Abstract
Due to the constant increase in the number of plant diseases and the lack of available treatments, there has been a growing interest in plant extracts over the past few decades. Numerous studies suggest that plant extract molecules possess valuable antimicrobial activities, particularly against fungi and bacteria. This suggests that these biomaterials could potentially serve as attractive therapeutic options for the treatment of phytopathogen infections. In the present study, we investigated and analyzed the methanolic extract of Eryngium campestre L. whole plant extract using HPLC. The analysis revealed the presence of several polyphenolic constituents, with benzoic acid, catechol, quercetin, vanillic acid, resveratrol, naringenin, and quinol being the most abundant. The amounts of these constituents were determined to be 2135.53, 626.728, 579.048, 356.489, 323.41, 153.038, and 128.77 mg/kg, respectively. Furthermore, we isolated and identified different plant fungal and bacterial isolates from symptomatic potato plants, which were accessioned as Rhizoctonia solani (OQ880458), Fusarium oxysporum (OQ820156) and Fusarium solani (OQ891085), Ralstonia solanacearum (OQ878653), Dickeya solani (OQ878655), and Pectobacterium carotovorum (OQ878656). The antifungal activity of the extract was assessed using fungal growth inhibitions (FGI) at concentrations of 100, 200, and 300 µg/mL. The results showed that at the lowest concentration tested (100 µg/mL), the extract exhibited the highest effectiveness against R. solani with an FGI of 78.52%, while it was least effective against F. solani with an FGI of 61.85%. At the highest concentration tested, the extract demonstrated the highest effectiveness against R. solani and F. oxysporum, with FGIs of 88.89% and 77.04%, respectively. Additionally, the extract displayed a concentration-dependent inhibitory effect on all three bacterial pathogens. At the highest concentration tested (3000 µg/mL), the extract was able to inhibit the growth of all three bacterial pathogens, although the inhibition zone diameter varied. Among the bacterial pathogens, D. solani exhibited the highest sensitivity to the extract, as it showed the largest inhibition zone diameter at most of the extract concentrations. These findings highlight the potential of the E. campestre extract as a source of natural antimicrobial agents for controlling various plant pathogens. Consequently, it offers a safer alternative to the currently employed protective methods for plant disease management.
Keywords:
Eryngium; Ralstonia; potato; Rhizoctonia; 16SrRNA; methanol; polyphenol; HPLC; benzoic acid; ITS 1. Introduction
Solanum tuberosum L., commonly known as potato, belongs to the nightshade family and is ranked third in terms of global consumption, following wheat and rice. There has been a steady increase in potato output over the world [1]. Egypt is a significant contributor to global potato production. The country has a thriving potato industry and is known for its substantial cultivation and harvest. Potato production in Egypt plays a crucial role in the national economy and food security. The crop serves as a staple food for the population and is widely consumed across the country. Additionally, Egypt exports a significant amount of potatoes to various international markets, contributing to its economic growth and foreign exchange earnings [2]. Plant-killing microorganisms, or phytopathogens, are a major cause of decreased crop quality and yield. They do serious economic damage and threaten our ability to provide food for ourselves as a country and the world. For example, worldwide potato output losses average 17.2% [3]. Heatwaves and droughts are two examples of unfavorable climatic conditions that make crops more vulnerable to invasions by phytopathogens [1].
Several bacterial phytopathogens are known to affect potato tubers, including Pectobacterium carotovorum, which causes soft rot [4,5]; Ralstonia solanacearum, which leads to potato wilting; and Dickeya solani, which specifically affects potato plants and causes a disease known as blackleg. Blackleg is characterized by the rotting and wilting of the plant’s stem and foliage, leading to significant yield losses and quality deterioration in potato tubers. The bacterium is highly aggressive and can quickly spread within and between potato fields [6]. Fungal phytopathogens that are commonly associated with potato storage include Fusarium spp., which causes Fusarium dry rot, and Rhizoctonia solani, which causes black scurf as reported in sources [7,8].
Sound seed potato production is contingent upon preemptive measures, including the eradication of sources of infection, the cultivation of resilient cultivars, and the guarantee of appropriate seed potato preservation from the point of harvest to that of planting. The ideal storage parameters entail the preservation of a temperature range of 3–4 °C and humidity levels within the range of 90–95%. The aforementioned conditions serve the purpose of averting weight loss, alterations in chemical composition, unsuccessful germination, and, notably, microbial infestations [9], throughout a prolonged period of storage. While chemical fungicides have demonstrated efficacy against plant pathogens, their overuse has been linked to the development of pathogen resistance and potential ongoing adverse impacts on people’s health [10]. Currently, there is a global trend towards reducing the reliance on synthetic fungicides, driving the search for less hazardous and more ecologically sustainable substitutes to counteract plant diseases.
The implementation of botanical extracts, which encompass secondary metabolites originating from plant cells, is a viable environmentally conscious remedy. The chemical composition and concentration of these compounds exhibit variations contingent upon the botanical taxonomy, ecological factors, pathogenic incidence, time of harvest, and techniques of extraction. However, their biopesticidal properties generally exhibit broad-spectrum efficacy against various plant pathogens. Furthermore, they are biodegradable and have minimal environmental impact. According to sources [10,11], the utilization of plant extracts represents a viable substitute for synthetic fungicides in the management of post-harvest storage phytopathogens in potatoes. Bioactive secondary metabolic products such as flavonoids, phenolic acids, saponins, coumarins, and essential oils are thought to be responsible for the antioxidant and pharmacological actions of plants of the genus Eryngium [12]. Among the several chemicals found in the Eryngium genus, polyphenolic compounds have been singled out for special attention [13]. Due to their additional bioactivities as antiviral, antibacterial, antifungal, and anti-inflammatory agents, phenolics have recently attracted significant interest [14,15,16,17].
The pharmacological potential of Eryngium species has been demonstrated [18]. The phenolic acid R-(+)-3′-O--d-glucopyranosyl rosmarinic acid has been found by phytochemical analysis of plant species [19], flavonoids, namely, kaempferol and quercetin glycosides [20,21], triterpenoid saponins [22], and coumarin derivatives [23]. The perennial herb Eryngium campestre L., which belongs to the Apiaceae family and is employed in traditional medicine, is widely distributed over Asia, Europe, and Africa [24], In addition to its culinary and ethnomedical applications, this spice is commonly used in Turkey, Spain, Kosovo, and Italy [25,26]. An IC50 of 363 ng/mL was found for E. campestre flower extract as a potent inhibitor of glutathione-s-transferase [27]. The n-butanol extract of E. campestre aerial parts showed much stronger antiradical activity than the conventional medication diclofenac [28], with an IC50 value of 16.140 ng/mL. Roots and stems have been studied for their potential to provide flavonoids and flavonoacyl compounds [29,30,31]. The antioxidant activity of the methanol-derived extract of E. campestre and the extracted flavonols was found to be mild to significant in both DPPH radical scavenging and reducing strength experiments [31].
