CellsDeepNet: A Novel Deep Learning-Based Web Application for the Automated Morphometric Analysis of Corneal Endothelial Cells
Abstract
:1. Introduction
- i.
- Development of CellsDeepNet, a novel, fully automated, and real-time web application for objective quantification of CEC morphology for CEC pathology assessment.
- ii.
- A new pre-processing procedure for reducing the noise and enhancing image quality is proposed to make cell boundaries more visible. Firstly, the contrast of the corneal endothelium image is improved by transforming the values using CLAHE. Secondly, novel image denoising and smoothing algorithms are proposed based on 2DDD-TCWT and Butterworth Bandpass filter to minimize the noise and enhance the edges of the corneal endothelial cell boundaries. This is followed by applying the brightness level adjustment step using the moving average filter and the CLAHE to reduce the effects of non-uniform illumination.
- iii.
- An improved version of U-Net architecture was applied to precisely identify all CEC boundaries in the enhanced image regardless of the endothelial cell size. An effective training methodology supported with some well-appointed training techniques (e.g., data augmentation, dropout technique, etc.) is utilized to assess different U-Net structures (e.g., the number of layers, the number of filters per layer, etc.) to prevent the overfitting issues and enhance the generalization capability of the final trained model.
- iv.
- The performance and generalization capability of the proposed CellsDeepNet system was verified in corneal endothelium images captured using corneal confocal microscopy (CCM) by the Heidelberg Retina Tomograph 3 with the Rostock Cornea Module (HRT 3 RCM) and a specular microscope. The results demonstrated that the measurement of the CEC morphology by the CellsDeepNet system highly correlates to the manual annotation and outperforms current state-of-the-art automated approaches on precise endothelial cell segmentation.
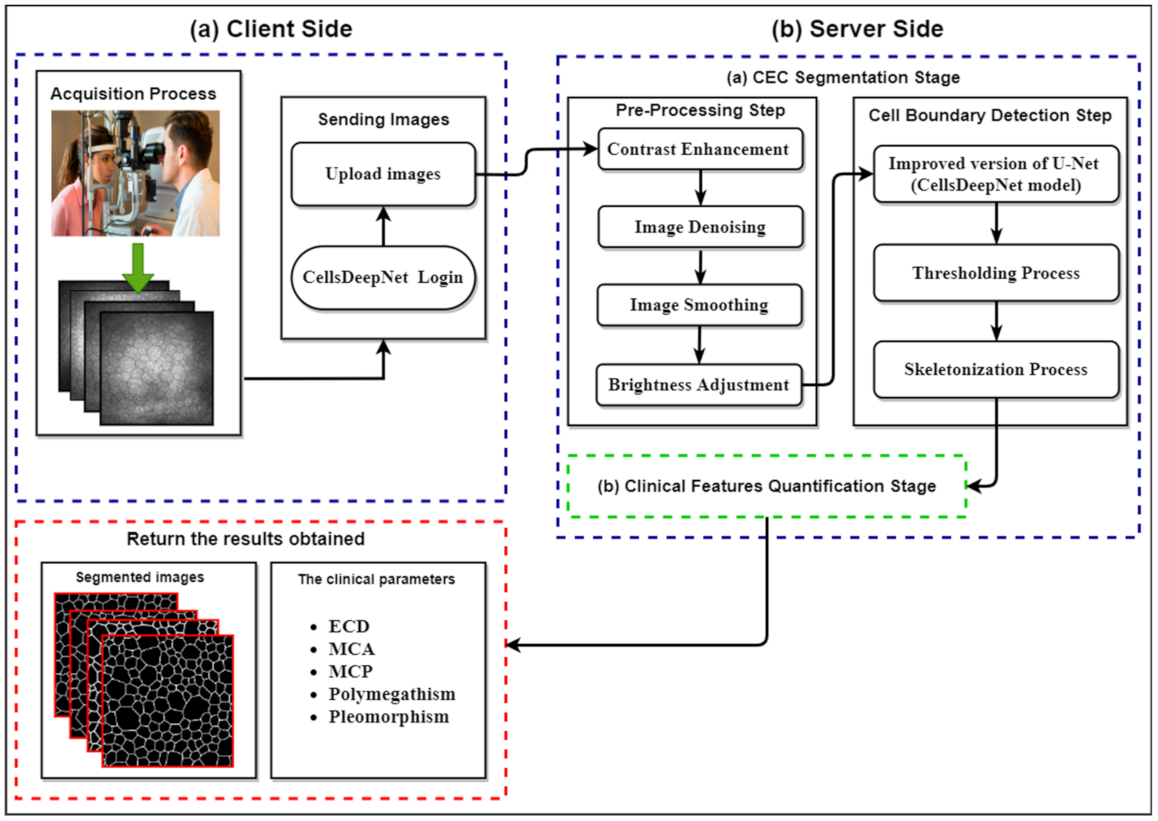
2. The Proposed CellsDeepNet System
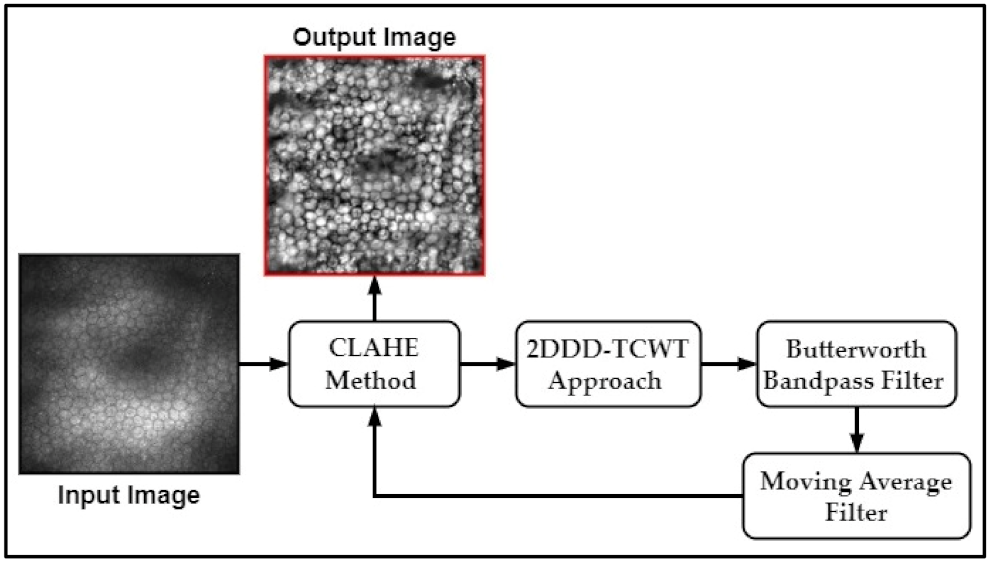
2.1. Pre-Processing Step
- (1)
- Divide the input image of size (M × M) pixels into non-overlapping tiles of size (8 × 8) pixels.
- (2)
- Estimate the histogram of each tile in the input image.
