits PI3K/NF-κB Signaling Pathway in Globotriaosylceramide-3-Negative Human Intestinal Epithelial Cells. J. Immunol. 2007, 178, 8168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.-H.; Park, H.K.; Park, J.S.; Yeom, J.S.; Lim, J.-Y.; Park, C.-H.; Woo, H.-O.; Youn, H.-S.; Jun, J.-S.; Ko, G.-H.; et al. Association between Gastric pH and Helicobacter pylori Infection in Children. Pediatr. Gastroenterol. Hepatol. Nutr. 2015, 18, 246–252. [Google Scholar] [CrossRef] [Green Version]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Sayd, T.; Peyron, M.-A.; Viet, S.; Santé-Lhoutellier, V. Impact of mastication on the gastric digestion of frankfurters: Case of elderly. Proceeding of the 62nd International Congress of Meat Science and Technology (ICoMST), Bangkok, Thailand, 14–19 August 2016. [Google Scholar]
- Harkouss, R.; Mirade, P.-S.; Gatellier, P. Development of a rapid, specific and efficient procedure for the determination of proteolytic activity in dry-cured ham: Definition of a new proteolysis index. Meat Sci. 2012, 92, 84–88. [Google Scholar] [CrossRef]
- Oliver, C.N.; Alin, B.W.; Moerman, E.J.; Goldstein, S.; Stadtman, E.R. Age-related changes in oxidized proteins. J. Biol. Chem. 1987, 262, 5488–5491. [Google Scholar] [CrossRef]
- Lynch, S.M.; Frei, B. Mechanisms of copper- and iron-dependent oxidative modification of human low-density lipoprotein. J. Lipid Res. 1993, 34, 1745–1751. [Google Scholar] [CrossRef]
- Stolze, K.; Dadak, A.; Liu, Y.; Nohl, H. Hydroxylamine and phenol-induced formation of methemoglobin and free radical intermediates in erythrocytes. Biochem. Pharmacol. 1996, 52, 1821–1829. [Google Scholar] [CrossRef]
