Abstract
Two-dimensional gel electrophoresis (2DE) is an important and well-established technical platform enabling extensive top-down proteomic analysis. However, the long-held but now largely outdated conventional concepts of 2DE have clearly impacted its application to in-depth investigations of proteomes at the level of protein species/proteoforms. It is time to popularize a new concept of 2DE for proteomics. With the development and enrichment of the proteome concept, any given “protein” is now recognized to consist of a series of proteoforms. Thus, it is the proteoform, rather than the canonical protein, that is the basic unit of a proteome, and each proteoform has a specific isoelectric point (pI) and relative mass (Mr). Accordingly, using 2DE, each proteoform can routinely be resolved and arrayed according to its different pI and Mr. Each detectable spot contains multiple proteoforms derived from the same gene, as well as from different genes. Proteoforms derived from the same gene are distributed into different spots in a 2DE pattern. High-resolution 2DE is thus actually an initial level of separation to address proteome complexity and is effectively a pre-fractionation method prior to analysis using mass spectrometry (MS). Furthermore, stable isotope-labeled 2DE coupled with high-sensitivity liquid chromatography-tandem MS (LC-MS/MS) has tremendous potential for the large-scale detection, identification, and quantification of the proteoforms that constitute proteomes.
1. Introduction
After several attempts to improve protein resolution by combining different electrophoretic methods [1,2,3,4,5,6,7], in 1975, O’Farrell published the combination of isoelectric focusing using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [8]. This combination resulted in a breakthrough in resolution where about 1000 spots could be resolved in one gel. This combination was named two-dimensional gel electrophoresis (2DE). Until quite recently, 2DE has largely been used to resolve and array proteins according to their isoelectric points (pI) and relative masses (Mr). The separated proteins, visualized as 2DE “spots” via the use of different staining reagents, thus form a two-dimensional protein map. The difference in spot volumes of matched gel-spots between two different given conditions, such as disease versus normal controls, is used to identify total alterations in protein abundance or changes in the abundance of certain post-translational modifications (PTM) with selective stains; this was primarily based on the conventional concept that 2DE is a high-resolution analytical tool yielding only one or two proteins per spot. Thus, the spot volume represented the combined abundance of each protein species. The difference in spot volumes between matched spots was said to indicate the protein abundance alterations between two different biological situations [9,10]. Excised protein spots of interest could then be subjected to in-gel digestion with trypsin (or other proteases) and identified using analyses coupling peptide mass fingerprint (PMF) or tandem mass spectrometry (MS/MS) and available databases. This has thus been one of the main analytical strategies to address the complexity of proteomes even before the concept of the proteome appeared in 1995 [11]. With continuous improvements to the overall methodology from a number of groups over the last 40 years, 2DE is the current gold standard for the optimal resolution of proteoforms that underlie proteome complexity; 2DE is thus a critical technical platform—indeed, the most well-established approach—for detailed top-down proteomic analyses. Methodological optimization began with improvements of reproducibility and resolution, reaching 10,000 spots in controlled running conditions and with very large gels (30 cm × 40 cm) [12,13]. The development and commercialization of immobilized pH gradient (IPG) strips [14], such as Bio-Rad and GE Healthcare products, significantly improved the ease and reproducibility of the isoelectric focusing (IEF) step, helping to establish the 2DE methodology in many laboratories all over the world. The multi-gel SDS-PAGE system was introduced to significantly improve reproducibility in the second dimension of resolution [15,16]; it was also established that non-linear (NL) pH 3–10 IPG strips were generally superior to the linear gradients and that a constant gel porosity in the second dimension was better than a gradient one with respect to the separation of proteoforms in the analysis of pituitary samples [15,16]. Meanwhile, some studies demonstrated that more spots were resolved with linear gradient IPG strip in the first dimension and there was a much better resolution with a gradient gel in the second dimension in analysis of serum samples [17,18]. 2DE protocols are also being continuously and rigorously refined [19,20,21,22,23] to improve the resolution and subsequent MS-identification, including the use of prefractionation [24] and third electrophoretic separations as a post-fractionation strategy [19], optimization of detergent composition in the solubilization buffer [20,25,26,27,28,29,30,31], pre-extraction of sample handling using automated frozen disruption [21], sampling of tissue with a picosecond-infrared laser (PIRL) technology for getting a better access to the original composition of proteoforms in a tissue [32], a routine “deep imaging” approach to resolve and detect even low-abundance species with 2DE [22], and the newer Coomassie formulation stain/wash protocol and UV-detection as the highest sensitivity protocol for in-gel protein detection, providing detection of intact proteoforms in the low-to-sub-femtomole range (i.e., comparable to the detection of peptides in routine MS analyses) [23]. Indeed, these improved 2DE methods have enabled a newfound respect for the true complexity of proteomes and for the depth of analysis necessary to genuinely understand molecular mechanisms and identify the best possible biomarkers. However, such analyses must go well beyond the still widely held conventional concept that there are only one to two proteins in each 2DE spot in the analysis of the complex proteome [33,34,35]. It is time for the field of proteomics to recognize the critical complementarity of different available approaches, as already emphasized in 2004 [36]. The pros and cons of each method must be fully understood and respected.
