QTL Mapping and GWAS Reveal the Genetic Mechanism Controlling Soluble Solids Content in Brassica napus Shoots
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurement of SSC
2.3. QTL Detection and GWAS of SSC
2.4. Prediction of Candidate Genes
3. Results
3.1. Statistical Analysis of the SSC in the AH and GWAS Populations
3.2. QTL Mapping of SSC
3.3. Genome-Wide Association Analysis of SSC
3.4. Candidate Genes Prediction for SSC
3.5. Selection of Superior Parents
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Hasan, K.; Tamanna, N.; Haque, M.A. Biochemical and histopathological profiling of Wistar rats treated with rapeseed (Brassica napus) oil. Food Sci. Hum. Well. 2017, 7, 77–82. [Google Scholar] [CrossRef]
- Huang, H.; Shi, Y.; Liu, T.; Zhou, Y. Oilseed-vegetable-dual-purpose rape key technology research and its application prospect analysis. Agric. Sci. 2014, 5, 1291–1295. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, G. Research status and development countermeasures of dual-use rape (in Chinese with an English abstract). J. Anhui Agric. Sci. 2007, 35, 9194–9195. [Google Scholar] [CrossRef]
- Wang, H. New-demand oriented oilseed rape industry developing strategy (in Chinese with an English abstract). Chin. J. Oil Crops Sci. 2018, 40, 613–617. [Google Scholar] [CrossRef]
- Mandrich, L.; Caputo, E. Brassicaceae-derived anticancer agents: Towards a green approach to beat cancer. Nutrients 2020, 12, 868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podsdek, A. Natural antioxidants and antioxidant capacity of Brassica vegetables: A review. LWT-Food Sci. Technol. 2007, 40, 1–11. [Google Scholar] [CrossRef]
- Doorn, H.V.; Van, D.; Holst, G.V.; Raaijmakers-Ruijs, N.; Postma, E.; Groeneweg, B.; Jongen, W. The glucosinolates sinigrin and progoitrin are important determinants for taste preference and bitterness of Brussels sprouts. J. Sci. Food Agric. 1998, 78, 30–38. [Google Scholar] [CrossRef]
- Drewnowski, A.; Gomez-Carneros, C. Bitter taste, phytonutrients, and the consumer: A review. Am. J. Clin. Nutr. 2001, 72, 1424–1435. [Google Scholar] [CrossRef]
- Dinehart, M.E.; Hayes, J.E.; Bartoshuk, L.M.; Lanier, S.L.; Duffy, V.B. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake. Physiol. Behav. 2006, 87, 304–313. [Google Scholar] [CrossRef]
- Fenwick, G.R.; Griffiths, N.M.; Heaney, R.K. Bitterness in brussels sprouts (Brassica oleracea L. var. gemmifera): The role of glucosinolates and their breakdown products. J. Sci. Food Agric. 1983, 34, 73–80. [Google Scholar] [CrossRef]
- Engel, E.; Baty, C.; Le Corre, D.; Souchon, I.; Martin, N. Flavor-active compounds potentially implicated in cooked cauliflower acceptance. J. Agric. Food Chem. 2002, 50, 6459–6467. [Google Scholar] [CrossRef] [PubMed]