Germacrene D, campestrolide, germacrene B, myrcene, α-cadinol, spathulenol, eudesma-4(15)-7-dien-1-ol, and -cadinol were all found in essential oil extracted from E. campestre aerial parts, and all of them exhibited significant antibacterial efficacy against Gram-positive bacteria. Specifically, cadinane, guaiane, and oxygenated chemicals are thought to be produced by plants in the eastern Mediterranean basin (Egypt) [32,33].
In this study, the polyphenolic methanolic extract of E. campestre L. grown in Egypt was analyzed by HPLC, and its bioactivities were investigated for the first time against molecularly characterized potato fungal and bacterial pathogens, Fusarium oxysporum, Fusarium solani, Rhizoctonia solani, Ralstonia solanacearum, Dickeya solani, and Pectobacterium carotovorum.
2. Materials and Methods
2.1. Instrumentation
In our study, we utilized various instruments to conduct laboratory tests. These instruments included a dehumidifier (Carrier Global Corporation, Palm Beach Gardens, FL, USA), Agilent HPLC 1200 Series (Agilent Technologies Inc., Santa Clara, CA, USA), electric mill grinder (Bosch TrueMixx Pro, Jabalpur, India), rotary evaporator (SH-RE05L, SH Scientific Co., Ltd., Sejong-si, Republic of Korea), PCR machine (Techne Prime thermal cycler, Cole-Parmer, Vernon Hills, IL, USA), Leica DM750 microscope (Leica Microsystems GmbH, Wetzlar, Germany), Benchtop pH/mV meter (Hanna Instruments, Woonsocket, RI, USA), laboratory incubator (Binder GmbH, Tuttlingen, Germany), analytical balance scale model BXX 30 (BOECO GmbH + Co, Hamburg, Germany), vertical loading autoclave sterilizer LAC-S (LabTech S.R.L., Seoul, Republic of Korea), and T80 series UV-Vis spectrophotometer (PG Instruments Limited, Leicester, UK).
2.2. Chemicals
In the study, various chemicals were used to facilitate the experimental procedures. Sodium chloride, methanol (99% purity), ethanol (99% purity), cetyltrimethylammonium bromide (CTAB), isopropanol alcohol, isoamyl alcohol, dimethyl sulfoxide (DMSO), formic acid, acetonitrile, and chloroform were sourced in analytical grade from Fisher Scientific, located in Loughborough, UK. Kits and a standard mixture of phenolic and flavonoid compounds, including pyrogallol, quinol, gallic acid, catechol, p-hydroxy benzoic acid, catechin, chlorogenic acid, vanillic acid, caffeic acid, syringic acid, p-coumaric acid, benzoic acid, ferulic acid, rutin, ellagic acid, o-coumaric acid, resveratrol, cinnamic acid, quercetin, rosmarinic acid, naringenin, and myricetin, were sourced from the Merck Group in Darmstadt, Germany. All PCR reagents were obtained from iNtRON Biotechnology DR in Gyeonggi-do, Republic of Korea, and all microbiological media were purchased from HiMedia Laboratories Private Limited in Mumbai, Maharashtra, India.
2.3. Plant Material and Extractions Setting Up
Eryngium campestre L. plants were collected at coordinates 30.848269 (latitude) and 29.573051 (longitude) in Borg El-Arab City, Alexandria, Egypt, in dry and sandy areas including the desert region in the spring during the onset of their blooming phase. The plant samples were morphologically identified by the Floriculture, Ornamental Horticulture, and Garden Design Department, Faculty of Agriculture (El-Shatby), Alexandria University, Alexandria, Egypt. A voucher specimen (03-42-ALX) has been deposited at the Herbarium of the Department of Agricultural Botany, Faculty of Agriculture Saba Basha, Alexandria University, Alexandria, Egypt. All plant parts of the samples were subjected to 48 h of exposure to air (air drying was performed in a room supplied with a dehumidifier, 10% relative humidity at room temperature) before being pulverized into a fine powder using an electric mill grinder (Bosch TrueMixx Pro, Jabalpur, India). The powder was then refined and sieved using a 200-mesh stainless steel sieve (FILTRA VIBRACIÓN, Barcelona, Spain). Subsequently, a quantity of 100 g of the powdered substance was mixed with 1 L of methanol (99%). The extract was subjected to filtration using filter paper (Whatman no.1), and the solvent was evaporated and the extract concentrated under vacuum using a rotary evaporator (SH-RE05L, SH Scientific Co., Ltd., Sejong-si, Republic of Korea) at temperature 40 °C. The yielded extract was calculated in percentage using the formula:
Yield of extract (%) = (Weight of the dried extracted material obtained after the extraction process and solvent removal/Weight of dried plant material in powdered form before the extraction) × 100. The extract was then stored in a refrigerator at 4 °C until its intended use [34].
2.4. Phytopathogenic Fungi
2.4.1. Isolation Trails
Samples of potato tubers affected by stem-end rot, dry rot, and black scurf diseases were collected from commercial potato-cultivated fields. The infected tubers showed circular lesions with sharply defined margins, slightly sunken and covered with white fungal growth on the stem-end portions of the potatoes. Additionally, these infected tubers displayed internal rot with light to dark brown or black discoloration, which was typically dry. To transfer the infected tubers to the lab for further examination, they were first packed in sterile disposable polythene bags. The contaminated tubers were chopped into pieces between 1 and 1.5 cm in size and then sterilized with 0.1% sodium hypochlorite for 60 s. The parts were disassembled, washed three times in double-distilled water, and dried. After cutting potatoes into small pieces, we cultured them in potato dextrose agar medium in Petri dishes for 5 days at 26 ± 2 °C. Mycelial growth from the potato dextrose agar plates was utilized to inoculate new potato dextrose agar plates to generate a pure culture [35].
2.4.2. DNA Isolation, PCR Amplification, Sequence Analysis, and Phylogenetic Tree
Fungal DNA extraction is a necessary first step in any kind of molecular study. For DNA extraction, the CTAB protocol [36] with some modifications was applied. Mycelium was cultured from fungi on potato dextrose agar (PDA, potato dextrose agar) for 3–5 days at 28 °C. The newly grown mycelium weighed between 50 and 100 mg and was transferred into a 2 mL microcentrifuge tube containing 500 µL of CTAB buffer. Following incubation at a temperature of 65 °C for 60 min with periodic agitation, the tube underwent thorough vortexing. After the incubation period, a solution of 500 µL of chloroform-isoamyl alcohol (24:1) was introduced, and the resultant blend was subjected to centrifugation at 10,000 rpm for 10 min, with concurrent vortexing for 1 min. Following the transfer of the aqueous phase to a distinct tube, a quantity of 500 µL of isopropanol was introduced to facilitate the precipitation of DNA. Following a centrifugation process lasting 10 min at a rate of 10,000 rpm, the supernatant was subsequently discarded. After the isolation process, the DNA pellet underwent a washing procedure utilizing a 70% ethanol solution, followed by desiccation under ambient conditions. The ultimate stages encompassed the dissolution of the DNA pellet in 50 µL of TE buffer, followed by its preservation at a temperature of −20 °C.