The term protein species [37,38,39], now often also referred to as a proteoform [40], introduced the view that protein species/proteoforms rather than proteins (i.e., the canonical amino acid sequence alone) are the basic units of a proteome. Protein species is a historically grown and chemically defined term [38,39]. Proteoform is a one-word and gene-centric term, which is currently widely accepted and already often used. Despite subtle original differences in the interpretations that led to the use of the terms “protein species” and proteoform, they are now most widely accepted as essentially conceptually identical [34,35,41], or proteoform is nearly identical with protein species. The products of a single gene actually represent a set of proteoforms, each of which has a specific pI and Mr, and can thus be resolved using 2DE [34,35]. Here, the term protein species/proteoform is further clearly defined as a primary amino acid sequence + PTMs + spatial conformation + cofactors + binding partners + localization + a clearly defined function (Figure 1). Furthermore, multiple variable transcripts (e.g., splicing variants) correspond to a single gene, and each transcript can be translated into a primary amino acid sequence, each of which may then also consist of multiple proteoforms. Overall, then, proteoforms derived from the same gene can be distributed within different 2DE spots according to variations in pI and Mr, and a given spot may be expected to contain multiple proteoforms derived from different genes. A visible given spot might thus consist of many tiny spots barely resolved from each other, and certainly not visually distinguishable with current technology. Furthermore, while there have been different processes used to estimate the size of the human proteome [42,43] with values ranging from at least one million proteoforms in a given cell type [44], up to even ≈1 billion proteoforms [45], it should come as no surprise that each gel spot may contain many proteoforms with the same or very similar pI and Mr. While there has been some early evidence consistent with this state of complexity [19,44,46,47,48], a recent study has unequivocally confirmed this phenomenon [33,34], finding that every spot in a 2DE map of a complex native human proteome contains on average fifty to several hundred proteoforms. Many if not all proteins derived from the same gene are distributed within different 2DE spots, and most species in each gel spot are of low-abundance [34]. Some studies also used a cutting 2D gel pixel (10 mm × 6.66 mm × 1 mm) strategy and identified an average of five proteoforms corresponding to each gene in HePG2 cells [49] and an average of four proteoforms corresponding to each gene in glioblastoma cells [50]. These findings, made possible with high sensitivity mass spectrometers, have now completely broken through the conventional concept of only one to two proteins in each 2DE spot in the analysis of complex proteomes [35]. If stable isotope labeling, such as isobaric tags for relative and absolute quantification (iTRAQ), tandem mass tags (TMT), and stable isotope labeling of amino acids in cell culture (SILAC), are incorporated into the 2DE-LC-MS methodology, then stable isotope-labeling 2DE-LC-MS/MS is expected to be an extremely powerful approach for the large-scale detection, identification, and quantification of proteoforms [35]. An alternative approach might be 2DE-LC-MS/MS using label-free quantification; although this would require a much larger number of LC-MS/MS runs, it is devoid of the issues associated with some of the pre-labelling techniques and would be immediately implementable for any sample type.
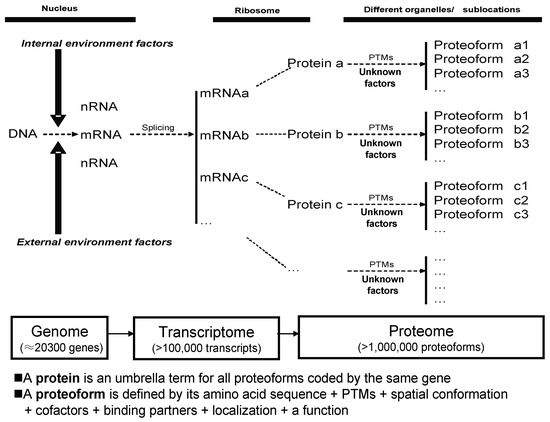
Figure 1.
Relationship of proteoform, protein, and proteome. PTM: Post-translational modifications. Modified from Zhan et al. [35], with permission from Hapres publisher open access publication, copyright 2018.
In summary 2DE-LC-MS/MS provides the most in-depth analysis of proteomes currently available. Further optimization is still possible and reasonably straightforward to achieve. Therefore, innovation in the conception and practice of 2DE, as well as in high-throughput/high-resolution LC-MSMS, will be essential to effectively address the real complexity of proteomes.
2. Definition of Spots and Pixels
Generally, a 2DE spot is a visible dot in a 2DE map, which is generated by 2DE-resolved proteoforms at a certain location (i.e., grid reference of pI and Mr). A spot is most generally visualized using stains, such as Coomassie [33] or silver [51], or using fluorescent stains, such as Sypro Ruby and Flamingo [52,53,54] (Figure 2A). Since a resolving gel has a certain thickness (commonly 1 mm), a given spot has a volume described by a Gaussian distribution, with spot height × πδxδy (OD·IU2) [16]. Spot height is the peak value of the Gaussian spot, and its unit is optical density (OD). δx is the standard deviation of the Gaussian distribution of the spot in the x-axis direction in image units (IU). δy is the standard deviation of the Gaussian distribution in the y-axis direction (also in IU). One IU = 100 micrometers (100 μm) = 0.1 mm (0.1 mm). The units of the spot volume are OD × IU2. One must note that a specific proteoform will resolve to a single very tiny spot that is resolved from others. However, any proteoforms with very similar pI and Mr will migrate to the identical position in the gel. A given visible spot on a 2DE gel can thus be understood as a tight and overlapping group of very tiny spots [35].
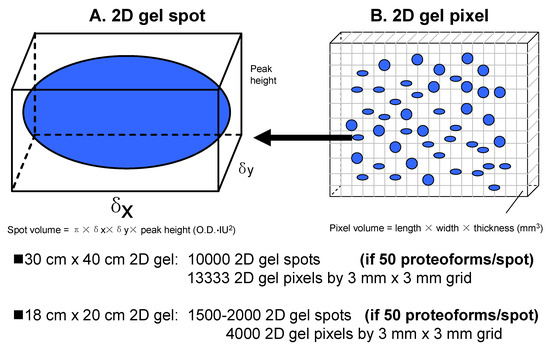
Figure 2.
The model of gel spots and gel pixels in a 2DE map. (A) The model of a 2D gel spot; (B) The model of a 2D gel pixel.