- Vislisel, J.M.; Schafer, F.Q.; Buettner, G.R. A simple and sensitive assay for ascorbate using a plate reader. Anal. Biochem. 2007, 365, 31–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonifacie, A.; Aubry, L.; Gatellier, P.; Santé-Lhoutellier, V.; Théron, L. Determination of nitroso-compounds in food products. MethodsX 2021, 8, 101289. [Google Scholar] [CrossRef]
- Hornsey, H.C. The colour of cooked cured pork. I.—Estimation of the Nitric oxide-Haem Pigments. J. Sci. Food Agric. 1956, 7, 534–540. [Google Scholar] [CrossRef]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Russell, V. Lenth Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.6.3. 2021. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 1 September 2021).
- Bax, M.-L.; Buffière, C.; Hafnaoui, N.; Gaudichon, C.; Savary-Auzeloux, I.; Dardevet, D.; Santé-Lhoutellier, V.; Rémond, D. Effects of Meat Cooking, and of Ingested Amount, on Protein Digestion Speed and Entry of Residual Proteins into the Colon: A Study in Minipigs. PLoS ONE 2013, 8, e61252. [Google Scholar] [CrossRef] [PubMed]
- Gobert, M.; Rémond, D.; Loonis, M.; Buffière, C.; Santé-Lhoutellier, V.; Dufour, C. Fruits, vegetables and their polyphenols protect dietary lipids from oxidation during gastric digestion. Food Funct. 2014, 5, 2166–2174. [Google Scholar] [CrossRef]
