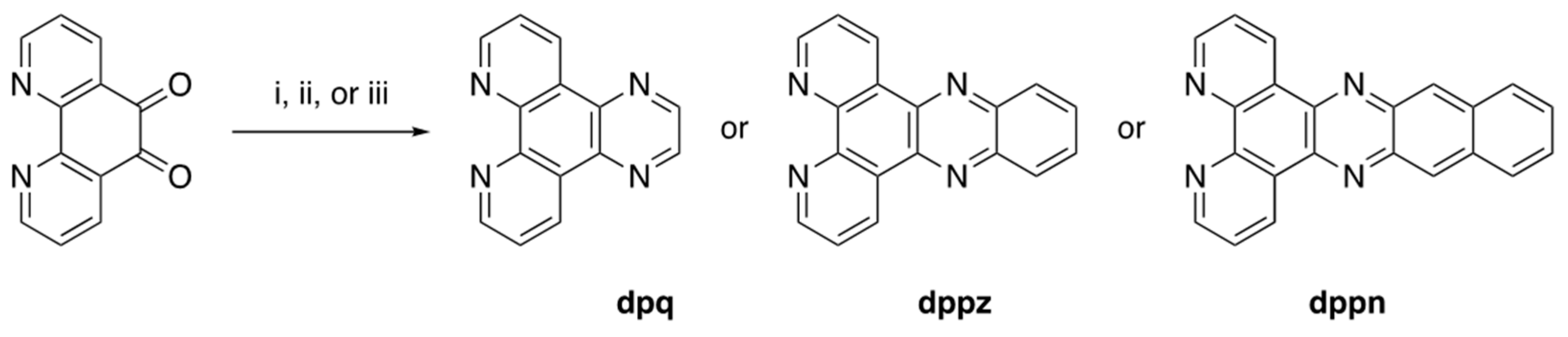
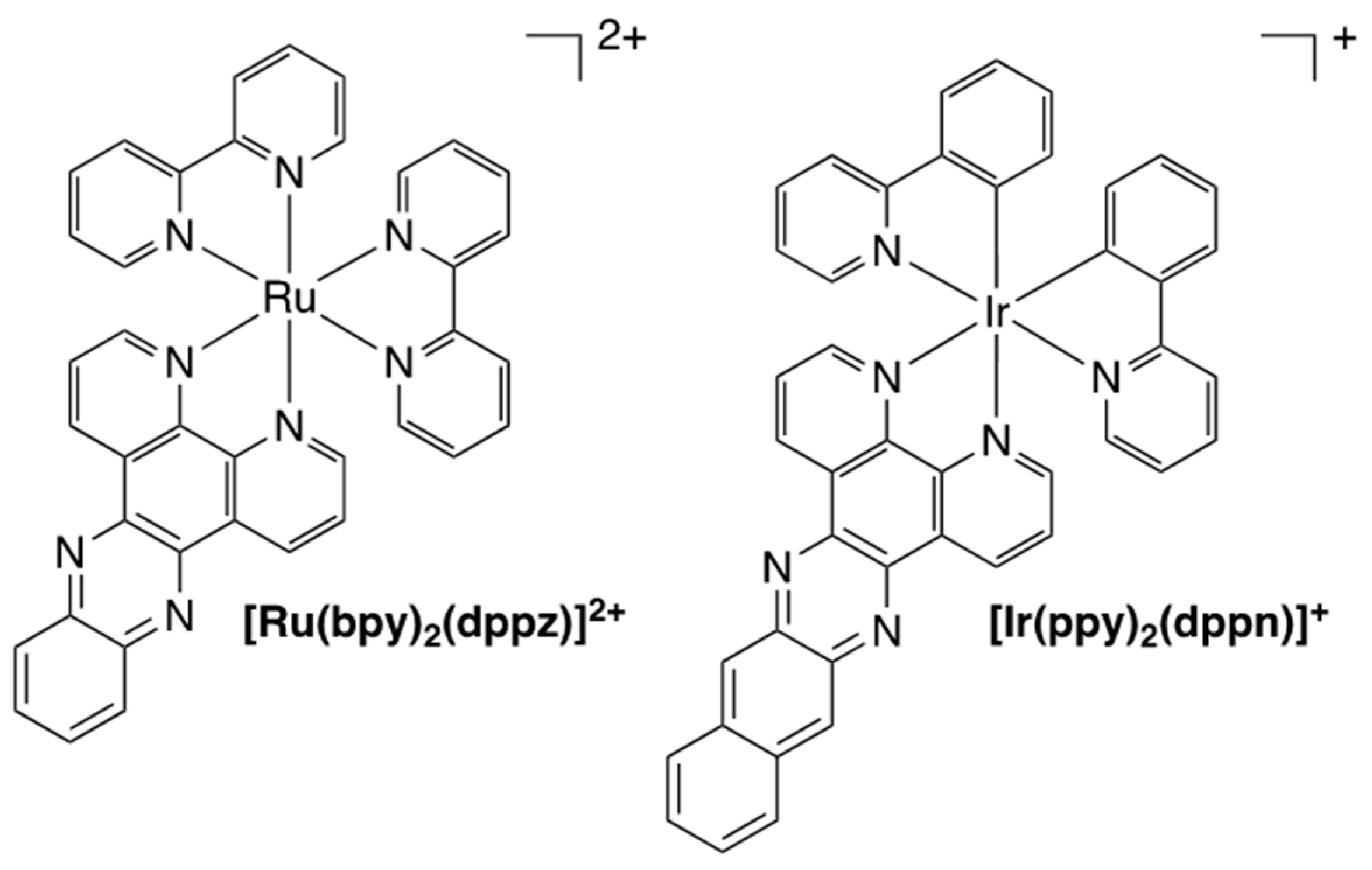
Crystal Structures of DNA Intercalating Agents Dipyrido[3,2-f:2′,3′-h]quinoxaline (dpq), (Benzo[i]dipyrido[3,2-a:2′,3′c]phenazine (dppn), and [Ir(ppy)2(dppn)][PF6] (Where Hppy = 2-Phenylpyridine)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis
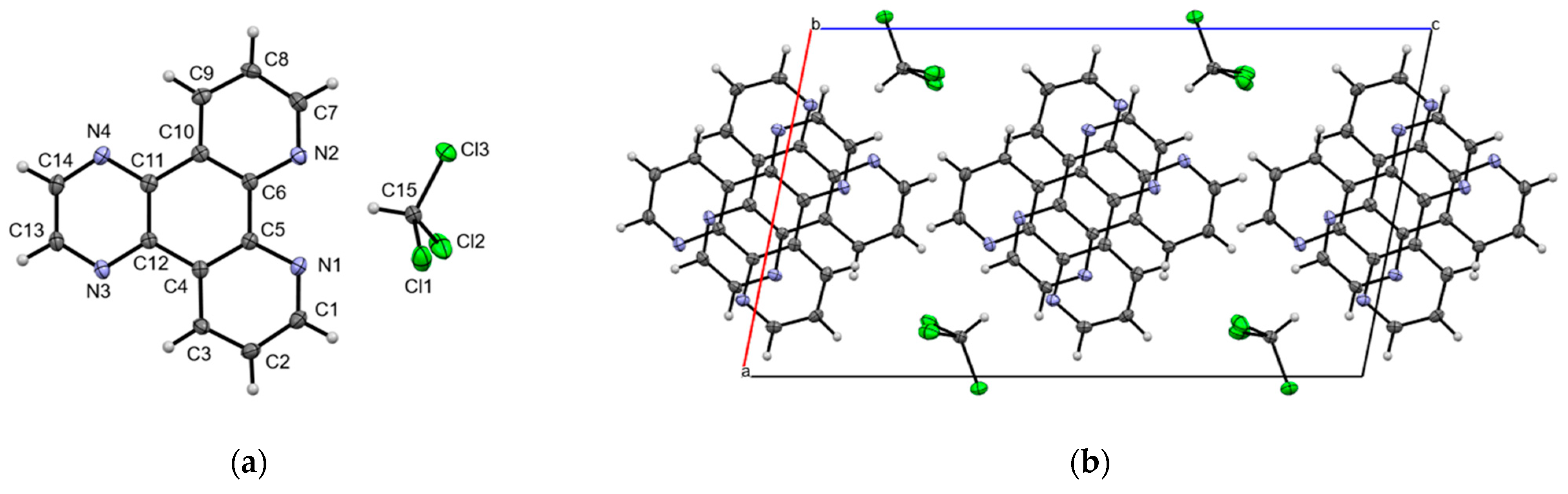
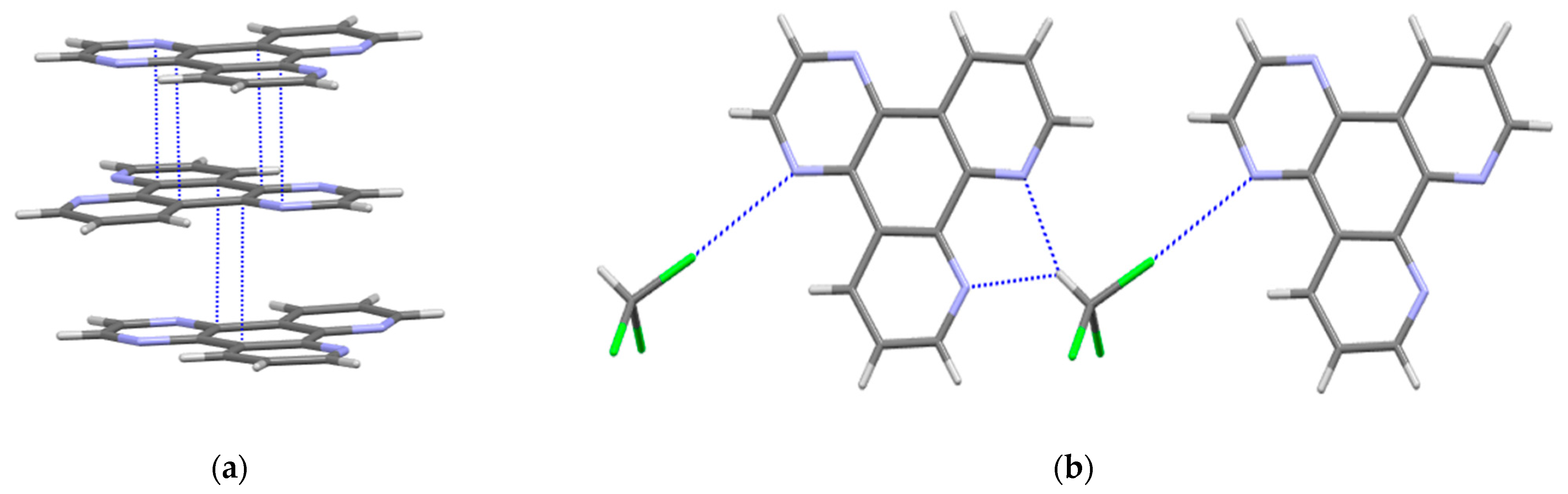
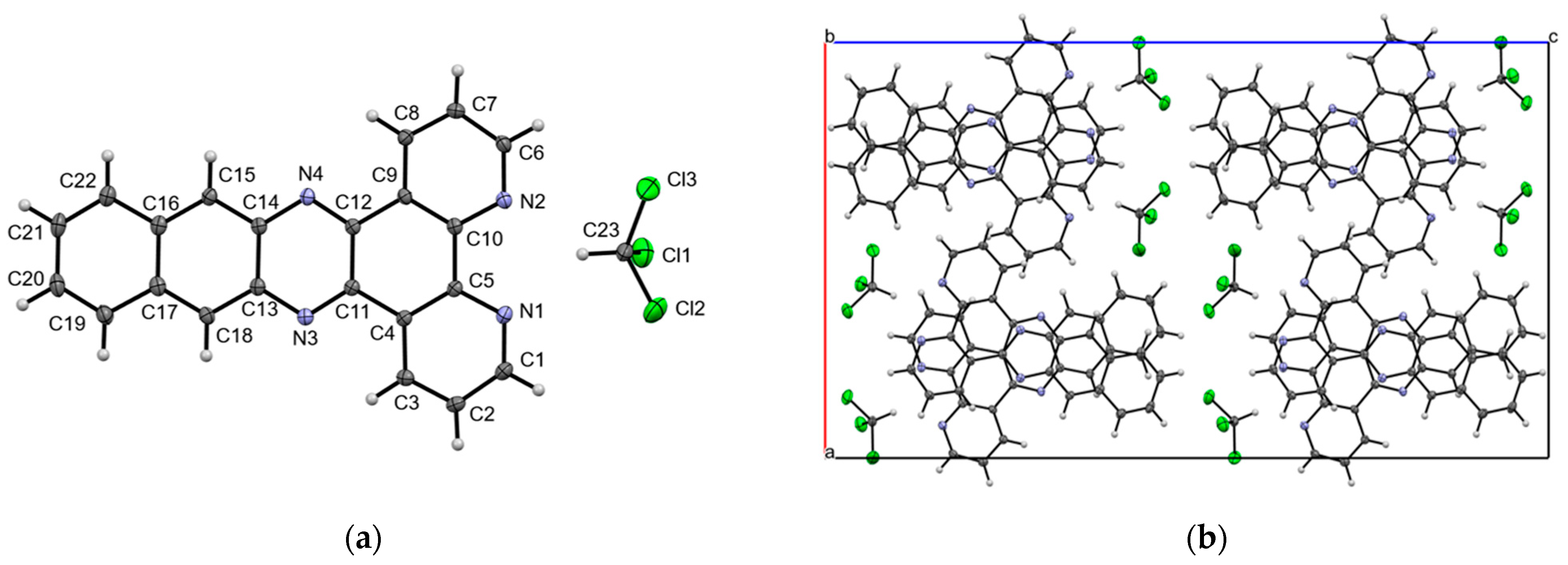
2.2. Crystal Structure of dpq·CHCl3
2.3. Crystal Structure of dppn·CHCl3
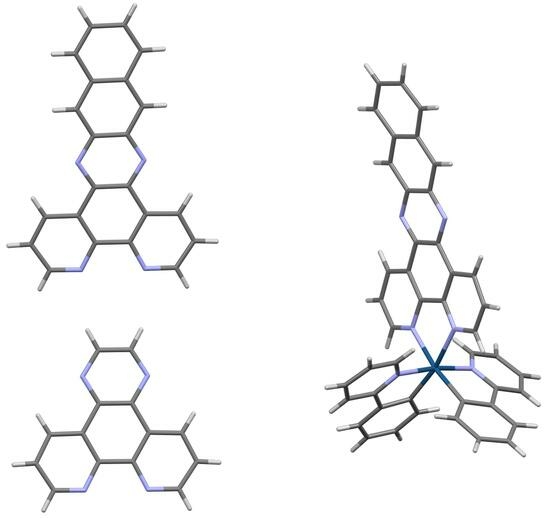
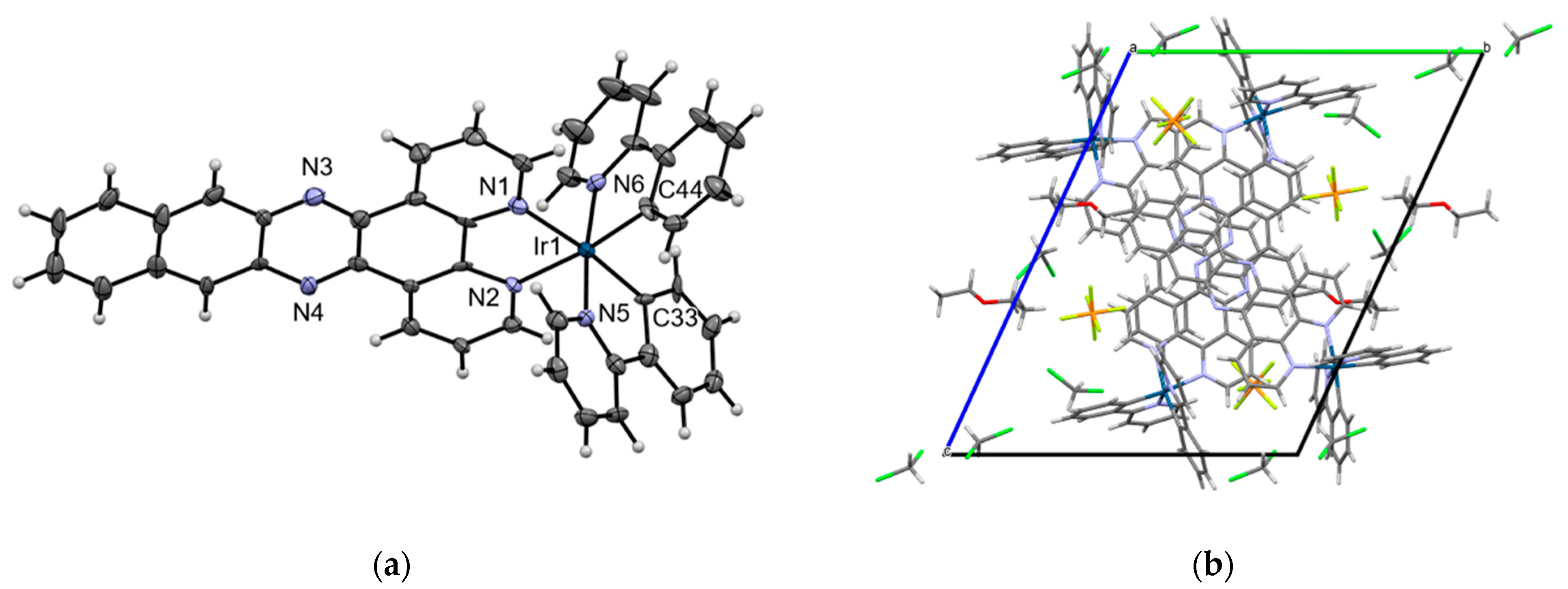
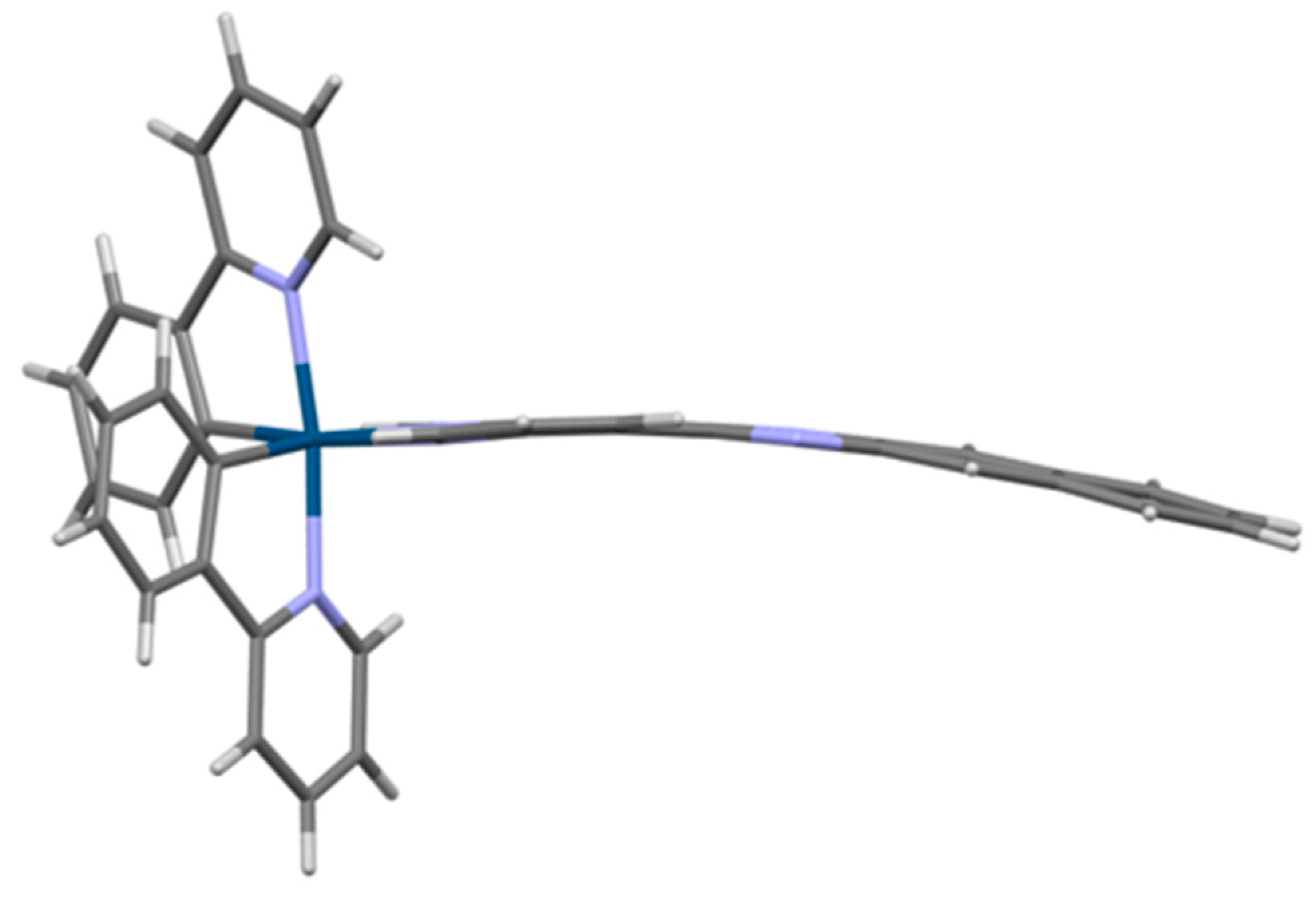
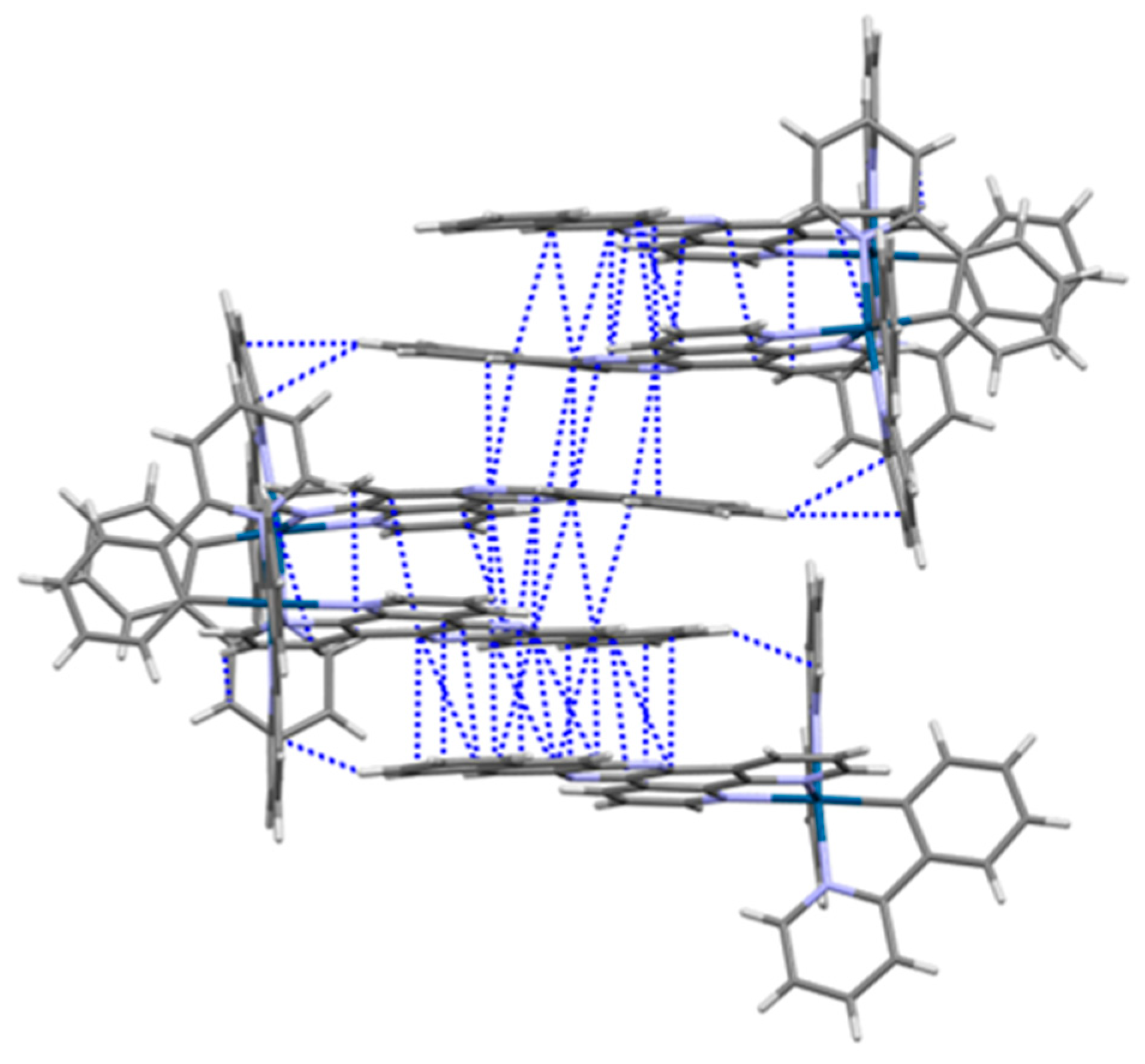
2.4. Crystal Structure of [Ir(ppy)2(dppn)][PF6] · 0.875 CH2Cl2, 0.25 C4H10O
3. Materials and Methods
3.1. Reagents and General Procedures
3.2. Synthesis of dpq (Adapted from the Literature [3])
3.3. Synthesis of dppn (Adapted from the Literature [2])
3.4. Synthesis of [Ir(ppy)2(dppn)][PF6] (Adapted from the Literature [12])
3.5. X-ray Crystallography
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Georgiades, S.N.; Vilar, R. Interaction of metal complexes with nucleic acids. Annu. Rep. Sect. A (Inorg. Chem.) 2010, 106, 481–503. [Google Scholar] [CrossRef]
- Foxon, S.P.; Green, C.; Walker, M.G.; Wragg, A.; Adams, H.; Weinstein, J.A.; Parker, S.C.; Meijer, A.J.H.M.; Thomas, J.A. Synthesis, Characterization, and DNA Binding Properties of Ruthenium(II) Complexes Containing the Redox Active Ligand Benzo[i]dipyrido[3,2-a:2′,3′-c]phenazine-11,16-quinone. Inorg. Chem. 2012, 51, 463–471. [Google Scholar] [CrossRef]