Every protein staining method has its own sensitivity (i.e., limit of detection) [23,42,55,56,57]. Spot detection thus correlates directly with the total amount of proteoforms present and the optical density of a spot does not give any information about the amount of any of the single components. In reality, the dynamic abundance range of proteoforms is very large, ≈12 orders of magnitude [58,59]. Considering the inherent complexity of proteomes, detection sensitivity, as well as the theoretical plates of separation for a given gel, it can be expected that spot-free regions of the gel contain rare proteoforms because many low-abundance proteoforms do not reach the detection limit of the protein-staining methods used. Therefore, one might cut the 2DE gel into multiple different pixel-sized pieces using a grid (e.g., 3 mm × 3 mm) for subsequent MS analysis [60] (Figure 2B). A gel pixel has a volume, with length (e.g., 3 mm) × width (e.g., 3 mm) × thickness (commonly 1 mm). Of course, the pixel size should be optimized according to the workload and detection sensitivity of the available 2DE-LC-MS/MS platform. Here, we initially assume a pixel size of 3 mm × 3 mm × 1 mm; it also might be 1 mm × 1 mm × 1 mm, 2 mm × 2 mm × 1 mm, 3 mm × 5 mm × 1 mm, or other sizes. For example, a big pixel size 10 mm × 6.66 mm × 1 mm (a total of 96 sections from a 2D gel 8 cm × 8 cm × 1 mm) was used to analyze HePG2 cells with the achievement of 20,462 proteoforms encoded by 3774 genes [49], and glioblastoma cells with the achievement of 16,012 proteoforms encoded by 4050 genes [50]. A pixel contains all proteoforms with a very similar pI and Mr in a given grid area [61,62]. Such a vision of a molecular scanner [60] was not realized until now because even with the available LC-MS techniques, this would be a very complex, time-consuming, and expensive cataloging exercise, but is ultimately necessary if we are aiming to decipher the proteome at the level of proteoforms.
3. Relationship between Proteoform, Protein, and Proteome
The apparent completion of the analysis of the human genome [63] has resulted in the identification of about 20,300 genes (although there are caveats to this estimate [64,65,66]), and has driven researchers into the era of transcriptomics and proteomics to study phenotypes and their potential linkage back to the genome. Due to RNA splicing and other factors, many transcripts are often derived from a single gene [67,68,69]; thus, at least 100,000 transcripts are contained in the human transcriptome [44,70]. Many of the newly synthesized proteins, after leaving the ribosome, are then processed, e.g., by proteolytic cleavage of a signal peptide and modified by one or more post-translational modifications (PTMs); at this point there are more than 400 known PTMs [70]. After being modified, the proteoform reaches its specific subcellular location and selectively interacts with surrounding molecules, sometimes engaging in a complex formation, to carry out its specific biological function. This often transient final structural and functional form of a protein is a protein species/proteoform [38,39,40]. Thus, a protein is in reality a set of proteoforms [35]. Each proteoform is the specific and ultimate form of a protein. Proteoforms, but not proteins, are the basic units of any proteome, and are the long-range final functional effectors of a gene (Figure 1). Given sufficient abundance, each proteoform, with its specific pI and Mr, can be resolved and detected using 2DE, and identified with coupled MS/MS.
4. A 2DE Spot Contains Many Proteoforms Derived from Different Genes
Many proteoforms with very similar pI and Mr exist in proteomes and can comigrate into a 2DE pixel. Different proteoforms in a single 2DE pixel usually have significant abundance differences. With the use of high sensitivity mass spectrometers, these low-abundance proteoforms can be detected, identified, and quantified. A recent study experimentally confirmed this principle, where every 2DE spot contained about 50 proteoforms on average, sometimes several hundred of them at maximum [33,34] (Figure 3). This completely breaks through the conventional concept of assuming only one to two proteins (which in reality are different proteoforms) in each gel spot in a 2DE map, although, in some gel spots of a 2DE map, several proteins had already been found [19,46,48,50,61]. In 2013, Thiede et al. [47] reported that 50% of the spots contained more than one protein (most spots: 2–3 proteins/spot, and only one spot: 22 proteins/spot). In 2015, the Zhan research group first reported that every identified spot contained over fifty proteoforms [33], and then in 2018, confirmed that every 2DE spot analyzed contained over fifty and even up to several hundred proteoforms [34]. Together, these results completely changed the traditional concept of 2DE for proteome analysis, which was addressed in detail in 2018 [35].
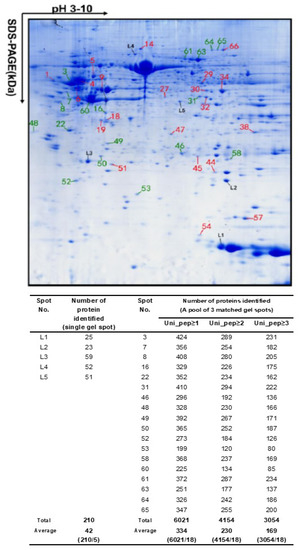
Figure 3.
An average of over 50 to several hundred protein species were identified in every two-dimensional gel electrophoresis (2DE) spot of the human glioblastoma proteome. The glioblastoma proteome was resolved using 18 cm pH 3–10 non-linear immobilized pH gradient gel (NL IPG) strips and 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in the second dimension, and stained with Coomassie Blue. Spots L1–L5 came from one gel for tandem mass spectrometry (MS/MS) analysis. Each spot labeled with a red or green number was combined from three matched spots from parallel gels for MS/MS analysis. Modified from Zhan X. et al. [34] with permission from Wiley-VCH.
5. Proteoforms from One Gene are Distributed within Different 2DE Spots
Many proteoforms arising from the same gene, but each with a specific pI and Mr, are expected to be resolved into different 2DE spots [71,72,73,74]. For example, (i) 24 human growth hormone (hGH) proteoforms (each GH proteoform identified with more than 3 peptides) derived from the same GH gene are present in 24 different spots in the 2DE map of the normal human pituitary proteome (Figure 4) [71]; (ii) 6 human prolactin (hPRL) proteoforms derived from the same hPRL gene are present in 6 different spots in the pituitary proteome 2DE map [72]; (iii) 59 HSP27 proteoforms derived from the same HSP27 gene are present in 59 different gel spots in the 2DE map of the human myocardium proteome [73]; (iv) 52 HSP70 proteoforms derived from the same HSP70 gene, 24 gamma-enolase-2 proteoforms derived from the same gamma enolase-2 gene, and 17 lactate dehydrogenase 2B proteoforms derived from the lactate dehydrogenase 2B gene are all present in different gel spots in the 2DE maps of the mouse brain proteome [74]; and (v) 28 and 29 proteoforms of lamin A/C and vimentin, respectively, have been identified in a 2DE map of Hela cells [47].