- (3)
- Compute the clipping threshold value to redistribute pixels of each tile.
- (4)
- The histogram equalization is estimated for each tile of redistributed pixels.
- (5)
- The center pixel is computed of each tile, and then all pixels within a tile are produced using an optimized bilinear interpolation function to eliminate boundary artefacts.
- (1)
- The forward 2DDD-TCWT is applied to the input CEC image to determine the wavelet sub-bands coefficients.
- (2)
- The level of the noise variance in the input image is evaluated.
- (3)
- The non-linear shrinkage function is applied to compute the threshold value.
- (4)
- The soft thresholding technique is adopted based of the thresholding value computed in step 3.
- (5)
- Finally, the denoised image is obtained by applying the inverse 2DDD-TCWT.
2.2. Endothelial Cell Boundary Detection Step
2.2.1. Network Architecture
2.2.2. The Training Methodology
- (1)
- Divide the dataset into 3 different sets: training set, validation set, and test set.
- (2)
- Choose an initial network architecture and a combination of training parameters.
- (3)
- Train the selected network architecture in step 2 using the training set.
- (4)
- Use the validation set to assess the performance of the selected network architecture through the training progression.
- (5)
- Repeat steps 3 through 4 by employing N = 100 epochs.
- (6)
- Choose the best-trained model with the smallest validation error.
- (7)
- Report the performance of the best model using the testing set.
2.3. Clinical Features Quantification Stage
- MCD is computed by dividing the number of CECs in the selected ROI (otherwise entire segmented image) on the total size of the selected ROI (otherwise entire segmented image) as follows:Herein, to accurately estimate the MCD, only the cells on the two adjacent borders of the selected ROI are included, discarding them on other borders.
- Polymegathism also called Coefficient of Variation (CV), is utilized to define the difference in the endothelial cell’s area. Increasing the standard deviation () value of the MCA can result in an imprecise estimate for the MCD. Therefore, increased polymegathism results in an imprecise estimation of MCA [6]. Polymegathism is computed as follows:where is the STD of the cell area divided by the MCA.