- Sicard, J.; Mirade, P.-S.; Portanguen, S.; Clerjon, S.; Kondjoyan, A. Simulation of the gastric digestion of proteins of meat bolus using a reaction–diffusion model. Food Funct. 2018, 9, 6455–6469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mennah-Govela, Y.A.; Singh, R.P.; Bornhorst, G.M. Buffering capacity of protein-based model food systems in the context of gastric digestion. Food Funct. 2019, 10, 6074–6087. [Google Scholar] [CrossRef]
- Hamosh, M.; Henderson, T.R.; Hamosh, P. Gastric Lipase and Pepsin Activities in the Developing Ferret: Nonparallel Development of the Two Gastric Digestive Enzymes. J. Pediatr. Gastroenterol. Nutr. 1998, 26, 162–166. [Google Scholar] [CrossRef]
- Large, T.M.; Walk, S.T.; Whittam, T.S. Variation in Acid Resistance among Shiga Toxin-Producing Clones of Pathogenic Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 2493–2500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, S.; Tasara, T.; Hummerjohann, J.; Stephan, R. An Overview of Molecular Stress Response Mechanisms in Escherichia coli Contributing to Survival of Shiga Toxin–Producing Escherichia coli during Raw Milk Cheese Production. J. Food Protect. 2011, 74, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Thanassi, D.G.; Cheng, L.W.; Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997, 179, 2512–2518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [Green Version]
- Hamner, S.; McInnerney, K.; Williamson, K.; Franklin, M.J.; Ford, T.E.E. Bile Salts Affect Expression of Escherichia coli O157:H7 Genes for Virulence and Iron Acquisition, and Promote Growth under Iron Limiting Conditions. PLoS ONE 2013, 8, e74647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandell, R.L.; Bernstein, C. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: Implications for co-lon cancer. Nut. Cancer 1991, 16, 227–238. [Google Scholar] [CrossRef]