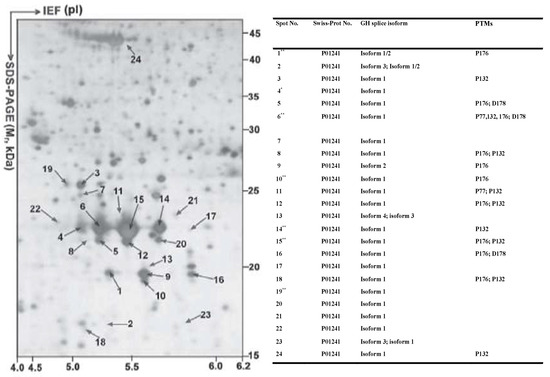
Figure 4.
A total of 24 hGH proteoforms are found in the 2DE map of the human pituitary proteome. The human pituitary proteome was resolved as described in Figure 3 and silver stained. IEF: isoelectric focusing. Modified from Zhan X. et al. [71] with permission from Wiley-VCH.
6. Most Proteoforms in a 2DE Spot are of Low-Abundance
With an upper estimate of over 1 million proteoforms in a given cell type of the human organism [44], the dynamic range of these proteoforms is estimated to be up to 1012 [58,59]. The deep-dive study confirmed that most proteoforms are of low-abundance among the about fifty-to-several hundred protein species found in a single given 2DE spot (Table 1) [34]. The important correlate of this is that that species most easily identified (i.e., having the best sequence coverage) is also the most abundant in any given spot and thus also most likely to be the species changing in abundance when differences in spot density are assessed (i.e., between experimental conditions) [47,74]. Overall, the results thus clearly demonstrate that 2DE in combination with LC-MS/MS is a robust, state-of-the-art, top-down approach to resolve and detect low-abundance proteoforms in complex proteomes, but also to carry out routine comparative proteomic analyses.

Table 1.
Most of proteoforms in a 2D gel spot have a low abundance or extremely low abundance in the analysis of the human glioblastoma proteome.
7. Stable Isotope-Labeled 2DE-LC-MS/MS for the Large-Scale Analysis of Proteoforms
Having established that 2DE-LC-MS/MS is the most effective available method to address the sheer complexity of proteomes via the large-scale resolution, detection, and identification of intact proteoforms in the analysis of the complex proteome [34], what are the current limits of the approach without additional refinements? Approximately 10,000 2DE spots can be resolved using a 40 cm tube gel in the first dimension of IEF separation and a 40 cm × 30 cm gel in the second dimension of separation [12], or 13,333 gel pixels can be obtained using a 3 mm × 3 mm grid for such a gel. Even for an 18 cm × 20 cm 2DE gel with an 18 cm pH 3–10 nonlinear (NL) IPG strip, commonly 1500–2000 2DE spots or 4000 gel pixels (3 mm × 3 mm grid) are routinely achievable (Figure 2). Similarly, capitalizing on more recent refinements to a 2DE protocol, including separate resolutions for the soluble and membrane sub-proteomes, high-sensitivity near-IR detection of proteoforms, 3D separations of pI extremes and regions of protein hyper-abundance, and deep imaging, a total of 2500–3000 spots were detected using a mini-gel format (i.e., 7 cm IPG strips and 82 mm × 58 mm × 1 mm SDS-PAGE); using a conservative estimate of 50 intact proteoforms per spot [34], this indicates a total proteoform resolution of ≈125,000–150,000 species, which is many times the resolution of any other currently available analytical approach. Estimates of proteoform resolution using large format 2DE thus extend from approximately 500,000 to a million or more proteoforms, values that are unlikely to be achieved by approaches using bottom-up only (which nonetheless do not routinely identify proteoforms) or alternate “in-line MS” top-down analyses that still require front-end separations, deal poorly with hydrophobic proteins, and do not permit any large-scale analysis of proteoforms larger than ≈40 kDa [75,76,77,78,79,80]. The best published detection using capillary zone electrophoresis and in-line top-down MS is 5700 proteoforms from 860 proteins [81,82]. Nonetheless, both the refinements to the 2DE protocol (i.e., 3D separations and deep imaging) in combination with subsequent high-sensitivity bottom-up MS/MS (LC-MS/MS) (i.e., OrbiTrap mass spectrometers) provide for the routine identification of a much larger number of proteoforms at very different levels of abundance (Figure 5). Thus, even considering the important technical advancements contributed by bottom-up approaches over the last decade, 2DE-LC-MS/MS is opening a new horizon that is central to the necessary deep analysis of proteomes at the level of proteoforms.
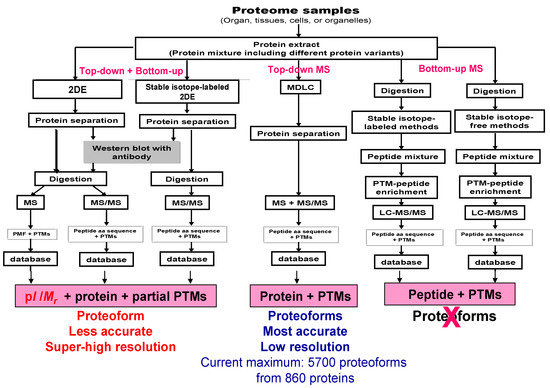
Figure 5.
Comparison of methods to study proteoforms. 2DE is a top-down analysis. MS/MS analysis after enzymatic digestion is a “selective” bottom-up analysis. 2DE—two-dimensional gel electrophoresis, MDLC—multiple dimensional liquid chromatography, MS—mass spectrometry, MS/MS—tandem mass spectrometry, PTM—post-translational modification, LC—liquid chromatography, PMF—peptide mass fingerprint. Modified from Zhan X. et al. [70] with permission from Elsevier publisher open access publication, copyright 2018.