- Pleomorphism, also called Hexagonality Coefficient (HC), is computed by dividing the number of CECs with a roughly hexagonal form (neighbouring with 6 cells) Chexagonal on the overall number of CECs in the selected ROI (otherwise entire segmented image) Cimage as follows:
3. Experimental Results
3.1. Datasets Description
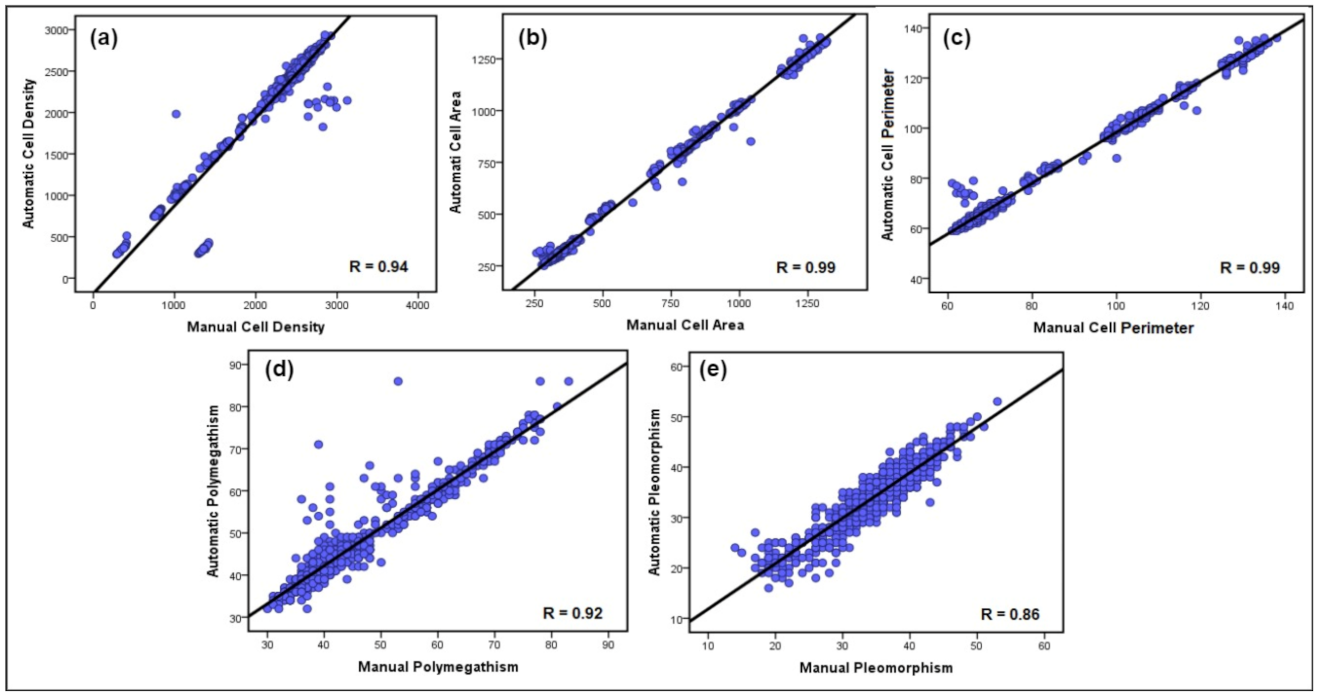
- Manchester Corneal Confocal Microscopy Dataset: The MCCM dataset contains a total of 1010 images of CECs acquired using the Heidelberg Retinal Tomograph III Rostock Cornea Module (Heidelberg Engineering GmbH, Heidelberg, Germany). This device uses a 670 nm red wavelength diode laser, which is a class I laser and therefore does not pose any ocular safety hazard. A 63× objective lens with a numerical aperture of 0.9 and a working distance relative to the applanating cap (TomoCap, Heidelberg Engineering GmbH, Heidelberg, Germany) of 0.0 to 3.0 mm was used. The images produced using this lens are (400 μm × 400 μm) with a (15°×15°) field of view and 10 μm/pixel transverse optical resolution. The cornea was locally anaesthetized by instilling 1 drop of 0.4% benoxinate hydrochloride (Chauvin Pharmaceuticals, Chefaro, UK), and Viscotears (Carbomer 980, 0.2%, Novartis, UK) was used as the coupling agent between the cornea and the TomoCap as well as between the TomoCap and the objective lens. The patients were asked to place their chin on the chin rest and press their forehead against the forehead support. They were asked also to fixate with the eye not being examined on an outer fixation light to enable examination of the central cornea. Images of the endothelial cells were captured using the “section” mode. Multiple images were taken from the endothelium immediately posterior to the posterior stroma. On CCM, endothelial cells are identified as a polygonal shape with bright cell bodies and dark borders. During image acquisition, 2–3 representative sharp images were selected by filtering out blurred images, pressure lines, or dark shadows caused by the pressure applied between the TomoCap and cornea or out of focus images. Some samples of unprocessed corneal endothelium images are presented in Figure 2. It is worth noting that the corneal endothelium images used in this dataset are very challenging poor-quality images compared to those used in the previous works. A major challenge for carrying out this research using this dataset was the unavailability of ground-truth images and the reference value measurements for all five clinical parameters. To obtain a manual version from this database, a freely available application, named GNU Image Manipulation Program (GIMP) was utilized by an expert ophthalmologist from the University of Manchester to manually detect endothelial cell boundaries and then generate a binary image from particular ROIs to serve as a ground-truth image in the segmentation evaluation and a manual estimation of the reference values of the clinical parameters, as shown in Figure 10.
- Endothelial Cell Alizarine Dataset: This dataset is composed of 30 corneal endothelium images captured by an IPCM (CK 40, Chroma Technology Corp, Windham, VT, USA) at (200 × magnification) and an analogue camera (SSC-DC50AP, Sony, Tokyo, Japan) [23]. These images were taken from 30 porcine eyes stained with alizarine and stored in the JPEG format of resolution (576 × 768) pixels. The ground truth images representing the borders of the endothelial cells traced manually by an expert ophthalmologist are also provided. Each image contains approximately 232 detected cells on average (ranging from 188 to 388 cells), along with an average cell area of 272.76 pixels. This dataset is freely available at http://bioimlab.dei.unipd.it, accessed on 10 December 2021.