- Deshpande, M.S.; Kumbhar, A.S.; Näther, C. Stabilization of acyclic water tetramer in a copper(ii) malonate framework structure. Dalton Trans. 2010, 39, 9146–9152. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, K.; Senthil Murugan, K.; Thangamuniyandi, P.; Sakthinathan, S. Synthesis, Micellization Behaviour, DNA/RNA Binding and Biological Studies of a Surfactant Cobalt(III) Complex With Dipyrido[3,2-a:2′,4′-c](6,7,8,9-tetrahydro)phenazine. J. Fluoresc. 2014, 24, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, M.L.; La Ganga, G.; Nastasi, F.; Puntoriero, F. Ru(II)-Dppz Derivatives and Their Interactions with DNA: Thirty Years and Counting. Appl. Sci. 2021, 11, 3038. [Google Scholar] [CrossRef]
- Friedman, A.E.; Chambron, J.C.; Sauvage, J.P.; Turro, N.J.; Barton, J.K. A molecular light switch for DNA: Ru(bpy)2(dppz)2+. J. Am. Chem. Soc. 1990, 112, 4960–4962. [Google Scholar] [CrossRef]
- Greguric, I.; Aldrich-Wright, J.R.; Collins, J.G. A 1H NMR Study of the Binding of Δ-[Ru(phen)2DPQ]2+ to the Hexanucleotide d(GTCGAC)2. Evidence for Intercalation from the Minor Groove. J. Am. Chem. Soc. 1997, 119, 3621–3622. [Google Scholar] [CrossRef]
- Collins, J.G.; Sleeman, A.D.; Aldrich-Wright, J.R.; Greguric, I.; Hambley, T.W. A 1H NMR Study of the DNA Binding of Ruthenium(II) Polypyridyl Complexes. Inorg. Chem. 1998, 37, 3133–3141. [Google Scholar] [CrossRef]
- Collins, J.G.; Aldrich-Wright, J.R.; Greguric, I.D.; Pellegrini, P.A. Binding of the Δ- and Λ-Enantiomers of [Ru(dmphen)2dpq]2+ to the Hexanucleotide d(GTCGAC)2. Inorg. Chem. 1999, 38, 5502–5509. [Google Scholar] [CrossRef]
- O’Donoghue, K.A.; Kelly, J.M.; Kruger, P.E. Unusual photophysical switching in a Ru(ii) diimine DNA probe caused by amide functionalisation. Dalton Trans. 2004, 2004, 13–15. [Google Scholar] [CrossRef]