- Poppel, G.V.; Verhoeven, D.T.; Verhagen, H.; Goldbohm, R.A. Brassica vegetables and cancer prevention. Epidemiology and mechanisms. Adv. Exp. Med. Biol. 1999, 472, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Bazzano, L.A.; Jiang, H.; Ogden, L.G.; Loria, C.M.; Suma, V.; Leann, M.; Whelton, P.K. Fruit and vegetable intake and risk of cardiovascular disease in US adults: The first national health and nutrition examination survey epidemiologic follow-up study. Am. J. Clin. Nutr. 2002, 76, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkerk, R.; Schreiner, M.; Krumbein, A.; Ciska, E.; Dekker, M. Glucosinolates in Brassica vegetables: The influence of the food supply chain on intake, bioavailability and human health. Mol. Nutr. Food Res. 2009, 53, 219–265. [Google Scholar] [CrossRef] [PubMed]
- Beck, T.K.; Jensen, S.; Bjoern, G.K.; Kidmose, U. The masking effect of sucrose on perception of bitter compounds in Brassica vegetables. J. Sens. Stud. 2014, 29, 190–200. [Google Scholar] [CrossRef]
- Suwanarit, A.; Sestapukdee, M. Stimulating effects of foliar k-fertilizer applied at the appropriate stage of development of maize: A new way to increase yield and improve quality. Plant Soil 1989, 120, 111–124. [Google Scholar] [CrossRef]
- Gene, E.; Lester, J.L.; Jifon, D.J.; Makus. Impact of potassium nutrition on postharvest fruit quality: Melon (Cucumis melo L) case study. Plant Soil 2010, 335, 117–131. [Google Scholar] [CrossRef]
- Beckles, D.M. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit-ScienceDirect. Postharvest Biol. Technol. 2012, 63, 129–140. [Google Scholar] [CrossRef]
- Mata, M.; José, N.; Natera, M.; Rafael, J. Effect of growth regulators on the epicarp, mesocarp and total solublesolids of muskmelon (Cucumis melo L.) fruit cv. Edisto 47. Rev. Cient. UDO Agric. 2009, 9, 295–303. [Google Scholar]
- Viskeliene, A.; Samuoliene, G.; Karkleliene, R.; Viskelis, P.; Sasnauskas, A.; Duchovskis, P. Quality and developmental changes in white head cabbage (Brassica oleracea L.) and radish (Raphanus sativus L.) during winter storage. Zemdirbyste 2017, 104, 229–234. [Google Scholar] [CrossRef]
- Helland, H.S.; Leufvén, A.; Bengtsson, G.B.; Skaret, J.; Wold, A.B. Postharvest biology and technology. Postharvest Biol. Technol. 2016, 111, 150–160. [Google Scholar] [CrossRef]
- Fridman, E.; Liu, Y.; Carmel-Goren, L.; Gur, A.; Shoresh, M.; Pleban, T.; Eshed, Y.; Zamir, D. Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol. Genet. Genom. 2002, 266, 821–826. [Google Scholar] [CrossRef]
- Mallor, C.; Balcells, M.; Mallor, F.; Sales, E. Genetic variation for bulb size, soluble solids content and pungency in the Spanish sweet onion variety Fuentes de Ebro. Response to selection for low pungency. Plant Breed. 2011, 130, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Samykanno, K.; Pang, E.; Marriott, P.J. Genotypic and environmental effects on flavor attributes of ‘Albion’ and ‘Juliette’ strawberry fruits. Sci. Hortic.-Amst. 2013, 164, 633–642. [Google Scholar] [CrossRef]