To ensure the best possible quantitative proteome coverage and the fullest possible sequence and PTM coverage of any given proteoforms, in order to link specific proteoforms to specific biological roles in different physiological or pathological states, further refinements are necessary. The most immediately obvious again capitalize on the complementarity of available methods. The first, quantitative approach would be to quantify each proteoform using stable isotope-labeling of native proteomes before 2DE-LC-MS/MS analysis [35]. For example, SILAC reagents can be used to label all the proteoforms in cultured cells under different conditions, followed by 2DE-LC-MS/MS analysis (Figure 5) [47]. An alternative method, more broadly applicable to the range of sample types encountered in proteomic analyses (i.e., tissues, body fluids), might be stable isotope-labeling of the extracted proteoforms [83,84,85,86], again followed by 2DE-LC-MS/MS analysis (Figure 5 and see below).
One must, however, realize that while SILAC-2DE-LC-MS/MS works well in cell culture [47], it is not applicable to tissue extracts or biological fluids. Alternatively, commercially available isobaric tags (iTRAQ or TMT) [83,84,85,86] or isotope-coded affinity tags (ICAT) can be used to label protein amine groups (i.e., the N-terminus and lysine side-chains) or cysteines, respectively, followed by 2DE-LC-MS/MS analysis [83,84,85,86,87]. However, currently, caveats regarding labeling conditions and the potential for non-quantitative labeling of species must be taken into account; in this regard, such deep proteome analyses may be the most effective test of how such reactive labeling approaches actually quantitatively represent the full dynamic range of native proteomes (i.e., are very low abundance species represented as effectively as high abundance species or is such labeling dominated by the law of mass action?). Isotope-free methods are another choice to quantify proteins with 2DE-LC-MS/MS, including label-free methods [75,88], selected reaction monitoring (SRM) [89], and sequential window acquisition of all theoretical mass spectra (SWATH) [90]. Label-free quantitative mass spectrometry (LFQ-MS) may be a good option with data-dependent acquisition (DDA) [91,92] to compare changes in the abundance of a specific proteoform between two different treatments or physiological conditions; this is perhaps the most straightforward of approaches considering the prior resolution of proteoforms by 2DE. However, for such an approach to be fully effective, new very complex algorithms will be needed considering the multiple dimensions of resolution (i.e., 2DE dimension, LC-MS dimension, and MS/MS dimension). Nevertheless, such an approach would best capitalize on the complementarity of available analytical methods, such as data-independent acquisition (DIA)-based SWATH methods [90].
8. Conclusions
Proteoforms are the basic units of the proteome, the critical players defining molecular mechanisms, and the best possible biomarkers; they are thus of central importance across the spectrum of biological, biomedical, agricultural, and environmental research [35,41,44,76,93]. 2DE has undergone significant innovations, both in practice and concept, and its recent further state-of-the-art combination with LC-MS/MS analyses of the highest available sensitivity has firmly established 2DE-LC-MS/MS as the most rigorous approach to deep, quantitative proteome analysis at the proteoform level. The applicability of SILAC-2DE-LC/MS has already been shown in the search for proteoforms changing in abundance during apoptosis [47]. In the future, such routine quantitative analyses will be further complemented using established labelled and label-free approaches. 2DE-based Western blot with a given protein antibody coupled with LC-MS/MS can also effectively reduce the number of 2DE-derived gel samples to be analyzed with LC-MS/MS as it can detect proteoforms of a given protein in a 2DE matrix [33,72,94]. Overall, the beauty and power of this coupled 2DE-LC-MS/MS methodology is its inherent capacity to capitalize on the combination of refined top-down and bottom-up analytical approaches. The capacity for genuine deep proteome analysis at the critical proteoform level is now a definite reality.
Author Contributions
X.Z. (Xianquan Zhan) conceived the concept, collected and analyzed references, designed and made the figures and tables, wrote and critically revised the manuscript, and was responsible for the financial support and corresponding works. B.L. participated in the collection of references and preparation of figures. X.Z. (Xiaohan Zhan) participated in the collection and analysis of references, and assisted in the editing of the English language. J.R.C., H.S., and P.R.J. assisted in the further development of the ideas, integration of critical research literature, and manuscript editing. All authors approved the final manuscript.