- O’Donoghue, K.; Penedo, J.C.; Kelly, J.M.; Kruger, P.E. Photophysical study of a family of [Ru(phen)2(Medpq)]2+ complexes in different solvents and DNA: A specific water effect promoted by methyl substitution. Dalton Trans. 2005, 2005, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.K.-W.; Chung, C.-K.; Zhu, N. Nucleic Acid Intercalators and Avidin Probes Derived from Luminescent Cyclometalated Iridium(III)–Dipyridoquinoxaline and –Dipyridophenazine Complexes. Chem. Eur. J. 2006, 12, 1500–1512. [Google Scholar] [CrossRef] [PubMed]
- Holmlin, R.E.; Yao, J.A.; Barton, J.K. Dipyridophenazine Complexes of Os(II) as Red-Emitting DNA Probes: Synthesis, Characterization, and Photophysical Properties. Inorg. Chem. 1999, 38, 174–189. [Google Scholar] [CrossRef]
- Stoeffler, H.D.; Thornton, N.B.; Temkin, S.L.; Schanze, K.S. Unusual Photophysics of a Rhenium(I) Dipyridophenazine Complex in Homogeneous Solution and Bound to DNA. J. Am. Chem. Soc. 1995, 117, 7119–7128. [Google Scholar] [CrossRef]
- Metcalfe, C.; Webb, M.; Thomas, J.A. A facile synthetic route to bimetallic ReI complexes containing two dppz DNA intercalating ligands. Chem. Commun. 2002, 2002, 2026–2027. [Google Scholar] [CrossRef]
- Lo, K.K.-W.; Tsang, K.H.-K. Bifunctional Luminescent Rhenium(I) Complexes Containing an Extended Planar Diimine Ligand and a Biotin Moiety. Organometallics 2004, 23, 3062–3070. [Google Scholar] [CrossRef]
- Herebian, D.; Sheldrick, W.S. Synthesis and DNA binding properties of bioorganometallic (η5-pentamethylcyclopentadienyl)iridium(iii) complexes of the type [(η5-C5Me5)Ir(Aa)(dppz)]+ (dppz = dipyrido[3,2-a:2′,3′-c]phenazine, n = 1–3), with S-coordinated amino acids (Aa) or peptides. J. Chem. Soc. Dalton Trans. 2002, 2002, 966–974. [Google Scholar] [CrossRef]