3.2. Experiments on MCCM Dataset
3.3. Experiments on ECA Dataset
3.4. Comparison Study
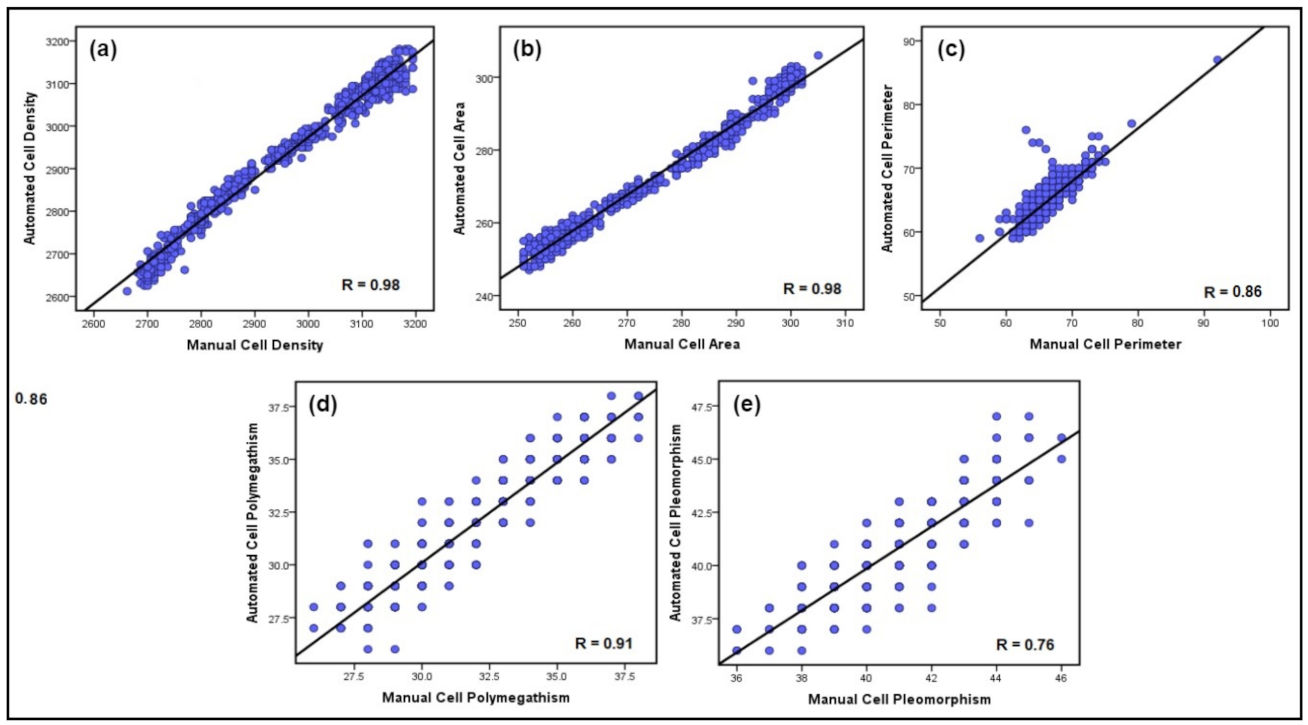
3.5. Discussion
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Fahdawi, R.S.; Qahwaji, A.R.S.; Al-Waisy, S.; Ipsopn, S. An Automatic Corneal Subbasal Nerve Registration System Using FFT and Phase Correlation Techniques for an Accurate DPN Diagnosis. In Proceedings of the 2015 IEEE International Conference on Computer and Information Technology; Ubiquitous Computing and Communications; Dependable, Autonomic and Secure Computing; Pervasive Intelligence and Computing, Liverpool, UK, 26–28 October 2015; pp. 1035–1041. [Google Scholar] [CrossRef]
- Al-Fahdawi, S.; Qahwaji, R.; Al-Waisy, A.S.; Ipson, S.; Ferdousi, M.; Malik, R.A.; Brahma, A. A fully automated cell segmentation and morphometric parameter system for quantifying corneal endothelial cell morphology. Comput. Methods Programs Biomed. 2018, 160, 11–23. [Google Scholar] [CrossRef]
- Al-Fahdawi, S.; Qahwaji, R.; Al-Waisy, A.S.; Ipson, S.; Malik, R.A.; Brahma, A.; Chen, X. A fully automatic nerve segmentation and morphometric parameter quantification system for early diagnosis of diabetic neuropathy in corneal images. Comput. Methods Programs Biomed. 2016, 135, 151–166. [Google Scholar] [CrossRef] [Green Version]
- Gavet, Y.; Pinoli, J.-C. Comparison and Supervised Learning of Segmentation Methods Dedicated to Specular Microscope Images of Corneal Endothelium. Int. J. Biomed. Imaging 2014, 2014, 704791. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Kamran, S.; Akhtar, N.; Ponirakis, G.; Al-Muhannadi, H.; Petropoulos, I.N.; Al-Fahdawi, S.; Qahwaji, R.; Sartaj, F.; Babu, B.; et al. Corneal Confocal Microscopy detects a Reduction in Corneal Endothelial Cells and Nerve Fibres in Patients with Acute Ischemic Stroke. Sci. Rep. 2018, 8, 17333. [Google Scholar] [CrossRef]
- McCarey, B.E.; Edelhauser, H.F.; Lynn, M.J. Review of Corneal Endothelial Specular Microscopy for FDA Clinical Trials of Refractive Procedures, Surgical Devices and New Intraocular Drugs and Solutions. Cornea 2008, 27, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ruggeri, A.; Grisan, E.; Jaroszewski, J. A new system for the automatic estimation of endothelial cell density in donor corneas. Br. J. Ophthalmol. 2005, 89, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Gain, P.; Thuret, G.; Kodjikian, L.; Gavet, Y.; Turc, P.H.; Theillere, C.; Acquart, S.; Le Petit, J.C.; Maugery, J.; Campos, L. Automated tri-image analysis of stored corneal endothelium. Br. J. Ophthalmol. 2002, 86, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Doughty, M.J.; Aakre, B.M. Further analysis of assessments of the coefficient of variation of corneal endothelial cell areas from specular microscopic images. Clin. Exp. Optom. 2008, 91, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Foracchia, M.; Ruggeri, A.M. Cell contour detection in corneal endothelium in-vivo microscopy. In Proceedings of the 22nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 23–28 July 2000; Volume 2, pp. 1033–1035. [Google Scholar] [CrossRef]
- Foracchia, M.; Ruggeri, A.M. Corneal Endothelium Cell Field Analysis by means of Interacting Bayesian Shape Models. In Proceedings of the 2007 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; Volume 62035–60387, pp. 6035–6038. [Google Scholar] [CrossRef]