- Tecle, I.Y.; Menda, N.; Buels, R.M.; Knaap, E.; Mueller, L.A. solQTL: A tool for QTL analysis, visualization and linking to genomes at SGN database. BMC Bioinform. 2010, 11, 525. [Google Scholar] [CrossRef] [Green Version]
- Etienne, C.; Rothan, C.; Moing, A.; Plomion, C.; Bodénès, C.; Svanella-Dumas, L.; Cosson, P.; Pronier, V.; Monet, R.; Dirlewanger, E. Candidate genes and QTLs for sugar and organic acid content in peach [Prunus persica (L.) Batsch]. Theor. Appl. Genet. 2002, 105, 145–159. [Google Scholar] [CrossRef]
- Paterson, A.H.; Damon, S.; Hewitt, J.D.; Zamir, D.; Tanksley, S.D. Mendelian factors underlying quantitative traits in tomato: Comparison across species, generations, and environments. Genetics 1991, 127, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Kongjaimun, A.; Somta, P.; Tomooka, N.; Kaga, A.; Vaughan, D.A.; Srinives, P. QTL mapping of pod tenderness and total soluble solid in yardlong bean [Vigna unguiculata (L.) Walp. subsp. unguiculata cv.-gr. sesquipedalis]. Euphytica 2013, 189, 217–223. [Google Scholar] [CrossRef]
- Castro, P.; Lewers, K.S. Identification of quantitative trait loci (QTL) for fruit-quality traits and number of weeks of flowering in the cultivated strawberry. Mol. Breed. 2016, 36, 138. [Google Scholar] [CrossRef]
- Wang, X.; Sun, L.; Li, W.; Peng, M.; Chen, F.; Zhang, W.; Sun, C.; Chen, S.; Hua, W.; Zhang, J. Dissecting the genetic mechanisms of waterlogging tolerance in Brassica napus through linkage mapping and a genome-wide association study. Ind. Crops Prod. 2020, 147, 112269. [Google Scholar] [CrossRef]
- Afzal, F.; Li, H.; Gul, A.; Subhani, A.; Ali, A.; Mujeeb-Kazi, A.; Ogbonnaya, F.; Trethowan, R.; Xia, X.; He, Z.; et al. Genome-wide analyses reveal footprints of divergent selection and drought adaptive traits in synthetic-derived wheats. G3 (Bethesda) 2019, 9, 1957–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samayoa, L.; Malvar, R.; Olukolu, B.A.; Holland, J.B.; Butrón, A. Genome-wide association study reveals a set of genes associated with resistance to the mediterranean corn borer (Sesamia nonagrioides L.) in a maize diversity panel. BMC Plant Biol. 2015, 15, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Xian, X.; Xu, X.; Qu, C.; Liu, L. Genome-wide association mapping of seed coat color in Brassica napus. J. Agric. Food Chem. 2017, 65, 5229–5237. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, B.; Xu, K.; Yan, G.; Qiao, J.; Li, J.; Li, H.; Li, L.; Xiao, X.; Zhang, T.; et al. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci. 2016, 242, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hu, K.; Zhang, Z.; Guan, C.; Chen, S.; Wei, H.; Li, J.; Wen, J.; Yi, B.; Shen, J. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 2015, 23, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Fikere, M.; Barbulescu, D.M.; Malmberg, M.M.; Spangenberg, G.C.; Cogan, N.O.I.; Daetwyler, H.D. Meta-analysis of GWAS in canola blackleg (Leptosphaeria maculans) disease traits demonstrates increased power from imputed whole-genome sequence. Sci. Rep. 2020, 10, 14300. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, A.; Qian, M.; Sun, B.; Mahmood, U.; Lu, K. Genome-wide association study identifies novel loci and candidate genes for drought stress tolerance in rapeseed. Oil Crop Sci. 2021, 6, 12–22. [Google Scholar] [CrossRef]
- Pal, L.; Sandhu, S.K.; Bhatia, D. Genome-wide association study and identification of candidate genes for seed oil content in Brassica napus. Euphytica 2021, 217, 66. [Google Scholar] [CrossRef]
- Kiran, A.; Wakeel, A.; Snowdon, R.; Friedt, W.; Léon, J. Genetic dissection of root architectural traits by QTL and genome-wide association mapping in rapeseed (Brassica napus). Plant Breed. 2019, 138, 184–192. [Google Scholar] [CrossRef]
- Sun, F.; Liu, J.; Hua, W.; Sun, X.; Wang, X.; Wang, H. Identification of stable QTLs for seed oil content by combined linkage and association mapping in Brassica napus. Plant Sci. 2016, 252, 388–399. [Google Scholar] [CrossRef]
- Wang, X.; Yu, K.; Li, H.; Qi, P.; Feng, C.; Zhang, W.; Chen, S.; Hu, M.; Zhang, J. High-density SNP map construction and QTL identification for the apetalous character in Brassica napus L. Front. Plant Sci. 2015, 6, 1164. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Zhang, W.; Yu, K.; Sun, L.; Gao, J.; Zhou, X.; Peng, Q.; Fu, S.; Hu, M.; Long, W.; et al. Unconditional and conditional QTL analyses of seed fatty acid composition in Brassica napus L. BMC Plant Biol. 2018, 18, 49. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.; Wang, X.; Li, W.; Sun, L.; Peng, Q.; Chen, F.; Zhang, W.; Guan, R.; Zhang, J. Identification and physical mapping of QTLs associated with flowering time in Brassica napus L. Euphytica 2019, 215, 152. [Google Scholar] [CrossRef]
- Sun, L.J.; Wang, X.; Yu, K.; Li, W.; Peng, Q.; Chen, F.; Zhang, W.; Fu, S.; Xiong, D.; Chu, P.; et al. Mapping of QTLs controlling seed weight and seed-shape traits in Brassica napus L. using a high-density SNP map. Euphytica 2018, 214, 228. [Google Scholar] [CrossRef]
- Sun, C.; Chen, F.; Chen, S.; Peng, Q.; Zhang, W.; Yi, B.; Zhang, J.; Fu, T. Genome-wide association analysis of seed number per angle in Brassica napus L. Acta Agron. Sin. 2020, 46, 147–153. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, T.; Cao, W.; Ma, C.; Pan, S.; Xu, X. Evaluation of nutrition and sensory quality of flowering Chinese cabbage based on principal component and cluster analysis (in Chinese with an English abstract). Food Ferment. Ind. 2020, 46, 253–258. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.; Gaffney, P.; Zeng, Z. Windows QTL Cartographer 2.0; Department of Statics, North Carolina State University: Raleigh, NC, USA, 2007. [Google Scholar]