Funding
X.Z. (Xianquan Zhan) was supported by the Hunan Provincial Hundred Talent Plan (to X.Z.), the Xiangya Hospital Funds for Talent Introduction (to X.Z.), the China “863” Plan Project (Grant No. 2014AA020610-1 to X.Z.), the National Natural Science Foundation of China (Grant No. 81272798 and 81572278 to X.Z.), and the Hunan Provincial Natural Science Foundation of China (Grant No. 14JJ7008 to X.Z.). J.R.C. acknowledges the support of the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN-2019-04324).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| 2DE | two-dimensional gel electrophoresis |
| DDA | data-dependent acquisition |
| DIA | data-independent acquisition |
| hGH | human growth hormone |
| hPRL | human prolactin |
| IEF | isoelectric focusing |
| IPG | immobilized pH gradient gel |
| ICAT | isotope-coded affinity tags |
| iTRAQ | isobaric tags for relative and absolute quantification |
| IU | image unit |
| LC | liquid chromatography |
| Mr | relative molecular mass |
| MS | mass spectrometry |
| MS/MS | tandem mass spectrometry |
| OD | optical density |
| pI | isoelectric point |
| PTM | post-translational modification |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SILAC | stable isotope labeling of amino acids in cell culture |
| SRM | selected reaction monitoring |
| SWATH | sequential window acquisition of all theoretical mass spectra |
| TMT | tandem mass tags |
References
- Smithies, O.; Poulik, M.D. Two-dimensional electrophoresis of serum proteins. Nature 1956, 177, 1033. [Google Scholar] [CrossRef] [PubMed]
- Raymond, S. Acrylamide gel electrophoresis. Ann. N. Y. Acad. Sci. 1964, 121, 350–365. [Google Scholar] [CrossRef] [PubMed]
- Kaltschmidt, E.; Wittmann, H.G. Ribosomal proteins. VII. Two-dimensional polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal. Biochem. 1970, 36, 401–412. [Google Scholar] [CrossRef]
- MacGillivray, A.J.; Rickwood, D. The heterogeneity of mouse-chromatin nonhistone proteins as evidenced by two-dimensional polyacrylamide-gel electrophoresis and ion-exchange chromatography. Eur. J. Biochem. 1974, 41, 181–190. [Google Scholar] [CrossRef]
- Klose, J. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 1975, 26, 231–243. [Google Scholar]
- Scheele, G.A. Two-dimensional gel analysis of soluble proteins. Charaterization of guinea pig exocrine pancreatic proteins. J. Biol. Chem. 1975, 250, 5375–5385. [Google Scholar]
- Iborra, F.; Buhler, J.M. Protein subunit mapping. A sensitive high resolution method. Anal. Biochem. 1976, 74, 503–511. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar]
- Zhan, X.; Desiderio, D.M. Heterogeneity analysis of the human pituitary proteome. Clin. Chem. 2003, 49, 1740–1751. [Google Scholar] [CrossRef]
- Zhan, X.; Desiderio, D.M.; Wang, X.; Zhan, X.; Guo, T.; Li, M.; Peng, F.; Chen, X.; Yang, H.; Zhang, P.; et al. Identification of the proteomic variations of invasive relative to non-invasive non-functional pituitary adenomas. Electrophoresis 2014, 35, 2184–2194. [Google Scholar]
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Poljak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Klose, J.; Kobalz, U. Two-dimensional electrophoresis of proteins: An updated protocol and implications for a functional analysis of the genome. Electrophoresis 1995, 16, 1034–1059. [Google Scholar] [CrossRef] [PubMed]
- Zimny-Arndt, U.; Schmid, M.; Ackermann, R.; Jungblut, P.R. Classical proteomics: Two-dimensional electrophoresis/MALDI mass spectrometry. Methods Mol. Biol. 2009, 492, 65–91. [Google Scholar] [PubMed]
- Görg, A.; Obermaier, C.; Boguth, G.; Harder, A.; Scheibe, B. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 2000, 21, 1037–1053. [Google Scholar] [CrossRef]
- Zhan, X.; Desiderio, D.M. Differences in the spatial and quantitative reproducibility between two second-dimensional gel electrophoresis systems. Electrophoresis 2003, 24, 1834–1846. [Google Scholar] [CrossRef]
- Zhan, X.; Desiderio, D.M. Spot volume vs. amount of protein loaded onto a gel: A detailed, statistical comparison of two gel electrophoresis systems. Electrophoresis 2003, 24, 1818–1833. [Google Scholar] [CrossRef]
- D’Silva, A.M.; Hyett, J.A.; Coorssen, J.R. A routine ‘top-down’ approach to analysis of the human serum proteome. Proteomes 2017, 5, 13. [Google Scholar] [CrossRef]
- D’Silva, A.M.; Hyett, J.A.; Coorssen, J.R. Proteomic analysis of first trimester maternal serum to identify candidate biomarkers potentially predictive of spontaneous preterm birth. J. Proteom. 2018, 178, 31–42. [Google Scholar] [CrossRef]
- Butt, R.H.; Coorssen, J.R. Postfractionation for enhanced proteomic analyses: Routine electrophoretic methods increase the resolution of standard 2D-PAGE. J. Proteome Res. 2005, 4, 982–991. [Google Scholar] [CrossRef]
- Churchward, M.A.; Butt, R.H.; Lang, J.C.; Hsu, K.K.; Coorssen, J.R. Enhanced detergent extraction for analysis of membrane proteomes by two-dimensional gel electrophoresis. Proteome Sci. 2005, 3, 5. [Google Scholar] [CrossRef]
- Butt, R.H.; Coorssen, J.R. Pre-extraction sample handling by automated frozen disruption significantly improves subsequent proteomic analyses. J. Proteome Res. 2006, 5, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.P.; Prasad, K.A.; Padula, M.P.; Coorssen, J.R. Deep imaging: How much of the proteome does current top-down technology already resolve? PLoS ONE 2014, 9, e86058. [Google Scholar] [CrossRef] [PubMed]
- Noaman, N.; Abbineni, P.S.; Withers, M.; Coorssen, J.R. Coomassie staining provides routine (sub)femtomole in-gel detection of intact proteoforms: Expanding opportunities for genuine top-down proteomics. Electrophoresis 2017, 38, 3086–3099. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, P.; Klose, J. Composition and genetic variability of Heparin-Sepharose CL-6B protein fractions obtained from the solubilized proteins of mouse organs. Biochem. Genet. 1986, 24, 925–939. [Google Scholar] [CrossRef]