- Kus, J.V.; Gebremedhin, A.; Dang, V.; Tran, S.-L.; Serbanescu, A.; Barnett Foster, D. Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2011, 193, 4509–4515. [Google Scholar] [CrossRef] [Green Version]
- Majou, D.; Christieans, S. Mechanisms of the bactericidal effects of nitrate and nitrite in cured meats. Meat Sci. 2018, 145, 273–284. [Google Scholar] [CrossRef]
- Casey, P.; Condon, S. Synergistic lethal combination of nitrite and acid pH on a verotoxin-negative strain of Escherichia coli O157. Int. J. Food Microbiol. 2000, 55, 255–258. [Google Scholar] [CrossRef]
- Gardner, A.M.; Helmick, R.A.; Gardner, P.R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002, 277, 8172–8177. [Google Scholar] [CrossRef] [Green Version]
- Gomes, C.M.; Giuffrè, A.; Forte, E.; Vicente, J.B.; Saraiva, L.M.; Brunori, M.; Teixeira, M. A Novel Type of Nitric-oxide Reductase: Escherichia coli flavorubredoxin *. J. Biol. Chem. 2002, 277, 25273–25276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wonderen, J.H.; Burlat, B.; Richardson, D.J.; Cheesman, M.R.; Butt, J.N. The Nitric Oxide Reductase Activity of Cytochrome c Nitrite Reductase from Escherichia coli *. J. Biol. Chem. 2008, 283, 9587–9594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vine, C.E.; Cole, J.A. Nitrosative stress in Escherichia coli: Reduction of nitric oxide. Biochem. Soc. Trans. 