- Fabijańska, A. Corneal Endothelium Image Segmentation Using Feedforward Neural Network. In Proceedings of the Federated Conference on Computer Science and Information Systems, Prague, Czech Republic, 3–6 September 2017; Volume 11, pp. 629–637. [Google Scholar] [CrossRef] [Green Version]
- Nadachi, R.; Nunokawa, K. Automated Corneal Endothelial Cell Analysis. In Proceedings of the Fifth Annual IEEE Symposium on Computer-Based Medical Systems, Durham, NC, USA, 14–17 June 1992; pp. 450–457. [Google Scholar]
- Mahzoun, M.; Okazaki, K.; Mitsumoto, H.; Kawai, H.; Sato, Y.; Tamura, S.; Kani, K. Detection and Complement of Hexagonal Borders in Corneal Endothelial Cell lmage. Med. Imaging Technol. 1996, 14, 56–69. [Google Scholar]
- Sanchez-Marin, F.J. Automatic segmentation of contours of corneal cells. Comput. Biol. Med. 1999, 29, 243–258. [Google Scholar] [CrossRef]
- Ayala, G.M.E.D.H.; Mart, L.H. Granulometric moments and corneal endothelium status. Pattern Recognit. 2001, 34, 1219–1227. [Google Scholar] [CrossRef]
- Scarpa, F.; Ruggeri, A. Segmentation of Corneal Endothelial Cells Contour by Means of a Genetic Algorithm. In Proceedings of the Ophthalmic Medical Image Analysis Second International Workshop, OMIA 2015, Held in Conjunction with MICCAI 2015, Munich, Germany, 9 October 2015; pp. 25–32. [Google Scholar]
- Sharif, M.; Qahwaji, R.; Shahamatnia, E.; Alzubaidi, R.; Ipson, S.; Brahma, A. An efficient intelligent analysis system for confocal corneal endothelium images. Comput. Methods Programs Biomed. 2015, 122, 421–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, L.M.; Masters, B.R. Morphological image processing and network analysis of cornea endothelial cell images. In Proceedings of the SPIE, San Diego, CA, USA, 1 July 1992; Volume 1769, pp. 212–226. [Google Scholar] [CrossRef]
- Gavet, Y.; Ijean-Charles, P. Visual Perception Based Automatic Recognition of Cell Mosaics in Human Corneal Endothelium Microscopy Images Cornea : Vision And Quality. Image Anal. Stereol. 2008, 27, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Selig, B.; Vermeer, K.A.; Rieger, B.; Hillenaar, T.; Hendriks, C.L.L. Fully automatic evaluation of the corneal endothelium from in vivo confocal microscopy. BMC Med. Imaging 2015, 15, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurzynska, K. Deep Learning as a Tool for Automatic Segmentation of Corneal Endothelium Images. Symmetry 2018, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Ruggeri, A.; Scarpa, F.; de Luca, M.; Meltendorf, C.; Schroeter, J. A system for the automatic estimation of morphometric parameters of corneal endothelium in alizarine red-stained images. Br. J. Ophthalmol. 2010, 94, 643–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabijańska, A. Segmentation of corneal endothelium images using a U-Net-based convolutional neural network. Artif. Intell. Med. 2018, 88, 1–13. [Google Scholar] [CrossRef]
- Sasi, N.M.; Jayasree, V.K. Contrast Limited Adaptive Histogram Equalization for Qualitative Enhancement of Myocardial Perfusion Images. Engineering 2013, 05, 326–331. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y. Application of the CLAHE algorithm based on optimized bilinear interpolation in near infrared vein image enhancement. In Proceedings of the 2nd International Conference on Computer Science and Application Engineering, Hohhot, China, 22–24 October 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Raj, V.N.P.; Venkateswarlu, T.P. Denoising of Medical Images Using Dual Tree Complex Wavelet Transform. Procedia Technol. 2012, 4, 238–244. [Google Scholar] [CrossRef] [Green Version]