The molecular identification of fungal isolates was carried out using the polymerase chain reaction (PCR) technique [37]. The primer sequences were devised per the fungi identification norms, specifically targeting the internal transcribed spacer (ITS) region [38]. The PCR reaction mixture was composed of 12.5 µL of 2x PCR master mix, 1 µL of each primer (10 M), 2 µL of template DNA, and 8.5 µL of nuclease-free water. PCR amplification was conducted in a thermal cycler using the following parameters: an initial denaturation step at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 1 min. The final step involved elongation at 72 °C for 10 min. The PCR products underwent purification through a PCR purification kit and were subjected to sequencing via the Sanger sequencing method. The sequences that were acquired were subjected to comparison with the sequences present in the GenBank database through utilization of the BLAST algorithm, to identify the fungal species. The Clustal Omega program was utilized to align the sequences, and subsequently, a phylogenetic tree was constructed through the employment of the maximum-likelihood method. The MEGA 5 software [39] was utilized to conduct the phylogenetic analysis. The analysis of genetic sequences yielded significant insights into the diversity of the fungi and facilitated the identification of distinct fungal species.
2.4.3. Antifungal Activity of the Extract
The poising assay was applied to determine the antifungal activity of the extract as follows: The fungal culture medium was prepared by dissolving PDA in distilled water and sterilizing it by autoclaving. The fungal culture discs obtained from fresh isolates (0.3 cm) added to the fungal culture medium in the middle of the plates were amended with different extract concentrations to achieve the final desired concentrations (100, 200, and 300 µg/mL) by diluting the stock solution in the extract medium using a sterile pipette. The plates were incubated at a suitable temperature of 25 ± 2 °C for a week, or until the fungal mycelium (negative control) grows to fill the plate. The antifungal activity of the extract can be determined by measuring the diameter of the fungal mycelium in the presence and absence of the extract. The inhibition percentage can be calculated using the following formula: Inhibition (%) = [(P − K)/P] × 100, where P is the diameter of the fungal hyphae in the control group and K is the diameter of the fungal hyphae in the treatment group. The poising assay was repeated three times to obtain reliable results.
2.5. Phytopathogenic Bacteria
2.5.1. Isolation Methods
The isolation of potato-infected bacteria can be achieved using different methods. The dilution plating method is commonly used for the isolation of Ralstonia from soil samples. Potato samples are collected from potato-growing areas and cut into halves, and the infected parts are taken and serially diluted in sterile water. The diluted samples are then plated on selective media Kelman’s Tetrazolium Chloride (KTC) agar [40]. The plates are then incubated at 28–30 °C for 3–4 days, and the typical colonies of Ralstonia are picked and further characterized. The enrichment culture method is another commonly used method for the isolation of Dickeya and Pectobacteria species. Plant tissues showing typical symptoms of soft rot disease are collected and added to a selective enrichment Modified CVP (MCVP) broth. The broth is then incubated at 28–30 °C for 24–48 h. After incubation, the broth is streaked onto selective agar plates CVP agar, and the colonies are further characterized.
2.5.2. DNA Extraction, Performed PCR, Sequence Analysis, and Phylogenetic Tree
The GenElute™ Bacterial Genomic Kit (St. Louis, MI, USA) was used to extract bacterial DNA from the isolated bacterial isolates, following the manufacturer. The extracted DNA was quantified using a spectrophotometer and stored at −20 °C until it was used. PCR amplification was performed using primers designed to target bacterial DNA. The primers were based on conserved regions of the bacterial genome, with primers targeting the 16S rRNA gene being commonly employed for amplifying bacterial DNA [41]. The amplified PCR products were visualized through agarose gel electrophoresis. Subsequently, the purified PCR products underwent Sanger sequencing, which was carried out by a commercial sequencing facility. The resulting sequencing data were saved as FASTA files. The obtained sequencing data were subjected to analysis using various software packages, such as MEGA. The sequences were aligned with each other and known reference sequences, and identified using a suitable alignment algorithm.
2.5.3. Antibacterial Activity of the Extract
The method of antibacterial activity of the extract can be described as follows. The bacterial strains to be tested for antibacterial activity were grown on nutrient agar plates at 30 °C for 24 h. A single colony of each bacterial strain was then inoculated into the nutrient broth and grown overnight at 30 °C with shaking. The extract’s antibacterial efficacy was evaluated through the use of the disc diffusion technique. Discs of sterile filter paper measuring 5 mm in diameter were saturated with an extract solution at varying concentrations of 100, 200, 300, 400, 600, 800, 1000, 2000, and 3000 µg/mL. These discs were then positioned on the surface of agar plates that had been previously inoculated with bacterial strains. Subsequently, the plates were subjected to incubation at a temperature of 30 °C for 24 h. The diameters of the zones of inhibition surrounding the discs were measured and recorded. The extract’s antibacterial efficacy was quantified by the average measurement of the inhibition zones’ diameter. The outcomes were juxtaposed with a conventional antibiotic, specifically Augmentin 25 µg/disc, and subsequently interpreted.
2.6. HPLC Conditions
In this regard, an Agilent HPLC 1200 Series (Agilent Technologies Deutschland GmbH, Boblingen, Germany) was used to analyze polyphenolic compounds found in E. campestre L. extract, and the specifications, conditions, and operation program were illustrated as follows:
- HPLC column: C18 column (150 mm × 4.6 mm, 5 μm particle size)
- Mobile phase composition:
- Solvent A: Water with 0.1% formic acid
- Solvent B: Acetonitrile
- Gradient elution:
- Initial conditions: 95% A/5% B
- Time (min)/% A/% B:
- 0/95/5
- 10/85/15
- 15/75/25
- 20/50/50
- 25/40/60
- 30/20/80
- 35/5/95
- 40/5/95 (hold for 5 min)
- 45/95/5 (re-equilibration, hold for 5 min)
- Flow rate: 1.0 mL/min
- Detection wavelength: UV multi-wave detector
- Injection volume: 10 μL
- Column temperature: 25 °C
- Retention time calibration: Analyze a standard mixture of phenolic compounds including (pyrogallol, quinol, gallic acid, catechol, p-hydroxy benzoic acid, catechin, chlorogenic acid, vanillic acid, caffeic acid, syringic acid, p-coumaric acid, benzoic acid, ferulic acid, rutin, ellagic acid, o-coumaric acid, resveratrol, cinnamic acid, quercetin, rosmarinic acid, naringenin, and myricetin) to determine the retention times of the target compounds.
2.7. Statistical Analysis
The statistical analysis of the antifungal and antibacterial activity of the extract concentrations was conducted through a one-way ANOVA. The statistical analysis involved the application of the Least Significant Difference (LSD) test at a significance level of 0.05 to compare the means.
3. Results
3.1. Identification of the Fungal Isolates
The cultural and morphological analysis revealed distinct characteristics of Rhizoctonia solani, which was obtained from potato plants afflicted with the disease. The hyphae of the isolated strain exhibited slight melanization and had irregular shapes, accompanied by a brownish coloration of the sclerotia. The microscopic examination further revealed that the hyphae branched at a 90° angle, with hyphal constriction and septa formation occurring shortly after the branching point. Additionally, no clamp connections, conidia, or rhizomorphs were observed. The Fusarium solani strain isolated from infected potato roots displayed a variable range of white creamy mycelium, ranging from sparse to abundant. Macroconidia of the strain exhibited a sickle-shaped morphology with a slightly blunted apical end, typically having an average of 3 to 4 septa. In contrast, microconidia were plentiful, characterized by an oval to kidney-shaped appearance, and were formed in false heads on elongated monophialides.