2011, 39, 213–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoppler, M.; Schönbächler, A.; Meile, L.; Hurrell, R.F.; Walczyk, T. Ferritin-Iron Is Released during Boiling and In Vitro Gastric Digestion. J. Nutr. 2008, 138, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.M.; Svistunenko, D.A.; Wilson, M.T.; Hemmings, A.M.; Moore, G.R.; Le Brun, N.E. Bacterial iron detoxification at the molecular level. J. Biol. Chem. 2020, 295, 17602–17623. [Google Scholar] [CrossRef]
- Lau, C.K.Y.; Ishida, H.; Liu, Z.; Vogel, H.J. Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J. Bacteriol. 2013, 195, 46–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honikel, K.-O. The use and control of nitrate and nitrite for the processing of meat products. Meat Sci. 2008, 78, 68–76. [Google Scholar] [CrossRef]
- Bonifacie, A.; Promeyrat, A.; Nassy, G.; Gatellier, P.; Santé-Lhoutellier, V.; Théron, L. Chemical reactivity of nitrite and ascorbate in a cured and cooked meat model implication in nitrosation, nitrosylation and oxidation. Food Chem. 2021, 348, 129073. [Google Scholar] [CrossRef] [PubMed]
- Nagababu, E.; Rifkind, J.M. Routes for Formation of S-Nitrosothiols in Blood. Cell Biochem. Biophys. 2013, 67, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Gyaneshwar, P.; Paliy, O.; McAuliffe, J.; Popham, D.L.; Jordan, M.I.; Kustu, S. Sulfur and nitrogen limitation in Escherichia coli K-12: Specific homeostatic responses. J. Bacteriol. 2005, 187, 1074–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Lewis, R.S. Effect of pH and Metal Ions on the Decomposition Rate of S-nitrosocysteine. Ann. Biomed. Eng. 2007, 35, 1554–1560. [Google Scholar] [CrossRef] [PubMed]