- Fodor, I.K.; Kamath, C. Denoising Through Wavelet Shrinkage: An Empirical Study. J. Electron. Imaging 2001, 12, 151–161. [Google Scholar] [CrossRef]
- Naimi, H.; Adamou-Mitiche, A.B.H.; Mitiche, L. Medical image denoising using dual tree complex thresholding wavelet transform and Wiener filter. J. King Saud Univ. Comput. Inf. Sci. 2015, 27, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Ginley, B.; Lutnick, J.B.E.; Tomaszewski, P.J.; Sarder, E.; Govind, D. Glomerular Detection and Segmentation from Multimodal Microscopy Images using a Butterworth Band-Pass Filter; Université de Grenoble: Grenoble, France, 2018; p. 10581. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Agarwal, A.; Negahban, S.; Wainwright, M.J. Noisy matrix decomposition via convex relaxation: Optimal rates in high dimensions. Ann. Stat. 2012, 40, 1171–1197. [Google Scholar] [CrossRef] [Green Version]
- Sornam, M.; Kavitha, M.S.; Nivetha, M. Hysteresis thresholding based edge detectors for inscriptional image enhancement. In Proceedings of the IEEE International Conference on Computational Intelligence and Computing Research (ICCIC), Chennai, India, 15–17 December 2016; pp. 1–4. [Google Scholar] [CrossRef]
- Al-Waisy, A.S.; Qahwaji, R.; Ipson, S.; Al-Fahdawi, S.; Nagem, T.A.M. A multi-biometric iris recognition system based on a deep learning approach. Pattern Anal. Appl. 2017, 21, 783–802. [Google Scholar] [CrossRef] [Green Version]
- Tavakoli, M.; Malik, R.A. Corneal Confocal Microscopy: A Novel Non-invasive Technique to Quantify Small Fibre Pathology in Peripheral Neuropathies. J. Vis. Exp. 2011, 12, e2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, J.; Agrawal, S.; Vig, R. Integration of Clustering, Optimization and Partial Differential Equation Method for Improved Image Segmentation. Int. J. Image Graph. Signal Process. 2012, 4, 26–33. [Google Scholar] [CrossRef]
- Xue, W.; Zhang, L.; Mou, X.; Bovik, A. Gradient Magnitude Similarity Deviation: A Highly Efficient Perceptual Image Quality Index. IEEE Trans. Image Process. 2013, 23, 684–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P.; Member, S. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. IMAGE Process 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meilă, M. Comparing Clusterings–An Information Based Distance. J. Multivar. Anal. 2007, 98, 873–895. [Google Scholar] [CrossRef] [Green Version]
- Mallikarjuna, K.; Prasad, K.S.; Subramanyam, M.V. Image Compression and Reconstruction using Discrete Rajan Transform Based Spectral Sparsing. Int. J. Image Graph. Signal Process. 2016, 8, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.; Fowlkes, C.; Tal, D.; Malik, J. A database of human segmented natural images and its application to evaluating segmentation algorithms and measuring ecological statistics. In Proceedings of the Eighth IEEE International Conference on Computer Vision. ICCV 2001, Vancouver, BC, Canada, 7–14 July 2001; Volume 2, pp. 416–423. [Google Scholar]
- XIII Mediterranean Conference on Medical and Biological Engineering and Computing 2013. IFMBE Proc. 2014, 41, 658–661. [CrossRef] [Green Version]