PCR amplification was conducted on the extracted DNA of the isolates to authenticate their morphological characterization. The findings indicate that the amplification of PCR products within the size range of 600 to 650 bp was accomplished. The ITS region of the isolates was subjected to PCR amplification, followed by sequencing of the resulting nucleotide sequences. Subsequently, a BLAST search was performed to analyze the obtained sequences. The results of molecular identification validated the morphological identification of the examined isolates. Upon conducting a comparative analysis of the ITS region of the F. solani isolate (OQ891085) with sequences previously archived in the GenBank database, it was observed that the F. solani isolate from China (AB369507) exhibited the highest degree of genetic similarity (100%), as depicted in Figure 1. Furthermore, it was observed that the F. solani isolate exhibited intimate phylogenetic affinities with F. oxysporum (OQ820156) and other isolates (OQ551199 and MT957565).

Figure 1.
A dendrogram tree generated using the maximum likelihood method showing the genetic relationship between the Fusarium solani and Fusarium oxysporum isolates (indicated as raf-era12 and F15) and other Fusaria isolates available in GenBank (NCBI).
By comparing the obtained sequence of R. solani with other isolates stored in GenBank, it was found that the highest genetic similarity was 99.84% with R. solani isolates previously identified in China (accession numbers MK084688 and MH172659). On the other hand, the lowest nucleotide sequence similarity of 99.69% was observed between this R. solani isolate and isolates identified also in China (accession numbers MW397194, and KJ170349), and Turkey (accession number KC590605). Furthermore, the R. solani isolated in this study exhibited genetic differences when compared to other R. solani isolates previously identified and deposited in the NCBI database (Figure 2). The nucleotide sequence analysis conducted using BLAST revealed that the R. solani isolates examined in this study exhibited genetic dissimilarity from other isolates and were not previously documented in the GenBank database. Consequently, this R. solani isolate was assigned the accession number OQ880458 in GenBank. The identification of this new R. solani isolate suggests that it may possess heightened virulence and pose a greater threat to economically important crops.

Figure 2.
A dendrogram tree generated using the maximum likelihood method showing the genetic relatedness among the Rhizoctonia solani isolate (raf-2), with those of other R. solani isolates available in National Center for Biotechnology Information (NCBI) GenBank.
3.2. Antifungal Properties
The data in Table 1 shows the effect of Eryngium campestre whole plant extract on the growth inhibition of three fungal pathogens, namely Rhizoctonia solani, Fusarium oxysporum, and Fusarium solani. The growth inhibition was measured as a percentage, and different concentrations of the extract were used (100, 200, and 300 µg/mL). A negative control (NC) was also included in the experiment. The Eryngium campestre whole plant extract inhibited the growth of all three fungal pathogens in a dose-dependent manner. As the concentration of the extract increased, the growth inhibition percentage also increased for all three fungi. At the lowest concentration tested (100 µg/mL), the extract was most effective against R. solani, with a growth inhibition percentage of 78.52%. The extract was least effective against F. solani at this concentration, with a growth inhibition percentage of 61.85%. At the highest concentration tested (300 µg/mL), the extract was most effective against R. solani and F. oxysporum, with growth inhibition percentages of 88.89% and 77.04%, respectively. The extract was least effective against F. solani at this concentration, with a growth inhibition percentage of 68.52%. The statistical analysis showed that the effect of the extract on all three fungal pathogens was significant (p ≤ 0.05), with p-values of 0.0000 for all three fungi. This indicates that the observed growth inhibition was unlikely to be due to chance. The effect is dose-dependent, with higher concentrations of the extract resulting in greater growth inhibition. The results also suggest that the extract may be more effective against R. solani and F. oxysporum than against F. solani.

Table 1.
Effect of Eryngium campestre whole plant extract on three fungal pathogens.
3.3. Bacterial Strain Identification
The isolation trials conducted on potato tubers resulted in the identification of certain bacterial strains. The 16SrRNA gene of these strains was amplified and sequenced. The analysis of the amplified amplicons confirmed that all the isolated bacterial strains belonged to plant pathogenic bacteria species. Specifically, the identified strains were R. solanacearum, D. solani, and P. carotovorum. These bacterial isolates were assigned the accession numbers OQ878653, OQ878655, and OQ878656, respectively, for future reference and study.
3.4. Antibacterial Properties
The data in Table 2 shows the inhibitory effect of E. campestre whole plant extract on three bacterial pathogens, R. solanacerum, D. solani, and P. carotovorum, at different concentrations ranging from 100 to 3000 µg/mL. The inhibition zone diameter was measured in millimeters (mm), and the results were compared with positive and negative controls. The extract showed a concentration-dependent inhibitory effect on all three bacterial pathogens, as the inhibition zone diameter increased with increasing concentration. At the highest concentration tested (3000 µg/mL), the extract was able to inhibit the growth of all three bacterial pathogens, although the inhibition zone diameter varied. The positive control (Augmentin 25 µg/disc) showed the highest inhibitory effect on all three bacterial pathogens, with the largest inhibition zone diameter compared to all extract concentrations. The negative control showed no inhibitory effect on any of the bacterial pathogens. Among the three bacterial pathogens, D. solani showed the highest sensitivity to the extract, as it exhibited the largest inhibition zone diameter at most of the extract concentrations. R. solanacerum showed the lowest sensitivity to the extract, as it exhibited the smallest inhibition zone diameter at most of the extract concentrations. Overall, the data suggest that E. campestre whole plant extract has the potential to inhibit the growth of R. solanacerum, D. solani, and P. carotovorum, and may have a future application as a natural antimicrobial agent. However, additional research is required to elucidate the mode of operation and potential adverse effects of the extract, as well as its efficacy under different conditions.

Table 2.
Effect of Eryngium campestre whole plant extract on three bacterial pathogens.