- de La Pomélie, D.; Santé-Lhoutellier, V.; Gatellier, P. Mechanisms and kinetics of tryptophan N-nitrosation in a gastro-intestinal model. Food Chem. 2017, 218, 487–495. [Google Scholar] [CrossRef] [PubMed]
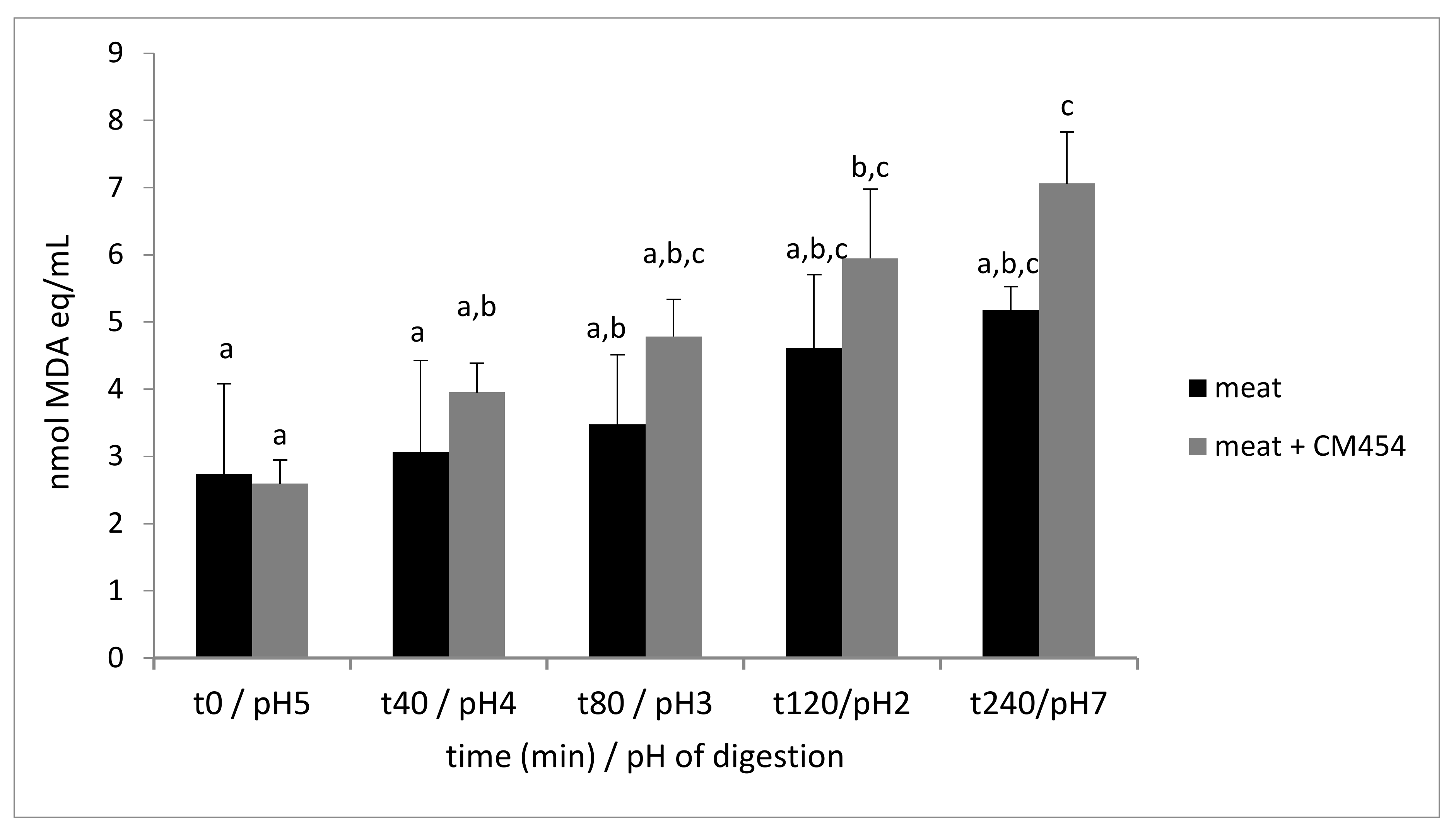
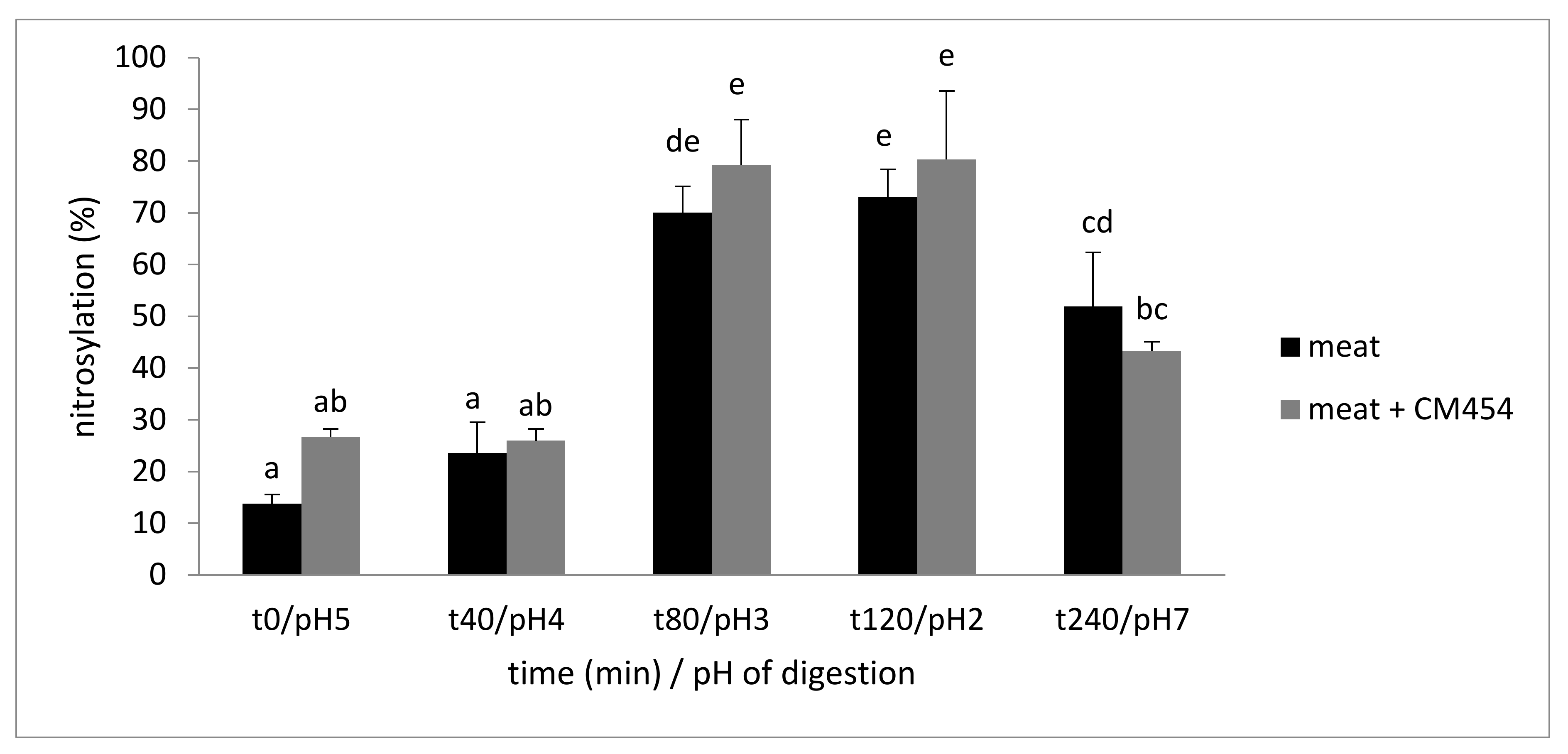
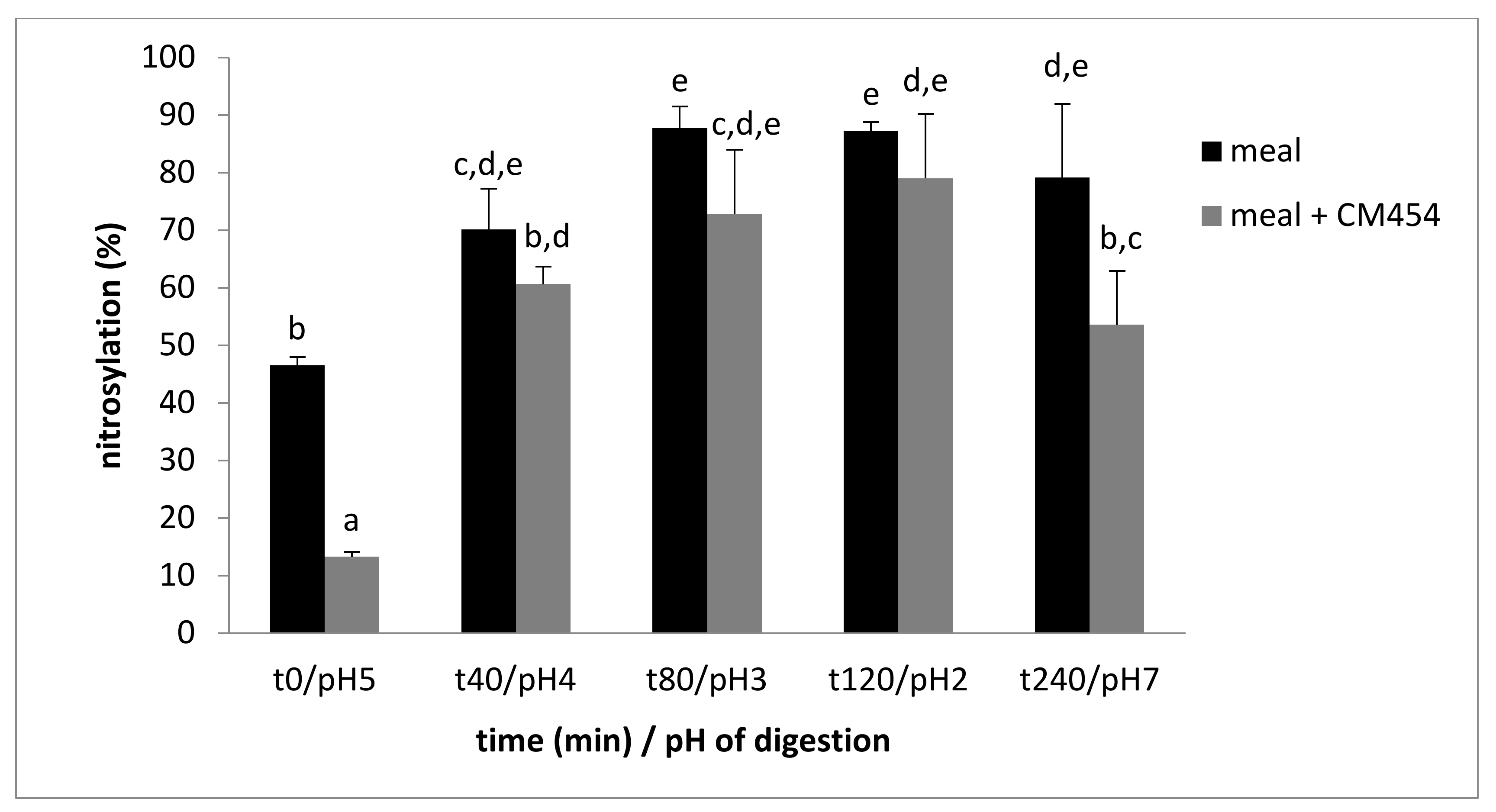
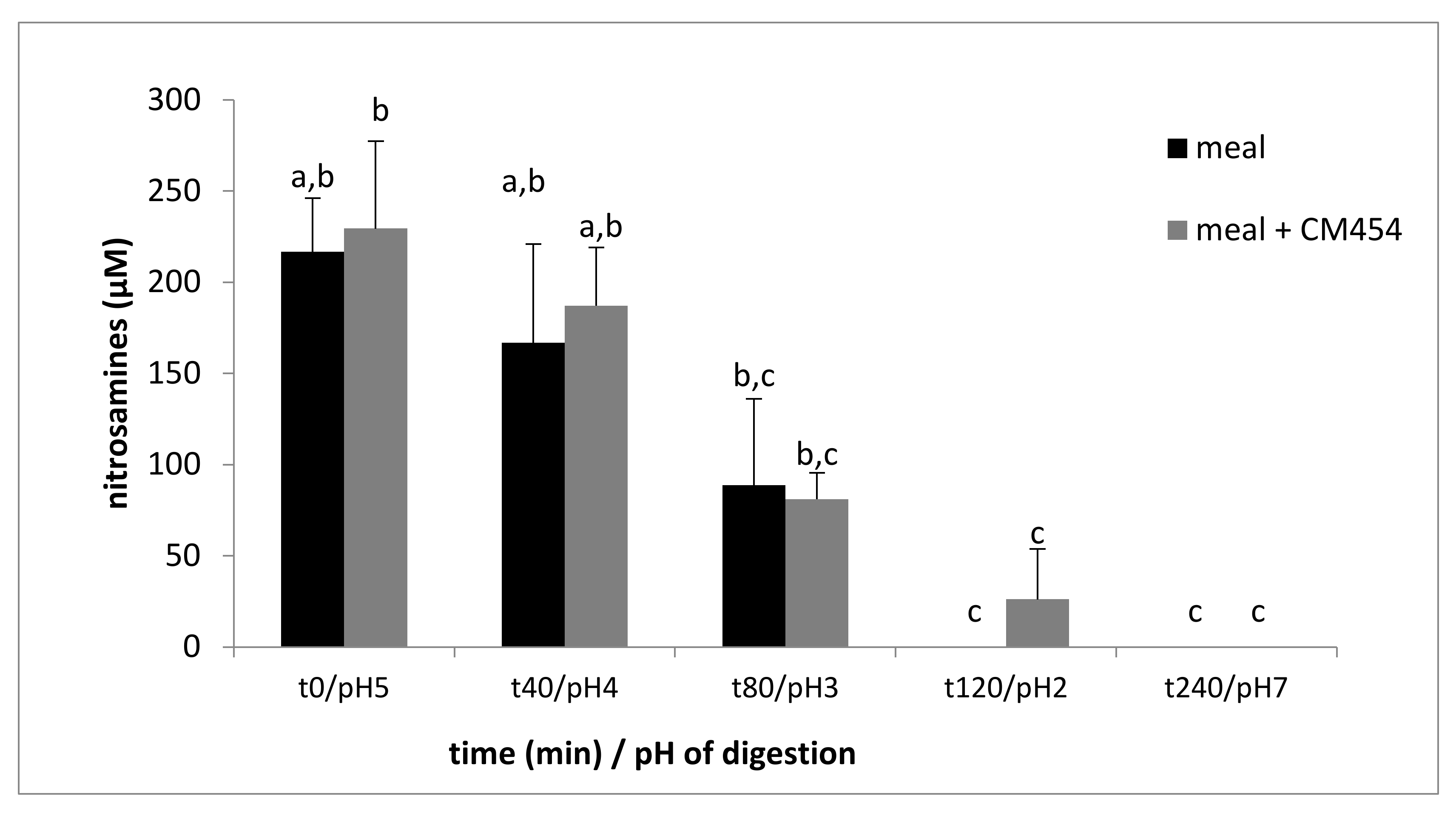
| Time min/pH | t0/pH5 | t40/pH4 | t80/pH3 | t120/pH2 | t240/pH7 |
|---|---|---|---|---|---|
| log CFU/mL | 8.0 ± 0.5 a | 7.9 ± 0.4 a | 7.2 ± 0.5 a,b | 6.7 ± 0.8 a,b | 6.7 ± 0.1 b |
| Absence of E Coli | Presence E coli | Time Effect | Inoculation Effect | Interaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time min /pH | t0/pH5 | t40/pH4 | t80/pH3 | t120/pH2 | t240/pH7 | t0/pH5 | t40/pH4 | t80/pH3 | t120/pH2 | t240/pH7 | |||
| free iron µM | 1.9 +/− 0.4 a | 2.0 +/− 0.4 a | 6.2 +/− 1.4 b | 6.9 +/− 1.4 b | 6.8 +/− 1.4 b | 2.0 +/− 0.4 a | 1.8 +/− 0.02 a | 7.4 +/− 0.7 b,c | 10.6 +/− 1.2 c | 15.3 +/− 2.3 d | p < 0.001 | p < 0.001 | p < 0.001 |
| Fe2+ µM | 0.21 +/− 0.20 a | 0.36 +/− 0.23 a | 0.74 +/− 0.62 a | 0.66 +/− 0.62 a | 0.56 +/− 0.43 a | 0.97 +/− 0.22 a | 1.30 +/− 0.36 a | 3.55 +/− 0.67 b | 6.36 +/− 1.82 c | 5.64 +/− 0.09 c | p < 0.001 | p < 0.001 | p < 0.001 |
| Fe3+ µM | 1.63 +/− 0.17 a | 1.67 +/− 0.30 a | 5.42 +/− 0.84 b | 6.20 +/− 0.84 b | 6.20 +/− 1.56 b | 1.02 +/− 0.41 a | 0.52 +/− 0.38 a | 3.89 +/− 0.24 b | 4.21 +/− 0.60 b | 9.69 +/− 2.40 c | p < 0.001 | NS | p < 0.001 |
| Time min/pH | t0/pH5 | t40/pH4 | t80/pH3 | t120/pH2 | t240/pH7 |
|---|---|---|---|---|---|
| log CFU/mL | 8.6 ± 0.3 a | 7.7 ± 0.5 a,b | 6.4 ± 0.4 b | 6.0 ± 0.5 b,c | 5.0 ± 0.5 c |