- Wang, X.; Wang, H.; Long, Y.; Li, D.; Yin, Y.; Tian, J.; Chen, L.; Liu, L.; Zhao, W.; Zhao, Y.; et al. Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS ONE 2013, 8, e80569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Wang, B.; Wang, X.; Hu, K.; Yi, B. Genome-wide association study dissecting the genetic architecture underlying the branch angle trait in rapeseed (Brassica napus L.). Sci. Rep. 2016, 6, 33673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Wang, B.; Yan, L.; Hu, K.; Liu, S.; Zhou, Y.; Guan, C.; Zhang, Z.; Li, J.; Zhang, J. Genome-wide association study provides insight into the genetic control of plant height in rapeseed (Brassica napus L.). Front. Plant Sci. 2016, 7, 1102. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Buckler, E.S. Tassel: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv 2014. [Google Scholar] [CrossRef]
- Tarkowski, U.P.; Tsirkone, V.G.; Osipov, E.M.; Beelen, S.; Strelkov, S.V. Crystal structure of Arabidopsis thaliana neutral invertase 2. Acta Crystallogr. F 2020, 76, 152–157. [Google Scholar] [CrossRef]
- Murakawa, M.; Shimojima, M.; Shimomura, Y.; Kobayashi, K.; Awai, K.; Ohta, H. Monogalactosyldiacylglycerol synthesis in the outer envelope membrane of chloroplasts is required for enhanced growth under sucrose supplementation. Front. Plant Sci. 2014, 5, 280. [Google Scholar] [CrossRef] [Green Version]
- Rottmann, T.M.; Fritz, C.; Lauter, A.; Schneider, S.; Fischer, C.; Danzberger, N.; Dietrich, P.; Sauer, N.; Stadler, R. Protoplast-esculin assay as a new method to assay plant sucrose transporters: Characterization of AtSUC6 and AtSUC7 sucrose uptake activity in Arabidopsis col-0 ecotype. Front. Plant Sci. 2018, 9, 430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottmann, T.; Klebl, F.; Schneider, S.; Kischka, D.; Ruscher, D.; Sauer, N.; Stadler, R. Sugar transporter STP7 specificity for l-arabinose and d-xylose contrasts with the typical hexose transporters STP8 and STP12. Plant Physiol. 2018, 176, 2330–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keurentjes, J.J.; Sulpice, R.; Gibon, Y.; Steinhauser, M.-C.; Fu, J.; Koornneef, M.; Stitt, M.; Vreugdenhil, D. Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol. 2008, 9, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oey, I.; Lille, M.; Loey, A.V.; Hendrickx, M. Effect of high-pressure processing on colour, texture and flavour of fruit- and vegetable-based food products: A review. Trends Food Sci. Technol. 2008, 19, 320–328. [Google Scholar] [CrossRef]