3.5. HPLC of the Yielded Extract
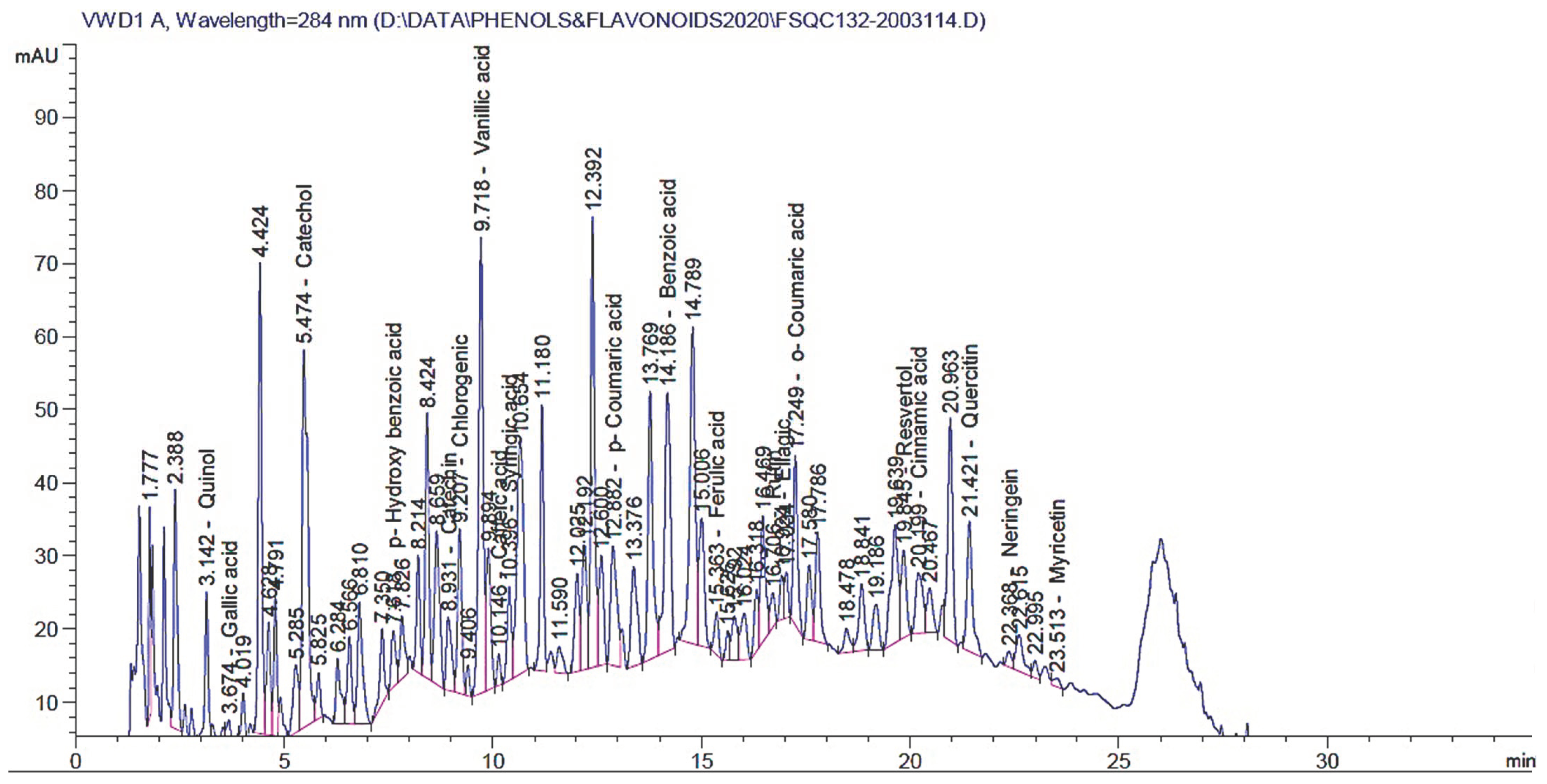
The yielded extract percentage was 6.39% for each 100 g of started powdered material. Table 3 and Figure 3 present the results of the HPLC analysis of the E. campestre extract. The table provides information on the retention time, the amount in mg/kg, and the identified compounds. The identified compounds in the extract include various types of phenolic acids, flavonoids, and other related compounds. The phenolic acids identified include gallic acid, p-hydroxybenzoic acid, chlorogenic acid, vanillic acid, caffeic acid, syringic acid, p-coumaric acid, ferulic acid, o-coumaric acid, and cinnamic acid. Flavonoids identified include rutin, naringenin, quercetin, myricetin, and kaempferol. Other identified compounds include pyrogallol, quinol, catechol, benzoic acid, and ellagic acid. The amounts of the identified compounds in the extract varied widely, with some compounds present in large quantities (e.g., benzoic acid and catechol) and others in smaller quantities (e.g., caffeic acid and kaempferol). The most abundant constituents were benzoic acid (2135.53 mg/kg), catechol (626.728 mg/kg), quercetin (579.048 mg/kg), vanillic acid (356.489 mg/kg), resveratrol (323.41 mg/kg), naringenin (153.038 mg/kg), and quinol (128.77 mg/kg).

Table 3.
HPLC analysis of Eryngium campestre extract.
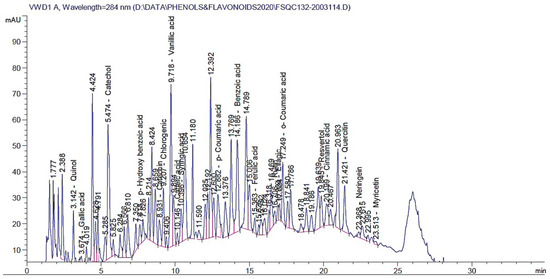
Figure 3.
HPLC chromatograms of identified polyphenolic compounds in the E. campestre methanolic extract.
4. Discussion
Our research assessed the antimicrobial efficacy of E. campestre extract as a potential biopesticide for protecting potatoes against plant pathogens in order to choose an extract based on its antimicrobial activity and economic feasibility. Furthermore, the chemical constituents of the extracts that exhibited the highest efficacy in suppressing the proliferation of plant pathogens were determined. These morphological findings of fusaria isolates align with previous studies conducted by Moni et al. [42] and Desvani et al. [43] who also reported similar characteristics of R. solani. The morphological characteristics of Fusaria species were the same as indicated by Ke et al. [44]. In this study, polymerase chain reaction (PCR) technology was employed for the diagnosis of F. solani and R. solani isolates. PCR is widely acknowledged for its high precision in identifying various organisms, including both pathogenic and non-pathogenic fungi such as F. solani, R. solani, Alternaria alternata, and Aspergillus spp. [45,46]. The nuclear ribosomal internal transcribed spacer (ITS) region serves as the official fungal barcode and is the most commonly analyzed genetic marker in the field of mycology. While the average length of the ITS region is approximately 550 base pairs (bp) across the fungal kingdom, it can significantly vary between different lineages [47]. This region consists of two variable spacers, known as ITS1 and ITS2, along with the highly conserved 5.8S ribosomal gene located in between. The 5.8S gene acts as a crucial connecting point for chimeric extension in mixed-template PCR, necessitating the inclusion of chimera control as a vital component in ITS-based mycological research [48].