| Absence of E Coli | Presence E coli | Time Effect | Inoculation Effect | Interaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time min/pH | t0/pH5 | t40/pH4 | t80/pH3 | t120/pH2 | t240/pH7 | t0/pH5 | t40/pH4 | t80/pH3 | t120/pH2 | t240/pH7 | |||
| Nitrite | 404 ± 28 a | 312 ± 70 a,b | 236 ± 43 b | 269 ± 71 a,b | 263 ± 68 a,b | 580 ± 58 c | 591 ± 64 c | 354 ± 35 a,b | 359 ± 40 a,b | 296 ± 47 a,b | p < 0.001 | p < 0.001 | p < 0.001 |
| Nitrate | 241 ± 34 a | 258 ± 30 a | 142 ± 23 a,b | 96 ± 52 b | 82 ± 56 b | 139 ± 30 a,b | 179 ± 80 a,b | 182 ± 41 a,b | 187 ± 86 a,b | 145 ± 76 a,b | p < 0.001 | NS | p < 0.001 |
| Total ascorbate | 0.15 ± 0.06 a | 1.06 ± 0.06 a | 1.06 ± 0.03 a | 1.06 ± 0.05 a | 0.54 ± 0.21 c | 1.26 ± 0.12 a | 1.04 ± 0.08 a | 0.96 ± 0.15 a,b | 0.69 ± 0.17 b,c | 0.52 ± 0.02 c | p < 0.001 | NS | p < 0.001 |
| Oxidized ascorbate | 0.63 ± 0.08 a,b | 0.61 ± 0.07 a,b | 0.99 ± 0.18 b | 1.02 ± 0.10 b | 0.49 ± 0.22 c | 1.28 ± 0.08 b | 0.99 ± 0.12 b | 1.09 ± 0.29 b | 0.66 ± 0.14 c | 0.54 ± 0.12 c | p < 0.001 | p < 0.001 | p < 0.001 |
| Reduced ascorbate | 0.51 ± 0.13 a | 0.44 ± 0.05 a | 0.08 ± 0.13 b | 0.06 ± 0.10 b | 0.04 ± 0.02 b | 0.006 ± 0.01 b | 0.05 ± 0.05 b | 0.006 ± 0.01 b | 0.033 ± 0.033 b | 0.034 ± 0.039 b | p < 0.001 | p < 0.001 | p < 0.001 |
| free iron µM | 2.26 ± 0.36 a,b | 2.29 ± 0.24 a,b | 3.77 ± 0.44 ac | 4.39 ± 0.31 ac | 3.94 ± 0.40 ac | 2.01 ± 0.39 a | 6.70 ± 0.82 b,c | 7.67 ± 2.26 c | 11.28 ± 3.60 c,d | 12.56 ± 2.51 d,e | p < 0.001 | p < 0.001 | p < 0.001 |