- Zhang, Y. A Multi-Branch Hybrid Transformer Networkfor Corneal Endothelial Cell Segmentation. 2021. Available online: http://arxiv.org/abs/2106.07557 (accessed on 1 March 2021).








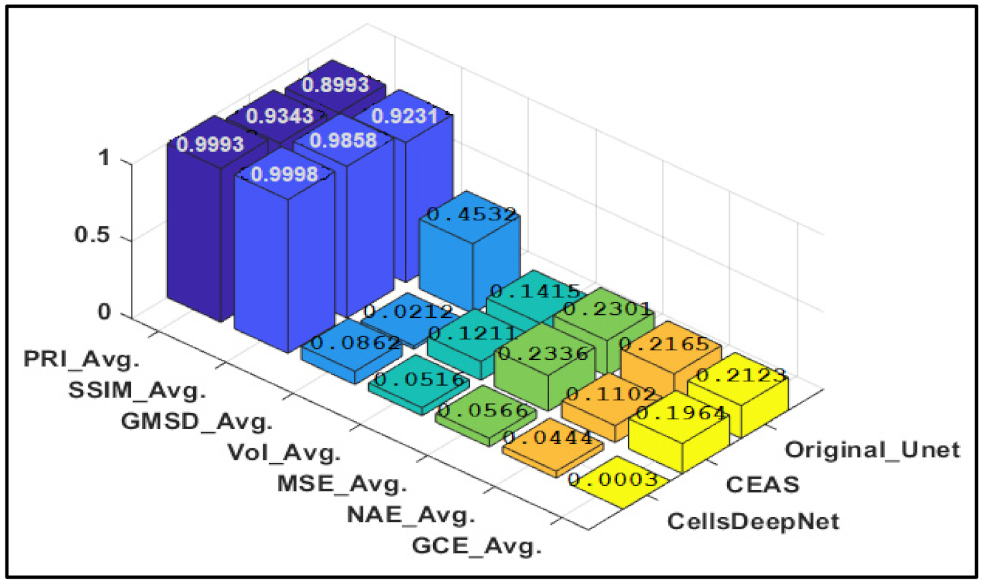






| Quantitative Measures | MCCM Dataset | ECA Dataset | ||
|---|---|---|---|---|
| With Image Pre-Processing | Without Image Pre-Processing | With Image Pre-Processing | Without Image Pre-Processing | |
| PRI | 0.9993 | 0.4956 | 0.9837 | 0.5053 |
| SSIM | 0.9998 | 0.4102 | 0.9768 | 0.4312 |
| VoI | 0.0516 | 0.7231 | 0.1032 | 0.8901 |
| GMSD | 0.0862 | 0.8231 | 0.0105 | 0.9212 |
| MSE | 0.0566 | 0.5342 | 0.0337 | 0.5231 |
| GCE | 0.0003 | 0.5223 | 0.0006 | 0.4345 |
| NAE | 0.0444 | 0.4532 | 0.0769 | 0.5301 |
| Manual | Auto. CEAS System | Auto. CellsDeepNet System | |||||
|---|---|---|---|---|---|---|---|
| MCD (Cells/mm2) | MCD (Cells/mm2) | Diff | Diff % | MCD (Cells/mm2) | Diff | Diff % | |
| Av. | 1898.08 | 1967.04 | 31.04 | 1.56 | 1984.88 | 13.2 | 0.66 |
| STD | 765.21 | 755.55 | 9.66 | 1.27 | 763.17 | 2.04 | 0.26 |
| Max | 3125 | 3226 | −101 | −3.18 | 2937 | 188 | 6.20 |
| Min | 1232 | 1280 | −48 | −3.82 | 1230 | 2 | 0.16 |
| MCA (µm2) | MCA (µm2) | Diff | Diff % | MCA (µm2) | Diff | Diff % | |
| Av. | 292.28 | 285 | 7.28 | 2.52 | 292.27 | 0.01 | 0.003 |
| STD | 41.77 | 44.45 | −2.68 | −6.21 | 41.76 | 0.01 | 0.02 |
| Max | 395 | 390 | 5 | 1.27 | 395 | 0 | 0 |
| Min | 229 | 233 | −4 | −1.73 | 229 | 0 | 0 |
| MCP (µm) | MCP (µm) | Diff | Diff % | MCP (µm) | Diff | Diff % | |
| Av. | 79.04 | 74.06 | 4.98 | 6.50 | 77.16 | 1.88 | 2.40 |
| STD | 20.55 | 21.67 | −1.12 | −5.30 | 20.93 | −0.38 | −1.83 |
| Max | 138 | 140 | −2 | −1.43 | 136 | 2 | 1.45 |
| Min | 61 | 63 | −2 | −3.22 | 59 | 2 | 3.33 |
| Polymegathism % | Polymegathism % | Diff | Diff % | Polymegathism % | Diff | Diff % | |
| Av. | 45.44 | 42.95 | 2.49 | 5.6 | 44.14 | 1.3 | 2.90 |
| STD | 11.14 | 12.68 | −1.54 | −12.93 | 10.48 | 0.66 | 6.10 |
| Max | 83 | 79 | 4 | 4.93 | 86 | −3 | −3.55 |
| Min | 30 | 34 | −4 | −12.5 | 32 | −2 | −6.45 |
| Pleomorphism % | Pleomorphism % | Diff | Diff % | Pleomorphism % | Diff | Diff % | |
| Av. | 34.78 | 32.5 | 2.28 | 6.77 | 34.19 | 0.59 | 1.71 |
| STD | 6.14 | 3.63 | 2.51 | 51.38 | 5.98 | 0.16 | 2.64 |
| Max | 53 | 56 | −3 | −5.50 | 53 | 0 | 0 |
| Min | 14 | 17 | −3 | −19.35 | 16 | −2 | −13.33 |
| Manual | Auto. CEAS System | Auto. CellsDeepNet System | |||||
|---|---|---|---|---|---|---|---|
| MCD (Cells/mm2) | MCD (Cells/mm2) | Diff | Diff % | MCD (Cells/mm2) | Diff | Diff % | |
| Av. | 2952 | 2925 | 27 | 0.91 | 2927 | 25 | 0.85 |
| STD | 161 | 157 | 4 | 2.51 | 158 | 3 | 1.88 |
| Max | 3194 | 3177 | 17 | 0.53 | 3181 | 13 | 0.40 |
| Min | 2662 | 2605 | 57 | 2.16 | 2612 | 50 | 1.89 |
| MCA (µm2) | MCA (µm2) | Diff | Diff % | MCA (µm2) | Diff | Diff % | |
| Av. | 274 | 269 | 5 | 1.84 | 272 | 2 | 0.73 |
| STD | 16.08 | 15.65 | 0.43 | 2.71 | 16.01 | 0.07 | 0.43 |
| Max | 305 | 300 | 5 | 1.65 | 306 | −1 | −0.32 |
| Min | 251 | 246 | 5 | 2.01 | 247 | 4 | 1.60 |
| MCP (µm) | MCP (µm) | Diff | Diff % | MCP (µm) | Diff | Diff % | |
| Av. | 66.68 | 65.58 | 1.1 | 1.66 | 65.20 | 1.48 | 2.24 |
| STD | 2.76 | 1.67 | 1.09 | 49.20 | 2.82 | −0.06 | −2.15 |
| Max | 92 | 90 | 2 | 2.19 | 87 | 5 | 5.58 |
| Min | 56 | 58 | −2 | −3.50 | 59 | −3 | −5.21 |
| Polymegathism % | Polymegathism % | Diff | Diff % | Polymegathism % | Diff | Diff % | |