- Barker, K.D.; Benoit, B.R.; Bordelon, J.A.; Davis, R.J.; Delmas, A.S.; Mytykh, O.V.; Petty, J.T.; Wheeler, J.F.; Kane-Maguire, N.A.P. Intercalative binding and photoredox behavior of [Cr(phen)2(dppz)]3+ with B-DNA. Inorg. Chim. Acta 2001, 322, 74–78. [Google Scholar] [CrossRef]
- Arounaguiri, S.; Maiya, B.G. Dipyridophenazine Complexes of Cobalt(III) and Nickel(II): DNA-Binding and Photocleavage Studies. Inorg. Chem. 1996, 35, 4267–4270. [Google Scholar] [CrossRef] [PubMed]
- Kulmaczewski, R.; Shepherd, H.J.; Cespedes, O.; Halcrow, M.A. A Homologous Series of [Fe(H2Bpz2)2(L)] Spin-Crossover Complexes with Annelated Bipyridyl Co-Ligands. Inorg. Chem. 2014, 53, 9809–9817. [Google Scholar] [CrossRef]
- Phillips, T.; Haq, I.; Meijer, A.J.H.M.; Adams, H.; Soutar, I.; Swanson, L.; Sykes, M.J.; Thomas, J.A. DNA Binding of an Organic dppz-Based Intercalator. Biochemistry 2004, 43, 13657–13665. [Google Scholar] [CrossRef]
- Zhernakov, M.A.; Sedykh, A.E.; Becker, J.; Maxeiner, M.; Müller-Buschbaum, K.; Shtyrlin, V.G. Three ytterbium(III) complexes with aromatic N-donors: Synthesis, structure, photophysical properties and thermal stability. Z. Für Anorg. Und Allg. Chem. 2022, 648, e202200230. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, X.; Cen, P.; Yang, H.; He, Z.; Guo, Y.; Tian, D.; Liu, X. Dual-sensitized Eu(III)/Tb(III) complexes exhibiting tunable luminescence emission and their application in cellular-imaging. Dalton Trans. 2022, 51, 3180–3187. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.; Dreyse, P.; Cortes-Arriagada, D.; Sundararajan, M.; Morgado, C.; Brito, I.; Roldan-Carmona, C.; Bolink, H.J.; Loeb, B. A comparative study of Ir(III) complexes with pyrazino[2,3-f][1,10]phenanthroline and pyrazino[2,3-f][4,7]phenanthroline ligands in light-emitting electrochemical cells (LECs). Dalton Trans. 2015, 44, 14771–14781. [Google Scholar] [CrossRef]