The presence of nine distinct phenolic acids, namely chlorogenic, ferulic, 3,4-dihydroxyphenylacetic, caffeic (which was notably absent in roots), protocatechuic, rosmarinic, syringic, 4-feruloylquinic, and 4-dihydroxybenzoic acids, was detected in E. campestre. Furthermore, a total of five flavonoids were detected, namely kaempferol, quercitrin (absent in shoots), rutoside (undetected in roots), quercetin, and astragalin [49]. Azizkhani and Sodanlo [50] reported that the methanol extract of E. campestre exhibited a total phenolic compound content of 120.5 ± 4.2 mg GAE/g of dry extract. In a study conducted by Güneş et al. [27], it was found that the flowers and leaves of E. campestre contained a total phenolic compound content of 116.69 mg and 109.62 mg GAE/mL of plant extract, respectively. In addition, it was found that the aboveground components of E. campestre produced monoglycosides of cis- and trans-p-coumaroyl-kaempferol. The quantification of total phenolic content (TPC) derived from the flowers and leaves of E. campestre yielded values of 116.69 mg and 109.62 mg GAE/mL of plant extract, respectively. Monoglycosides of cis- and trans-p-coumaroyl-kaempferol were extracted from the aerial components of E. campestre, according to a previous study [20]. According to a study by Paun et al. [51], the polyphenolic-rich extract of Eryngium planum contained several compounds including chlorogenic acid, p-coumaric acid, rutin, quercetin 3-β-D-glucoside, ferulic acid, rosmarinic acid, daidzein, luteolin, quercetol, genistein, kaempferol, and isorhamnetin. Additionally, another study reported the isolation of three phenolic acids, namely rosmarinic acid, chlorogenic acid, and caffeic acid, from the tissue culture of Eryngium planum [52].
Thiem et al. [53] conducted thin-layer chromatography (TLC) analysis on multiple shoots and roots obtained from in vitro-cultivated Eryngium planum. The results revealed that these plant parts produced significant quantities of phenolic acids, with a particular emphasis on rosmarinic acid. In a separate study by Zhang et al. [22], several compounds were isolated from the whole plant of Eryngium yuccifolium. These compounds included eryngiosides, kaempferol-3-O-(2,6-di-O-trans-p-coumaroyl)-β-d-glucopyranoside, caffeic acid, 21β-angeloyloxy-3β-[β-d-glucopyranosyl-(1→2)]-[β-d-xylopyranosyl-(1→3)]-β-d-glucuronopyranosyloxyolean-12-ene-15α,16α,22α,28-tetrol, and saniculasaponin III. In E. maritimum, ten phenolic acids were identified, including chlorogenic acid, ferulic acid, 3,4-dihydroxyphenylacetic acid, caffeic acid (not found in roots), protocatechuic acid, rosmarinic acid, syringic acid, vanillic acid, 4-feruloylquinic acid, and 4-dihydroxybenzoic acid. Additionally, three flavonoids were present: kaempferol, quercitrin, and rutoside. Similarly, in Eryngium planum, ten phenolic acids were found: chlorogenic acid, isochlorogenic acid, ferulic acid, 3,4-dihydroxyphenylacetic acid, caffeic acid, protocatechuic acid, rosmarinic acid, syringic acid, vanillic acid (not detected in roots), and 4-feruloylquinic acid. Additionally, three flavonoids were identified: kaempferol, quercitrin, and astragalin. It is worth noting that the detection and quantification of most of these acids, except for chlorogenic acid, caffeic acid, rosmarinic acid, and ferulic acid, were reported for the first time in the raw materials of these species [49]. Moreover, ethanolic extracts obtained from E. campestre leaves and roots exhibited moderate antibacterial activity against Staphylococcus aureus, weak activity against Bacillus subtilis, and good antifungal activity against Candida albicans, Candida glabrata, Trichophyton mentagrophytes, and Cryptococcus neoformans [30].
When conducting studies on the antimicrobial activity of plant extracts, several limitations should be considered. These limitations can impact the interpretation and generalizability of the findings. For example, the composition and potency of bioactive compounds in plant extracts can vary depending on factors such as plant species, geographical location, harvesting season, and extraction methods. This variability can affect the consistency and reproducibility of antimicrobial activity results. Another limitation is the lack of standardized extraction methods and quality control measures for plant extracts. This can introduce variability in the concentration and composition of bioactive compounds, making it challenging to compare results across studies or establish a direct correlation between specific compound concentrations and antimicrobial activity. Furthermore, antimicrobial activity studies often focus on a limited number of microbial strains or a specific group of pathogens. As a result, the results may not fully represent the broad spectrum of antimicrobial activity or the effectiveness of plant extracts against emerging or resistant strains. Additionally, the lack of standardized protocols for evaluating antimicrobial activity can lead to variations in testing methods, making it challenging to compare results across studies. Establishing standardized testing procedures would enhance the consistency and reliability of research outcomes. It is important for researchers to acknowledge these limitations and address them in future studies to ensure accurate interpretation and meaningful application of the findings. By considering these limitations and working towards standardization, researchers can improve the robustness and reliability of antimicrobial activity studies in the context of plant extracts.
Overall, the practical value and study implications of research on the antimicrobial activity of plant extracts span a wide range of applications, from agriculture to medicine and food preservation. Continued research in this field holds promise for developing sustainable and effective solutions to combat microbial infections and promote a healthier and more environmentally conscious approach to antimicrobial use.
5. Conclusions
In conclusion, the study investigated the efficacy of the methanolic extract of Eryngium campestre whole plant against phytopathogenic microorganisms. Different plant fungal and bacterial isolates, including Rhizoctonia solani, Fusarium oxysporum, F. solani, Ralstonia solanacearum, Dickeya solani, and Pectobacterium carotovorum, were identified and tested. The extract exhibited inhibitory effects on the growth of these pathogens, with the extent of inhibition varying based on concentration. Among the bacterial pathogens, Dickeya solani was the most sensitive to the extract. The extract was found to contain various polyphenolic compounds, including benzoic acid, catechol, quercetin, vanillic acid, resveratrol, naringenin, and quinol, which may contribute to its antimicrobial properties. These findings suggest that the E. campestre extract has the potential to serve as a natural antimicrobial agent for controlling plant pathogens, providing a safer alternative to current methods of plant disease management. Further research and development of this extract could lead to its practical application in agriculture to improve plant health and reduce the reliance on synthetic chemicals for disease control.