| Av. | 31.81 | 30.54 | 1.27 | 4.07 | 31.65 | 0.16 | 0.50 |
| STD | 2.85 | 2.68 | 0.17 | 6.14 | 2.83 | 0.02 | 0.70 |
| Max | 38 | 42 | −4 | −10 | 38 | 0 | 0 |
| Min | 26 | 29 | −3 | −10.9 | 26 | 0 | 0 |
| Pleomorphism % | Pleomorphism % | Diff | Diff % | Pleomorphism % | Diff | Diff % | |
| Av. | 40.84 | 37.5 | 3.34 | 8.52 | 40.68 | 0.16 | 0.39 |
| STD | 1.73 | 2.52 | −0.79 | −37.17 | 1.95 | −0.22 | −11.95 |
| Max | 46 | 44 | 2 | 4.44 | 47 | −1 | −2.15 |
| Min | 36 | 33 | 3 | 8.69 | 36 | 0 | 0 |
| Scarpa and Ruggeri [17] | Ruggeri et al. [23] | Poletti and Ruggeri [42] | CellsDeepNet System | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MCD (cells/mm2) | Diff | Diff % | Diff | Diff % | Diff | Diff % | Diff | Diff % | |
| Av. | 17.73 | 0.82 | −12 | 0.28 | 96.35 | 3.11 | 25 | 0.85 | |
| STD | 16.31 | 1.08 | −10 | 3.1 | 58.83 | 2.29 | 3 | 1.88 | |
| Max | 0 | 0 | −7 | 0.18 | 18 | 0.57 | 13 | 0.40 | |
| Min | 3.80 | 6.24 | 35 | 0.72 | 245 | 10.37 | 50 | 1.89 | |
| Polymegathism % | Diff | Diff % | Diff | Diff % | Diff | Diff % | Diff | Diff % | |
| Av. | 1.45 | 3.95 | 0.7 | 3.02 | 5.72 | 14.78 | 0.16 | 0.50 | |
| STD | 0.79 | 1.93 | 0.2 | 6.89 | 2.56 | 5.90 | 0.02 | 0.70 | |
| Max | 0 | 0 | 1.1 | 6.41 | 1.10 | 2.20 | 0 | 0 | |
| Min | 2.80 | 6.86 | −0.1 | 0.35 | 12.20 | 24.30 | 0 | 0 | |
| Diff | Diff % | Diff | Diff % | Diff | Diff % | Diff | Diff % | ||
| Pleomorphism % | Av. | 1.83 | 3.13 | −0.5 | 1.03 | 4.21 | 7.32 | 0.16 | 0.39 |
| STD | 1.33 | 2.27 | −0.9 | 23.37 | 2.72 | 4.57 | −0.22 | −11.95 | |
| Max | 0 | 0 | 2.2 | 5.62 | 0 | 0 | −1 | −2.15 | |
| Min | 3.80 | 6.24 | −4.2 | 7.47 | 11.80 | 16.21 | 0 | 0 | |
| Models | Measures | |||||
|---|---|---|---|---|---|---|
| DI | JA | F1 | SP | SE | MHD | |
| Fabija´nska [24] | 0.86 | - | - | - | - | - |
| Zhang et al. [43], | 0.78 | - | 0.82 | 0.94 | 0.87 | - |
| Nurzynska [22] | 0.94 | 0.94 | - | - | - | 0.14 |
| Orig. U-Net [31] | 0.77 | 0.79 | 0.81 | 0.96 | 0.81 | 0.22 |
| CellsDeepNet | 0.97 | 0.98 | 0.89 | 0.98 | 0.90 | 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Waisy, A.S.; Alruban, A.; Al-Fahdawi, S.; Qahwaji, R.; Ponirakis, G.; Malik, R.A.; Mohammed, M.A.; Kadry, S. CellsDeepNet: A Novel Deep Learning-Based Web Application for the Automated Morphometric Analysis of Corneal Endothelial Cells. Mathematics 2022, 10, 320. https://doi.org/10.3390/math10030320
Al-Waisy AS, Alruban A, Al-Fahdawi S, Qahwaji R, Ponirakis G, Malik RA, Mohammed MA, Kadry S. CellsDeepNet: A Novel Deep Learning-Based Web Application for the Automated Morphometric Analysis of Corneal Endothelial Cells. Mathematics. 2022; 10(3):320. https://doi.org/10.3390/math10030320
Chicago/Turabian StyleAl-Waisy, Alaa S., Abdulrahman Alruban, Shumoos Al-Fahdawi, Rami Qahwaji, Georgios Ponirakis, Rayaz A. Malik, Mazin Abed Mohammed, and Seifedine Kadry. 2022. "CellsDeepNet: A Novel Deep Learning-Based Web Application for the Automated Morphometric Analysis of Corneal Endothelial Cells" Mathematics 10, no. 3: 320. https://doi.org/10.3390/math10030320
APA StyleAl-Waisy, A. S., Alruban, A., Al-Fahdawi, S., Qahwaji, R., Ponirakis, G., Malik, R. A., Mohammed, M. A., & Kadry, S. (2022). CellsDeepNet: A Novel Deep Learning-Based Web Application for the Automated Morphometric Analysis of Corneal Endothelial Cells. Mathematics, 10(3), 320. https://doi.org/10.3390/math10030320