- Sarkar, T.; Kumar, A.; Sahoo, S.; Hussain, A. Mixed-Ligand Cobalt(III) Complexes of a Naturally Occurring Coumarin and Phenanthroline Bases as Mitochondria-Targeted Dual-Purpose Photochemotherapeutics. Inorg. Chem. 2021, 60, 6649–6662. [Google Scholar] [CrossRef]
- Travis, W.; Knapp, C.E.; Savory, C.N.; Ganose, A.M.; Kafourou, P.; Song, X.; Sharif, Z.; Cockcroft, J.K.; Scanlon, D.O.; Bronstein, H.; et al. Hybrid Organic-Inorganic Coordination Complexes as Tunable Optical Response Materials. Inorg. Chem. 2016, 55, 3393–3400. [Google Scholar] [CrossRef]
- Li, Z.; Leed, N.A.; Dickson-Karn, N.M.; Dunbar, K.; Turro, C. Directional charge transfer and highly reducing and oxidizing excited states of new dirhodium(II,II) complexes: Potential applications in solar energy conversion. Chem. Sci. 2014, 5, 727–737. [Google Scholar] [CrossRef]
- Chen, G.-J.; Zhou, Y.; Jin, G.-X.; Dong, Y.-B. [Dy(acac)3(dppn)]·C2H5OH: Construction of a single-ion magnet based on the square-antiprism dysprosium(III) ion. Dalton Trans. 2014, 43, 16659–16665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yue, S.; Li, B.; Fan, D. A series of [Cu(N-N)(P-P)]BF4 complexes: Luminescence quenching caused by electron-configuration transformation in excited state. Inorg. Chim. Acta 2012, 384, 225–232. [Google Scholar] [CrossRef]
- Kulmaczewski, R.; Halcrow, M.A. Structures and spin states of crystalline [Fe(NCS)2L2] and [FeL3]2+ complexes (L = an annelated 1,10-phenanthroline derivative). CrystEngComm 2016, 18, 2570–2578. [Google Scholar] [CrossRef]