Author Contributions
Conceptualization, methodology, software, and investigation, S.B., A.E.M., O.A.S., A.A., A.A.A.-A., R.N., Y.S. and S.I.B.; writing—original draft preparation, A.A. and S.I.B.; project administration, A.A. and A.A.A.-A.; funding acquisition, A.A.A.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Researchers Supporting Project number (RSP2023R505), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors express their sincere thanks to the City of Scientific Research and Technological Applications (SRTA-City) and the Faculty of Agriculture (Saba Basha), Alexandria University, Egypt, for providing the necessary research facilities. The authors would like to extend their appreciation to the Researchers Supporting Project number (RSP2023R505), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stokstad, E. The new potato. Science 2019, 363, 574–577. [Google Scholar] [CrossRef]
- Rabia, A.H.; Mohamed, A.; Abdelaty, E.F.; Shahin, S.F.; Yacout, D.M.M. Investigating Adaptation Strategies Developed by Potato Farmers to Cope with Climate Change Impacts in Egypt. Alex. Sci. Exch. J. 2021, 42, 871–881. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Muturi, P.; Yu, J.; Maina, A.N.; Kariuki, S.; Mwaura, F.B.; Wei, H. Bacteriophages isolated in China for the control of Pectobacterium carotovorum causing potato soft rot in Kenya. Virol. Sin. 2019, 34, 287–294. [Google Scholar] [CrossRef]
- Behiry, S.I.; Ashmawy, N.A.; Abdelkhalek, A.A.; Younes, H.A.; Khaled, A.E.; Hafez, E.E. Compatible- and incompatible-type interactions related to defense genes in potato elucidation by Pectobacterium carotovorum. J. Plant Dis. Prot. 2018, 125, 197–204. [Google Scholar] [CrossRef]
- Curland, R.D.; Mainello, A.; Perry, K.L.; Hao, J.; Charkowski, A.O.; Bull, C.T.; McNally, R.R.; Johnson, S.B.; Rosenzweig, N.; Secor, G.A. Species of Dickeya and Pectobacterium isolated during an outbreak of blackleg and soft rot of potato in northeastern and north Central United States. Microorganisms 2021, 9, 1733. [Google Scholar] [CrossRef]
- Trabelsi, B.M.; Abdallah, R.A.B.; Ammar, N.; Kthiri, Z.; Hamada, W. Bio-suppression of Fusarium wilt disease in potato using nonpathogenic potatoassociated fungi. J. Plant Pathol. Microbiol. 2016, 7, 2. [Google Scholar]
- Tapwal, A.; Garg, S.; Gautam, N.; Kumar, R. In vitro antifungal potency of plant extracts against five phytopathogens. Braz. Arch. Biol. Technol. 2011, 54, 1093–1098. [Google Scholar] [CrossRef]
- Pinhero, R.G.; Coffin, R.; Yada, R.Y. Post-harvest storage of potatoes. In Advances in Potato Chemistry and Technology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 339–370. [Google Scholar]
- Jiménez-Reyes, M.F.; Carrasco, H.; Olea, A.F.; Silva-Moreno, E. Natural compounds: A sustainable alternative to the phytopathogens control. J. Chil. Chem. Soc. 2019, 64, 4459–4465. [Google Scholar] [CrossRef]
- Šernaitė, L. Plant extracts: Antimicrobial and antifungal activity and appliance in plant protection. Sodinink. Daržinink 2017, 36, 58–68. [Google Scholar]
- Nebija, F.; Stefkov, G.; Karapandzova, M.; Stafilov, T.; Panovska, T.K.; Kulevanova, S. Chemical characterization and antioxidant activity of Eryngium campestre L., Apiaceae from Kosovo. Maced. Pharm. Bull. 2009, 55, 22–32. [Google Scholar] [CrossRef]
- Küpeli, E.; Kartal, M.; Aslan, S.; Yesilada, E. Comparative evaluation of the anti-inflammatory and antinociceptive activity of Turkish Eryngium species. J. Ethnopharmacol. 2006, 107, 32–37. [Google Scholar] [CrossRef]
- Gugliucci, A.; Bastos, D.H.M. Chlorogenic acid protects paraoxonase 1 activity in high density lipoprotein from inactivation caused by physiological concentrations of hypochlorite. Fitoterapia 2009, 80, 138–142. [Google Scholar] [CrossRef]
- Behiry, S.I.; Soliman, S.A.; Al-Askar, A.A.; Alotibi, F.O.; Basile, A.; Abdelkhalek, A.; Elsharkawy, M.M.; Salem, M.Z.M.; Hafez, E.E.; Heflish, A.A. Plantago lagopus extract as a green fungicide induces systemic resistance against Rhizoctonia root rot disease in tomato plants. Front. Plant Sci. 2022, 13, 966929. [Google Scholar] [CrossRef]
- Behiry, S.I.; Philip, B.; Salem, M.Z.M.; Amer, M.A.; El-Samra, I.A.; Abdelkhalek, A.; Heflish, A. Urtica dioica and Dodonaea viscosa leaf extracts as eco-friendly bioagents against Alternaria alternata isolate TAA-05 from tomato plant. Sci. Rep. 2022, 12, 16468. [Google Scholar] [CrossRef]
- Elbanoby, N.E.; El-Settawy, A.A.A.; Mohamed, A.A.; Salem, M.Z.M. Phytochemicals derived from Leucaena leucocephala (Lam.) de Wit (Fabaceae) biomass and their antimicrobial and antioxidant activities: HPLC analysis of extracts. Biomass Convers. Biorefinery 2022, 1–17. [Google Scholar] [CrossRef]
- Wang, P.; Su, Z.; Yuan, W.; Deng, G.; Li, S. Phytochemical constituents and pharmacological activities of Eryngium L.(Apiaceae). Pharm. Crops 2012, 3, 99–120. [Google Scholar] [CrossRef]
- Le Claire, E.; Schwaiger, S.; Banaigs, B.; Stuppner, H.; Gafner, F. Distribution of a new rosmarinic acid derivative in Eryngium alpinum L. and other Apiaceae. J. Agric. Food Chem. 2005, 53, 4367–4372. [Google Scholar] [CrossRef]
- Hohmann, J.; Pall, Z.; Günther, G.; Mathe, I. Flavonolacyl glycosides of the aerial parts of Eryngium campestre. Planta Med. 1997, 63, 96. [Google Scholar] [CrossRef]
- Kholkhal, W.; Ilias, F.; Bekhechi, C.; Bekkara, F.A. Eryngium maritimum: A rich medicinal plant of polyphenols and flavonoids compounds with antioxidant, antibacterial and antifungal activities. Curr. Res. J. Biol. Sci. 2012, 4, 437–443. [Google Scholar]
- Zhang, Z.; Li, S.; Ownby, S.; Wang, P.; Yuan, W.; Zhang, W.; Beasley, R.S. Phenolic compounds and rare polyhydroxylated triterpenoid saponins from Eryngium yuccifolium. Phytochemistry 2008, 69, 2070–2080. [Google Scholar] [CrossRef]
- Erdelmeier, C.A.J.; Sticher, O. Coumarin Derivatives from Eryngium campestre1. Planta Med. 1985, 51, 407–409. [Google Scholar] [CrossRef]
- Ingram, M. Species Account: Eryngium Campestre; Bot. Soc. Br. Isles: Durham, UK, 2006. [Google Scholar]
- Baytop, T. Türkiye’de Bitkilerle Tedavi–Geçmişten Bugüne, 2nd ed.; Nobel Tıp Basımevi: İstanbul, Türkiye, 1999; pp. 348–349. [Google Scholar]