- Santos, F.M.; Albuquerque, T.; Gaspar, L.M.; Dias, J.M.L.; Castro, E.; Sousa, J.P.; Paradela, A.; Tomaz, C.T.; Passarinha, L.A. Refinement of two-dimensional electrophoresis for vitreous proteome profiling using an artificial neural network. Anal. Bioanal. Chem. 2019, 411, 5115–5126. [Google Scholar] [CrossRef] [PubMed]
- Posch, A. Sample preparation guidelines for two-dimensional electrophoresis. Arch. Physiol. Biochem. 2014, 120, 192–197. [Google Scholar] [CrossRef]
- Kočevar, N.; Grazio, S.F.; Komel, R. Two-dimensional gel electrophoresis of gastric tissue in an alkaline pH range. Proteomics. 2014, 14, 311–321. [Google Scholar] [CrossRef]
- Moche, M.; Albrecht, D.; Maaß, S.; Hecker, M.; Westermeier, R.; Büttner, K. The new horizon in 2D electrophoresis: New technology to increase resolution and sensitivity. Electrophoresis 2013, 34, 1510–1518. [Google Scholar] [CrossRef]
- Zabel, C.; Klose, J. High-resolution large-gel 2DE. Methods Mol. Biol. 2009, 519, 311–338. [Google Scholar]
- Afjehi-Sadat, L.; Lubec, G. Identification of enzymes and activity from two-dimensional gel electrophoresis. Nat. Protoc. 2007, 2, 2318–2324. [Google Scholar] [CrossRef]
- Khoudoli, G.A.; Porter, I.M.; Blow, J.J.; Swedlow, J.R. Optimisation of the two-dimensional gel electrophoresis protocol using the Taguchi approach. Proteome Sci. 2004, 2, 6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kwiatkowski, M.; Wurlitzer, M.; Krutilin, A.; Kiani, P.; Nimer, R.; Omidi, M.; Mannaa, A.; Bussmann, T.; Bartkowiak, K.; Kruber, S.; et al. Homogenization of tissues via picosecond-infrared laser (PIRL) ablation: Giving a closer view on the in-vivo composition of protein species as compared to mechanical homogenization. J. Proteom. 2016, 134, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Li, J.; Guo, T.; Yang, H.; Li, M.; Sang, S.; Li, X.; Desiderio, D.M.; Zhan, X. Nitroproteins in human astrocytomas discovered by gel electrophoresis and tandem mass spectrometry. J. Am. Soc. Mass Spectrum. 2015, 26, 2062–2076. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Yang, H.; Peng, F.; Li, J.; Mu, Y.; Long, Y.; Cheng, T.; Huang, Y.; Li, Z.; Lu, M.; et al. How many proteins can be identified in a 2-DE gel spot within an analysis of a complex human cancer tissue proteome? Electrophoresis 2018, 39, 965–980. [Google Scholar] [CrossRef]
- Zhan, X.; Li, N.; Zhan, X.; Qian, S. Revival of 2DE-LC/MS in proteomics and its potential for large-scale study of human proteoforms. Med One 2018, 3, e180008. [Google Scholar]
- Schmidt, F.; Donahoe, S.; Hagens, K.; Mattow, J.; Schaible, U.E.; Kaufmann, S.H.E.; Aebersold, R.; Jungblut, P.R. Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope coded affinity tag technology. Mol. Cell. Proteom. 2004, 3, 24–42. [Google Scholar] [CrossRef]
- Jungblut, P.; Thiede, B.; Zimny-Arndt, U.; Müller, E.C.; Scheler, C.; Wittmann-Liebold, B.; Otto, A. Resolution power of two-dimensional electrophoresis and identification of proteins from gels. Electrophoresis 1996, 17, 839–847. [Google Scholar] [CrossRef]
- Jungblut, P.R.; Holzhütter, H.G.; Apweiler, R.; Schlüter, H. The speciation of the proteome. Chem. Cent. J. 2008, 2, 16. [Google Scholar] [CrossRef]
- Schlüter, H.; Apweiler, R.; Holzhütter, H.G.; Jungblut, P.R. Finding one’s way in proteomics: A protein species nomenclature. Chem. Cent. J. 2009, 3, 11. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L. Consortium for Top Down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Coorssen, J.R.; Yergey, A.L. Proteomics is analytical chemistry: Fitness-for-purpose in the application of top-down and bottom-up analyses. Proteomes 2015, 3, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N.; Lisitsa, A.V.; Zgoda, V.G.; Ponomarenko, E.A.; Archakov, A.I. 2DE-based approach for estimation of number of protein species in a cell. Electrophoresis 2014, 35, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The size of the human proteome: The width and depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef]
- Kelleher, N.L. A cell-based approach to the human proteome project. J. Am. Soc. Mass Spectrom. 2012, 23, 1617–1624. [Google Scholar] [CrossRef]
- Campostrini, N.; Areces, L.B.; Rappsilber, J.; Pietrogrande, M.C.; Dondi, F.; Pastorino, F.; Ponzoni, M.; Righetti, P.G. Spot overlapping in two-dimensional maps: A serious problem ignored for much too long. Proteomics 2005, 5, 2385–2395. [Google Scholar] [CrossRef]
- Thiede, B.; Koehler, C.J.; Strozynski, M.; Treumann, A.; Stein, R.; Zimny-Arndt, U.; Schmid, M.; Jungblut, P.R. High resolution quantitative proteomics of HeLa cells protein species using stable isotope labeling with amino acids in cell culture(SILAC), two-dimensional gel electrophoresis(2DE) and nano-liquid chromatograpohy coupled to an LTQ-OrbitrapMass spectrometer. Mol. Cell. Proteom. 2013, 12, 529–538. [Google Scholar]
- Lim, H.; Eng, J.; Yates, J.R., 3rd; Tollaksen, S.L.; Giometti, C.S.; Holden, J.F.; Adams, M.W.; Reich, C.I.; Olsen, G.J.; Hays, L.G. Identification of 2D-gel proteins: A comparison of MALDI/TOF peptide mass mapping to mu LC-ESI tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2003, 14, 957–970. [Google Scholar] [CrossRef]
- Naryzhny, S.N.; Maynskova, M.A.; Zgoda, V.G.; Ronzhina, N.L.; Kleyst, O.A.; Vakhrushev, I.V.; Archakov, A.I. Virtual-experimental 2DE approach in chromosome-centric human proteome project. J. Proteome Res. 2016, 15, 525–530. [Google Scholar] [CrossRef]