- Lin, R.-K.; Chiu, C.-I.; Hsu, C.-H.; Lai, Y.-J.; Venkatesan, P.; Huang, P.-H.; Lai, P.-S.; Lin, C.-C. Photocytotoxic Copper(II) Complexes with Schiff-Base Scaffolds for Photodynamic Therapy. Chem. Eur. J. 2018, 24, 4111–4120. [Google Scholar] [CrossRef]
- Foxon, S.P.; Metcalfe, C.; Adams, H.; Webb, M.; Thomas, J.A. Electrochemical and Photophysical Properties of DNA Metallo-intercalators Containing Ruthenium(II) Tris(1-pyrazolyl)methane Unit. Inorg. Chem. 2007, 46, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Lameijer, L.N.; Breve, T.G.; van Rixel, V.H.S.; Askes, S.H.C.; Siegler, M.A.; Bonnet, S. Effects of the Bidentate Ligand on the Photophysical Properties, Cellular Uptake, and (Photo)cytotoxicity of Glycoconjugates Based on the [Ru(tpy)(NN)(L)]2+ Scaffold. Chem. Eur. J. 2018, 24, 2709–2717. [Google Scholar] [CrossRef]
- Scharwitz, M.A.; Ott, I.; Geldmacher, Y.; Gust, R.; Sheldrick, W.S. Cytotoxic half-sandwich rhodium(III) complexes: Polypyridyl ligand influence on their DNA binding properties and cellular uptake. J. Organomet. Chem. 2008, 693, 2299–2309. [Google Scholar] [CrossRef]
- Yam, V.W.-W.; Lo, K.K.-W.; Cheung, K.-K.; Kong, R.Y.-C. Synthesis, photophysical properties and DNA binding studies of novel luminescent rhenium(I) complexes. X-ray crystal structure of [Re(dppn)(CO)3(py)](OTf). J. Chem. Soc. Chem. Commun. 1995, 1995, 1191–1193. [Google Scholar] [CrossRef]
- Hall, J.P.; O’Sullivan, K.; Naseer, A.; Smith, J.A.; Kelly, J.M.; Cardin, C.J. Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand. Proc. Natl. Acad. Sci. USA 2011, 108, 17610–17614. [Google Scholar] [CrossRef]