| Fe2+ µM | 1.79 ± 0.29 a | 0.66 ± 0.39 a | 1.73 ± 1.16 a,b | 0.71 ± 0.62 a | 0.35 ± 0.38 a | 2.12 ± 0.48 a,b | 6.06 ± 1.10 b | 5.71 ± 3.32 b | 9.38 ± 3.80 b,c | 10.02 ± 0.61 b,c | p < 0.001 | p < 0.001 | p < 0.001 |
| Fe3+ µM | 0.48 ± 0.46 a | 1.63 ± 0.56 a,b | 2.04 ± 0.94 a,b | 3.67 ± 0.37 b | 3.59 ± 0.35 b | 0.38 ± 0.19 a | 0.58 ± 0.33 a | 1.55 ± 1.42 a,b | 1.96 ± 0.95 a,b | 2.09 ± 2.21 a,b | p < 0.001 | p < 0.001 | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de La Pomelie, D.; Leroy, S.; Talon, R.; Ruiz, P.; Gatellier, P.; Santé-Lhoutellier, V. Investigation of Escherichia coli O157:H7 Survival and Interaction with Meal Components during Gastrointestinal Digestion. Foods 2021, 10, 2415. https://doi.org/10.3390/foods10102415
de La Pomelie D, Leroy S, Talon R, Ruiz P, Gatellier P, Santé-Lhoutellier V. Investigation of Escherichia coli O157:H7 Survival and Interaction with Meal Components during Gastrointestinal Digestion. Foods. 2021; 10(10):2415. https://doi.org/10.3390/foods10102415
Chicago/Turabian Stylede La Pomelie, Diane, Sabine Leroy, Régine Talon, Philippe Ruiz, Philippe Gatellier, and Véronique Santé-Lhoutellier. 2021. "Investigation of Escherichia coli O157:H7 Survival and Interaction with Meal Components during Gastrointestinal Digestion" Foods 10, no. 10: 2415. https://doi.org/10.3390/foods10102415
APA Stylede La Pomelie, D., Leroy, S., Talon, R., Ruiz, P., Gatellier, P., & Santé-Lhoutellier, V. (2021). Investigation of Escherichia coli O157:H7 Survival and Interaction with Meal Components during Gastrointestinal Digestion. Foods, 10(10), 2415. https://doi.org/10.3390/foods10102415