- Cuerra, N.; Carrozzi, L.; Goni, M.G.; Roura, S.; Yommi, A. Quality characterization of celery (Apium graveolens L.) by plant zones and two harvest dates. J. Food Sci. 2010, 75, 327–332. [Google Scholar] [CrossRef]
- Kramchote, S.; Nakano, K.; Kanlayanarat, S.; Ohashi, S.; Takizawa, K.; Bai, G. Rapid determination of cabbage quality using visible and near-infrared spectroscopy. LWT-Food Sci. Technol. 2014, 59, 695–700. [Google Scholar] [CrossRef]
- Sun, J.; Li, Y.; Wu, X.; Dai, C.; Chen, Y. SSC prediction of cherry tomatoes based on IRIV-CS-SVR model and near infrared reflectance spectroscopy. J. Food Process Eng. 2018, 41, e12884. [Google Scholar] [CrossRef]
- Li, M.; Han, D.; Liu, W. Non-destructive measurement of soluble solids content of three melon cultivars using portable visible/near infrared spectroscopy. Biosyst. Eng. 2019, 188, 31–39. [Google Scholar] [CrossRef]
- Raman, R.; Allen, H.; Diffey, S.; Raman, H.; Martin, P.; Mckelvie, K. Localisation of quantitative trait loci for quality attributes in a doubled haploid population of wheat (Triticum aestivum L.). Genome 2009, 52, 701–715. [Google Scholar] [CrossRef]
- Chun, J.; Zeng, Z. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 1995, 140, 1111–1127. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010, 11, 459–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarka, M.; Akesson, M.; Beraldi, D.; Hernandez-Sanchez, J.; Hasselquist, D.; Bensch, S.; Hansson, B. A strong quantitative trait locus for wing length on chromosome 2 in a wild population of great reed warblers. Proc. R. Soc. B 2010, 277, 2361–2369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengt, H.; Hanna, S.; Martin, S.; Maja, T.; Suvi, P.; Maria, S.; Helena, W.; Dennis, H. Contrasting results from GWAS and QTL mapping on wing length in great reed warblers. Mol. Ecol. Resour. 2018, 18, 867–876. [Google Scholar] [CrossRef]
- Yabe, S.; Hara, T.; Ueno, M.; Enoki, H.; Iwata, H. Disagreement between results obtained from GWAS and biparental QTL mapping suggests epistatic interaction. In Proceedings of the International Plant and Animal Genome Conference XXII 2014, San Diego, CA, USA, 11–15 January 2014. [Google Scholar]
- Yativ, M.; Harary, I.; Wolf, S. Sucrose accumulation in watermelon fruits: Genetic variation and biochemical analysis. J. Plant Physiol. 2010, 167, 589–596. [Google Scholar] [CrossRef]
- Winter, H.; Huber, S.C. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 253–289. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Du, L.; Xie, J.; Yao, Y.; Sun, G. Expression patterns, activities and carbohydrate-metabolizing regulation of sucrose phosphate synthase, sucrose synthase and neutral invertase in pineapple fruit during development and ripening. Int. J. Mol. Sci. 2012, 13, 9460–9477. [Google Scholar] [CrossRef] [Green Version]