- Song, H.; Kaiser, J.T.; Barton, J.K. Crystal structure of Δ-[Ru(bpy)2dppz]2+ bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation. Nature Chem. 2012, 4, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Skórka, Ł.; Filapek, M.; Zur, L.; Małecki, J.G.; Pisarski, W.; Olejnik, M.; Danikiewicz, W.; Krompiec, S. Highly Phosphorescent Cyclometalated Iridium(III) Complexes for Optoelectronic Applications: Fine Tuning of the Emission Wavelength through Ancillary Ligands. J. Phys. Chem. C 2016, 120, 7284–7294. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]

| dpq·CHCl3 | dppn·CHCl3 | [Ir(ppy)2(dppn)][PF6] · 0.875 CH2Cl2, 0.25 C4H10O | |
|---|---|---|---|
| Formula | C15H9Cl3N4 | C23H13Cl3N4 | C45.88H32.25Cl1.75F6IrN6O0.25P |
| F. W. (g/mol) | 351.61 | 451.72 | 1070.73 |
| Temperature (K) | 100 (2) | 100 (2) | 100 (2) |
| Crystal System | Monoclinic | Orthorhombic | Triclinic |
| Space group | P21/c (no. 14) | Pbca (no. 61) | P-1 (no. 2) |
| a (Å) | 11.3166 (7) | 18.1564 (5) | 12.5328 (8) |
| b (Å) | 6.6154 (4) | 6.93200 (10) | 18.0553 (11) |
| c (Å) | 19.8106 (12) | 31.6298 (7) | 22.0638 (15) |
| α (°) | 90 | 90 | 112.989 (2) |
| β (°) | 101.289 (2) | 90 | 93.685 (2) |
| γ (°) | 90 | 90 | 103.869 (2) |
| Volume (Å3) | 1454.40 (15) | 3980.93 (15) | 4392.0 (5) |
| Z | 4 | 8 | 4 |
| D (calcd) (mg/m3) | 1.606 | 1.507 | 1.619 |
| μ, mm−1 | 0.630 | 0.479 | 3.249 |
| F (000) | 712 | 1840 | 2109 |
| Cryst. Size (mm) | 0.02 × 0.03 × 0.11 | 0.09 × 0.10 × 0.41 | 0.02 × 0.08 × 0.23 |
| θ range, ° | 3.05 to 25.74 | 2.58 to 28.42 | 1.81 to 25.80 |
| Reflns. collected | 39,390 | 37,847 | 85,424 |
| Indep. Reflns. | 2758 | 4971 | 16,732 |
| No. of parameters | 199 | 271 | 1198 |