- Soumia, B. Eryngium campestre L.: Polyphenolic and flavonoid compounds. applications to health and disease; In Polyphenols: Mechanisms of Action in Human Health and Disease; Academic Press: Cambridge, MA, USA, 2018; pp. 69–79. [Google Scholar]
- Güneş, M.G.; İşgör, B.S.; İşgör, Y.G.; Moghaddam, N.S.; Geven, F.; Yildirim, Ö. The effects of Eryngium campestre extracts on glutathione-s-transferase, glutathione peroxidase and catalase enzyme activities. Turk. J. Pharm. Sci. 2014, 11, 339–346. [Google Scholar]
- Bouzidi, S.; Benkiki, N.; Hachemi, M.; Haba, H. Investigation of In Vitro Antioxidant Activity and In Vivo Antipyretic and Anti-Inflammatory Activities of Algerian Eryngium campestre L. Curr. Bioact. Compd. 2017, 13, 340–346. [Google Scholar] [CrossRef]
- Kartnig, T. Flavonoids from the aerial parts of Eryngium campestre. Planta Med. 1993, 59, 285. [Google Scholar] [CrossRef]
- Thiem, B.; Goslinska, O.; Kikowska, M.; Budzianowski, J. Antimicrobial activity of three Eryngium L. species (Apiaceae). Herba Pol. 2010, 56, 52–58. [Google Scholar]
- Hawas, U.W.; El-Kassem, L.A.T.; Awad, H.M.; Taie, H.A.A. Anti-Alzheimer, antioxidant activities and flavonol glycosides of Eryngium campestre L. Curr. Chem. Biol. 2013, 7, 188–195. [Google Scholar] [CrossRef]
- Abd-Elmonem, A.R.; Shehab, N.G. Study of the volatile oil of Eryngium campestre L. growing in Egypt. Bull. Fac. Pharm. Cairo. Univ. 2008, 44, 3379–3388. [Google Scholar]
- Medbouhi, A.; Benbelaïd, F.; Djabou, N.; Beaufay, C.; Bendahou, M.; Quetin-Leclercq, J.; Tintaru, A.; Costa, J.; Muselli, A. Essential oil of Algerian Eryngium campestre: Chemical variability and evaluation of biological activities. Molecules 2019, 24, 2575. [Google Scholar] [CrossRef]
- Azizkhani, M.; Tooryan, F. Antioxidant and antimicrobial activities of rosemary extract, mint extract and a mixture of tocopherols in beef sausage during storage at 4C. J. Food Saf. 2015, 35, 128–136. [Google Scholar] [CrossRef]
- Aktaruzzaman, M.; Afroz, T.; Lee, Y.-G.; Kim, B.-S. Morphological and molecular characterization of Fusarium tricinctum causing postharvest fruit rot of pumpkin in Korea. J. Gen. Plant Pathol. 2018, 84, 407–413. [Google Scholar] [CrossRef]
- Möller, E.M.; Bahnweg, G.; Sandermann, H.; Geiger, H.H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992, 20, 6115. [Google Scholar] [CrossRef]
- Soliman, S.A.; Al-Askar, A.A.; Sobhy, S.; Samy, M.A.; Hamdy, E.; Sharaf, O.A.; Su, Y.; Behiry, S.I.; Abdelkhalek, A. Differences in Pathogenesis-Related Protein Expression and Polyphenolic Compound Accumulation Reveal Insights into Tomato–Pythium aphanidermatum Interaction. Sustainability 2023, 15, 6551. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Thomas, P.; Upreti, R. Significant effects due to peptone in Kelman medium on colony characteristics and virulence of Ralstonia solanacearum in tomato. Open Microbiol. J. 2014, 8, 95. [Google Scholar] [CrossRef]
- El-Gendi, H.; Al-Askar, A.A.; Király, L.; Samy, M.A.; Moawad, H.; Abdelkhalek, A. Foliar Applications of Bacillus subtilis HA1 Culture Filtrate Enhance Tomato Growth and Induce Systemic Resistance against Tobacco mosaic virus Infection. Horticulturae 2022, 8, 301. [Google Scholar] [CrossRef]
- Moni, Z.R.; Ali, M.A.; Alam, M.S.; Rahman, M.A.; Bhuiyan, M.R.; Mian, M.S.; Iftekharuddaula, K.M.; Latif, M.A.; Khan, M.A.I. Morphological and genetical variability among Rhizoctonia solani isolates causing sheath blight disease of rice. Rice Sci. 2016, 23, 42–50. [Google Scholar] [CrossRef]
- Desvani, S.D.; Lestari, I.B.; Wibowo, H.R.; Supyani; Poromarto, S.H. Hadiwiyono Morphological characteristics and virulence of Rhizoctonia solani isolates collected from some rice production areas in some districts of Central Java. In Proceedings of the International Conference on Science and Applied Science (ICSAS) 2018, Surakarta, Indonesia, 12 May 2018; Volume 2014, p. 020068. [Google Scholar]
- Ke, X.; Lu, M.; Wang, J. Identification of Fusarium solani species complex from infected zebrafish (Danio rerio). J. Vet. Diagn. Investig. 2016, 28, 688–692. [Google Scholar] [CrossRef]
- Al-Abedy, A.N.; Al-Fadhal, F.A.; Karem, M.H.; Al–Masoudi, Z.; Al-Mamoori, S.A. Genetic variability of different isolates of Rhizoctonia solani Kühn isolated from Iranian imported potato tubers (Solanum tuberosum L.). Int. J. Agric. Statatistic Sci. 2018, 14, 587–598. [Google Scholar]
- Khan, M.; Wang, R.; Li, B.; Liu, P.; Weng, Q.; Chen, Q. Comparative evaluation of the LAMP assay and PCR-based assays for the rapid detection of Alternaria solani. Front. Microbiol. 2018, 9, 2089. [Google Scholar] [CrossRef]
- Schocha, C.L.; Seifertb, K.A.; Huhndorfc, S.; Robertd, V.; Spougea, J.L.; Levesqueb, C.A.; Chenb, W.; Consortiuma, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Dickie, I.A. Does host plant richness explain diversity of ectomycorrhizal fungi? Re-evaluation of Gao et al. (2013) data sets reveals sampling effects. Mol. Ecol. 2014, 23, 992–995. [Google Scholar] [CrossRef]
- Kikowska, M.; Chanaj-Kaczmarek, J.; Derda, M.; Budzianowska, A.; Thiem, B.; Ekiert, H.; Szopa, A. The evaluation of phenolic acids and flavonoids content and antiprotozoal activity of Eryngium species biomass produced by biotechnological methods. Molecules 2022, 27, 363. [Google Scholar] [CrossRef]
- Azizkhani, M.; Sodanlo, A. Antioxidant activity of Eryngium campestre L., Froriepia subpinnata, and Mentha spicata L. polyphenolic extracts nanocapsulated in chitosan and maltodextrin. J. Food Process. Preserv. 2021, 45, e15120. [Google Scholar] [CrossRef]
- Paun, G.; Neagu, E.; Moroeanu, V.; Albu, C.; Savin, S.; Lucian Radu, G. Chemical and bioactivity evaluation of Eryngium planum and Cnicus benedictus polyphenolic-rich extracts. Biomed Res. Int. 2019, 2019, 3692605. [Google Scholar] [CrossRef]
- Kikowska, M.; Budzianowski, J.; Krawczyk, A.; Thiem, B. Accumulation of rosmarinic, chlorogenic and caffeic acids in in vitro cultures of Eryngium planum L. Acta Physiol. Plant. 2012, 34, 2425–2433. [Google Scholar] [CrossRef]
- Thiem, B.; Kikowska, M.; Krawczyk, A.; Więckowska, B.; Sliwinska, E. Phenolic acid and DNA contents of micropropagated Eryngium planum L. Plant Cell Tissue Organ Cult. 2013, 114, 197–206. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).