- Lytovchenko, A.; Fernie, A.R. Photosynthetic metabolism is severely impaired on the parallel reduction of plastidial and cytosolic isoforms of phosphoglucomutase. Plant Physiol. Biochem. 2003, 41, 193–200. [Google Scholar] [CrossRef]
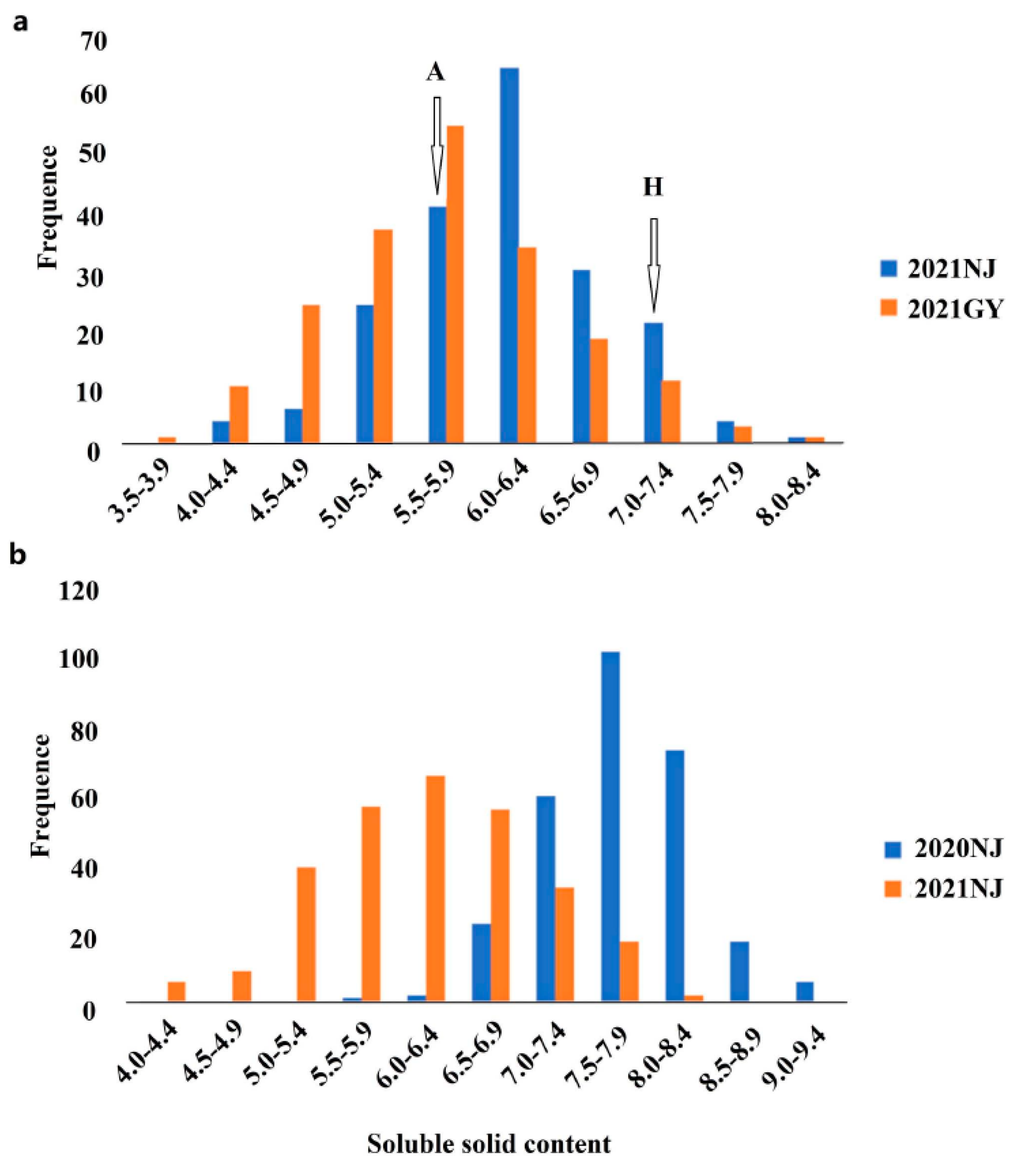
| Trait | Population | Environment | Mean ± SD | Mode (°Brix) | Min (°Brix) | Max (°Brix) | Variable Coefficient/CV(%) 1 | Broad-Sense Heritability/h2(%) 2 |
|---|---|---|---|---|---|---|---|---|
| Soluble solids content | Recombinant inbred linepopulation | 21 Nanjing | 6.10 ± 0.70 | 6.27 | 4.07 | 8.32 | 11.5 | 52.7 |
| 21 Guiyang | 5.68 ± 0.81 | 5.90 | 3.70 | 9.30 | 14.3 | |||
| GWASpopulation | 20 Nanjing | 7.73 ± 0.58 | 7.50 | 5.90 | 9.32 | 7.50 | 61.1 | |
| 21 Nanjing | 6.24 ± 0.80 | 6.56 | 4.14 | 8.38 | 12.80 |
| Population | QTL | Chromosome | Position | LOD 1 | Additive Effect 2 | R2 (%) 3 | SNP Interval | Physical Position (bp) | Environment |
|---|---|---|---|---|---|---|---|---|---|
| Recombinant inbred line population | qSSC/21NJ.A01-1 | A01 | 85.41 | 3.1 | 0.19 | 5.98 | Bn-A01-p21532512-BnGMS33 | 18,199,676–18,199,725 | 21 NJ |
| qSSC/21NJ.A01-2 | A01 | 104.51 | 4.0 | −0.22 | 8.19 | Bn-scaff_15879_1-p327427-Bn-scaff_15879_1-p715384 | 31,567,749–31,567,806 | 21 NJ | |
| qSSC/21NJ.A04-1 | A04 | 70.01 | 2.9 | −0.17 | 5.58 | Bn-A04-p17742799-Bn-A04-p18125679 | 18,471,893–18,471,942 | 21 NJ | |
| qSSC/21NJ.A09-1 | A09 | 72.71 | 5.0 | 0.22 | 9.78 | Bn-scaff_16361_1-p1749058-Bn-A09-p28925363 | 29,297,004–29,298,009 | 21 NJ | |
| qSSC/21GY.A04-1 | A04 | 4.11 | 3.9 | −0.74 | 12.92 | Bn-A04-p1274596-Bn-A04-p1900422 | 1,640,081–1,640,130 | 21 GY | |
| qSSC/21GY.A08-1 | A08 | 53.31 | 3.0 | −0.52 | 10.18 | Bn-A08-p18592850-Bn-A08-p19573947 | 16,352,113–16,352,162 | 21 GY |
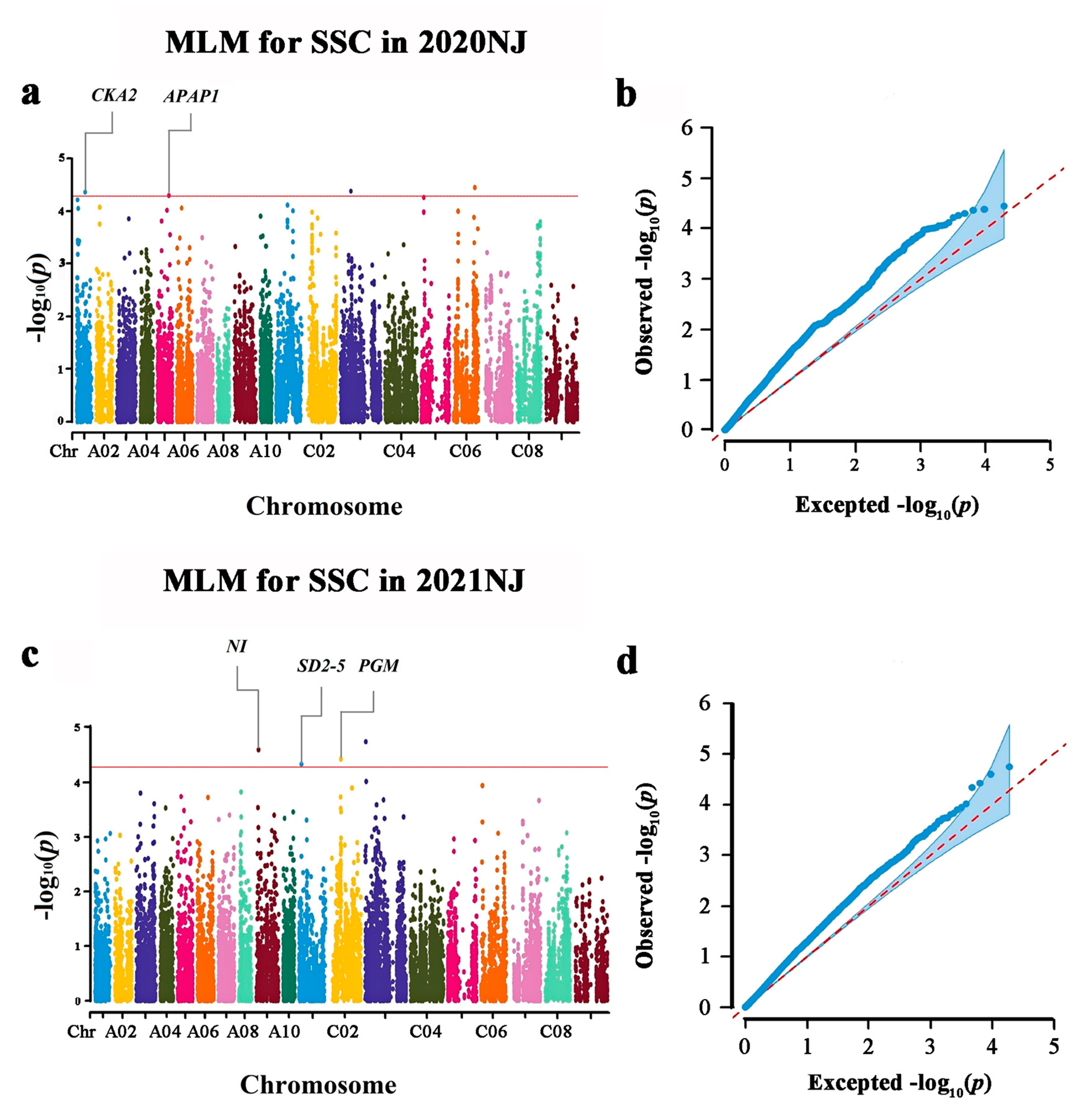
| Marker | Chromosome | Position (bp) | −lg(p) 1 | R2(%) 2 | Environment |
|---|---|---|---|---|---|
| Bn-A01-p15502589 | A01 | 12943376 | 4.36 | 9.39 | 20 NJ |
| Bn-A05-p19436126 | A05 | 17655794 | 4.30 | 9.22 | 20 NJ |
| Bn-A09-p3994437 | A09 | 3999318 | 4.60 | 9.69 | 21 NJ |
| Bn-scaff_15838_5-p335984 | C01 | 3176216 | 4.33 | 9.27 | 21 NJ |
| Bn-scaff_17566_1-p21523 | C02 | 16722668 | 4.42 | 9.48 | 21 NJ |
| Bn-scaff_16614_1-p1120223 | C03 | 971293 | 4.75 | 10.18 | 21 NJ |
| Bn-scaff_16352_1-p308563 | C03 | 15443140 | 4.38 | 9.36 | 20 NJ |
| Bn-scaff_16874_1-p411591 | C06 | 31817621 | 4.45 | 9.61 | 20 NJ |
| Marker | Rapeseed Gene | Physical Position | Homologs in A. thaliana | Functional Annotation |
|---|---|---|---|---|
| QTL region | ||||
| Bn-scaff_16361_1-p1749058-Bn-A09-p28925363 | BnaA09g41790D | A09:29,129,265–29,131,192 | AT4G34860 | plant neutral invertase family protein (NI) |
| GWAS region | ||||
| Bn-A01-p15502589 | BnaA01g21040D | A01:12,943,079–12,945,493 | AT3G50000 | casein kinase II, alpha chain 2 (CKA2) |