| R indices (I > 2σ(I)) | R1 = 0.0412 a wR2 = 0.0879 b | R1 = 0.0378 a wR2 = 0.0895 b | R1 = 0.0719 a wR2 = 0.1849 b |
| R indices (all data) | R1 = 0.0725 a wR2 = 0.1047 b | R1 = 0.0533 a wR2 = 0.0986 b | R1 = 0.0968 a wR2 = 0.2022 b |
| S | 1.034 | 1.022 | 1.105 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
James, M.; Shevlin, M.R.; Green, T.B.; Smart, M.M.; McMillen, C.D.; Pienkos, J.A. Crystal Structures of DNA Intercalating Agents Dipyrido[3,2-f:2′,3′-h]quinoxaline (dpq), (Benzo[i]dipyrido[3,2-a:2′,3′c]phenazine (dppn), and [Ir(ppy)2(dppn)][PF6] (Where Hppy = 2-Phenylpyridine). Inorganics 2023, 11, 353. https://doi.org/10.3390/inorganics11090353
James M, Shevlin MR, Green TB, Smart MM, McMillen CD, Pienkos JA. Crystal Structures of DNA Intercalating Agents Dipyrido[3,2-f:2′,3′-h]quinoxaline (dpq), (Benzo[i]dipyrido[3,2-a:2′,3′c]phenazine (dppn), and [Ir(ppy)2(dppn)][PF6] (Where Hppy = 2-Phenylpyridine). Inorganics. 2023; 11(9):353. https://doi.org/10.3390/inorganics11090353
Chicago/Turabian StyleJames, Marisa, Madelyn R. Shevlin, Thomas B. Green, Megan M. Smart, Colin D. McMillen, and Jared A. Pienkos. 2023. "Crystal Structures of DNA Intercalating Agents Dipyrido[3,2-f:2′,3′-h]quinoxaline (dpq), (Benzo[i]dipyrido[3,2-a:2′,3′c]phenazine (dppn), and [Ir(ppy)2(dppn)][PF6] (Where Hppy = 2-Phenylpyridine)" Inorganics 11, no. 9: 353. https://doi.org/10.3390/inorganics11090353
APA StyleJames, M., Shevlin, M. R., Green, T. B., Smart, M. M., McMillen, C. D., & Pienkos, J. A. (2023). Crystal Structures of DNA Intercalating Agents Dipyrido[3,2-f:2′,3′-h]quinoxaline (dpq), (Benzo[i]dipyrido[3,2-a:2′,3′c]phenazine (dppn), and [Ir(ppy)2(dppn)][PF6] (Where Hppy = 2-Phenylpyridine). Inorganics, 11(9), 353. https://doi.org/10.3390/inorganics11090353