| Bn-A05-p19436126 | BnaA05g23350D | A05:17,683,715–17,684,062 | AT3G45230 | hydroxyproline-rich glycoprotein family protein (APAP1) |
| Bn-A09-p3994437 | BnaA09g08200D | A09:3,996,167–3,999,048 | AT2G11810 | glycolipid biosynthetic process (MGD3) |
| Bn-scaff_15838_5-p335984 | BnaA09g08760D | A09:4,377,564–4,379,821 | AT4G02050 | sugar transporter protein 7 (STP7) |
| BnaC01g06180D | C01:3,255,144–3,256,301 | AT4G32300 | S-domain-2 5 (SD2-5) | |
| Bn-scaff_17566_1-p21523 | BnaC02g20320D | C02:16,723,975–16,724,687 | AT1G70820 | Phosphoglucomutase (PGM) |
| Population | Environment | Material Name | Soluble Solid Content (°Brix) | Average |
|---|---|---|---|---|
| Recombinant inbred line population | 21 NJ | AH124 | 8.32 | 7.54 |
| AH194 | 7.75 | |||
| AH174 | 7.55 | |||
| AH120 | 7.50 | |||
| AH162 | 7.45 | |||
| AH083 | 7.43 | |||
| AH193 | 7.42 | |||
| AH192 | 7.35 | |||
| AH117 | 7.30 | |||
| AH195 | 7.30 | |||
| 21 GY | AH169 | 9.30 | 7.60 | |
| AH173 | 8.10 | |||
| AH174 | 7.90 | |||
| AH164 | 7.80 | |||
| AH166 | 7.40 | |||
| AH180 | 7.40 | |||
| AH042 | 7.36 | |||
| AH188 | 7.00 | |||
| AH029 | 6.90 | |||
| AH121 | 6.80 | |||
| GWAS population | 20 NJ | L452 | 9.32 | 9.06 |
| L192 | 9.30 | |||
| L449 | 9.20 | |||
| L456 | 9.10 | |||
| L247 | 9.00 | |||
| L360 | 9.00 | |||
| L465 | 9.00 | |||
| L145 | 8.90 | |||
| L166 | 8.90 | |||
| L380 | 8.90 | |||
| 21 NJ | L166 | 8.38 | 7.93 | |
| L527 | 8.12 | |||
| L523 | 7.98 | |||
| L275 | 7.92 | |||
| L363 | 7.90 | |||
| L381 | 7.86 | |||
| L260 | 7.86 | |||
| L510 | 7.76 | |||
| L392 | 7.76 | |||
| L380 | 7.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Chen, F.; Zhao, X.; Pang, C.; Shi, R.; Liu, C.; Sun, C.; Zhang, W.; Wang, X.; Zhang, J. QTL Mapping and GWAS Reveal the Genetic Mechanism Controlling Soluble Solids Content in Brassica napus Shoots. Foods 2021, 10, 2400. https://doi.org/10.3390/foods10102400
Wu X, Chen F, Zhao X, Pang C, Shi R, Liu C, Sun C, Zhang W, Wang X, Zhang J. QTL Mapping and GWAS Reveal the Genetic Mechanism Controlling Soluble Solids Content in Brassica napus Shoots. Foods. 2021; 10(10):2400. https://doi.org/10.3390/foods10102400
Chicago/Turabian StyleWu, Xu, Feng Chen, Xiaozhen Zhao, Chengke Pang, Rui Shi, Changle Liu, Chengming Sun, Wei Zhang, Xiaodong Wang, and Jiefu Zhang. 2021. "QTL Mapping and GWAS Reveal the Genetic Mechanism Controlling Soluble Solids Content in Brassica napus Shoots" Foods 10, no. 10: 2400. https://doi.org/10.3390/foods10102400
APA StyleWu, X., Chen, F., Zhao, X., Pang, C., Shi, R., Liu, C., Sun, C., Zhang, W., Wang, X., & Zhang, J. (2021). QTL Mapping and GWAS Reveal the Genetic Mechanism Controlling Soluble Solids Content in Brassica napus Shoots. Foods, 10(10), 2400. https://doi.org/10.3390/foods10102400