- Naryzhny, S.N.; Maynskova, M.A.; Zgoda, V.G.; Ronzhina, N.L.; Novikova, S.E.; Belyakova, N.V.; Kleyst, O.A.; Legina, O.K.; Pantina, R.A. Proteomic profiling of highgrade glioblastoma using virtual-experimental 2DE. J. Proteom. Bioinform. 2016, 9, 158–165. [Google Scholar] [CrossRef]
- Zhan, X.; Desiderio, D.M. A reference map of a human pituitary adenoma proteome. Proteomics 2003, 3, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhan, X.; Li, M.; Li, G.; Zhang, P.; Xiao, Z.; Shao, M.; Peng, F.; Hu, R.; Chen, Z. Mitochondrial proteomics of nasopharyngeal carcinoma metastasis. BMC Med. Genom. 2012, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Arentz, G.; Weiland, F.; Oehler, M.K.; Hoffmann, P. State of the art of 2D DIGE. Proteom. Clin. Appl. 2015, 9, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Centeno, D.; Rofidal, V.; Tauzin, M.; Martin, O.; Sonamerer, N.; Rossignol, M. Different impact of staining procedures using visible stains and fluorescent dyes for large-scale investigation of proteomes by MALDI-TOF mass spectrometry. J. Proteome Res. 2006, 5, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Gauci, V.J.; Padula, M.P.; Coorssen, J.R. Coomassie blue staining for high sensitivity gel-based proteomics. J. Proteom. 2013, 90, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Liew, Y.K.; Neela, V.; Hamat, R.A.; Nordin, S.A.; Chong, P.P. Modified silver staining in 2DE improves protein detection even at extremely low sample concentration. Electrophoresis 2013, 34, 397–400. [Google Scholar] [CrossRef]
- Gauci, V.J.; Wright, E.P.; Coorssen, J.R. Quantitative proteomics: Assessing the spectrum of in-gel protein detection methods. J. Chem. Biol. 2011, 4, 3–29. [Google Scholar] [CrossRef]
- Swindell, W.R.; Remmer, H.A.; Sarkar, M.K.; Xing, X.; Barnes, D.H.; Wolterink, L.; Voorhees, J.J.; Nair, R.P.; Johnston, A.; Elder, J.T.; et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015, 7, 86. [Google Scholar] [CrossRef]
- Mehdi, A.M.; Patrick, R.; Bailey, T.L.; Bod’en, M. Predicting the dynamics of protein abundance. Mol. Cell. Proteom. 2014, 13, 1330–1340. [Google Scholar] [CrossRef]
- Bienvenut, W.V.; Sanchez, J.C.; Karmime, A.; Rouge, V.; Rose, K.; Binz, P.A.; Hochstrasser, D.F. Toward a clinical molecular scanner for proteome research: Parallel protein chemical processing before and during western blot. Anal. Chem. 1999, 71, 4800–4807. [Google Scholar] [CrossRef]
- Naryzhny, S. Inventory of proteoforms as a current challenge of proteomics: Some technical aspects. J. Proteom. 2019. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N. Towards the Full Realization of 2DE Power. Proteomes 2016, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Roest Crollius, H.; Jaillon, O.; Bernot, A.; Dasilva, C.; Bouneau, L.; Fischer, C.; Fizames, C.; Wincker, P.; Brottier, P.; Quétier, F.; et al. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet. 2000, 25, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Ewing, B.; Green, P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat. Genet. 2000, 25, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Juan, D.; Jungreis, I.; Kellis, M.; Martinez, L.; Rigau, M.; Rodriguez, J.M.; Vazquez, J.; Tress, M.L. Loose ends: Almost one in five human genes still have unresolved coding status. Nucleic Acids Res. 2018, 46, 7070–7084. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.; Seilhamer, J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997, 18, 533–537. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [CrossRef]
- Zhan, X.; Long, Y.; Zhan, X.; Mu, Y. Consideration of statistical vs. biological significances for omics data-based pathway network analysis. Med One 2017, 1, e170002. [Google Scholar]
- Zhan, X.; Long, Y.; Lu, M. Exploration of variations in proteome and metabolome for predictive diagnostics and personalized treatment algorithms: Innovative approach and examples for potential clinical application. J. Proteom. 2018, 188, 30–40. [Google Scholar] [CrossRef]
- Zhan, X.; Giorgianni, F.; Desiderio, D.M. Proteomics analysis of growth hormone isoforms in the human pituitary. Proteomics 2005, 5, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Yang, Y.; Li, N.; Cheng, T.; Wang, X.; Liu, J.; Li, X.; Desiderio, D.M.; Zhan, X. Prolactin variants in human pituitaries and pituitary adenomas identified with two-dimensional gel electrophoresis and mass spectrometry. Front. Endocrinol. 2018, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Scheler, C.; Müller, E.C.; Stahl, J.; Müller-Werdan, U.; Salnikow, J.; Jungblut, P.R. Identification and characterization of heat shock protein 27 protein species in human myocardial two-dimensional electrophoresis patterns. Electrophoresis 1997, 18, 2823–2831. [Google Scholar] [CrossRef] [PubMed]
- Klose, J.; Nock, C.; Herrmann, M.; Stühler, K.; Marcus, K.; Blüggel, M.; Krause, E.; Schalkwyk, L.C.; Rastan, S.; Brown, S.D.; et al. Genetic analysis of the mouse brain proteome. Nat. Genet. 2002, 30, 385–393. [Google Scholar] [CrossRef]
- Szabo, Z.; Szomor, J.S.; Foeldi, I.; Janaky, T. Mass spectrometry-based label free quantification of gel separated proteins. J. Proteom. 2012, 75, 5544–5553. [Google Scholar] [CrossRef]
- Schaffer, L.V.; Millikin, R.J.; Miller, R.M.; Anderson, L.C.; Fellers, R.T.; Ge, Y.; Kelleher, N.L.; LeDuc, R.D.; Liu, X.; Payne, S.H.; et al. Identification and quantification of proteoforms by mass spectrometry. Proteomics 2019, 19, e1800361. [Google Scholar] [CrossRef]
- Li, H.; Wongkongkathep, P.; Van Orden, S.L.; Ogorzalek Loo, R.R.; Loo, J.A. Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J. Am. Soc. Mass Spectrom. 2014, 25, 2060–2068. [Google Scholar] [CrossRef]
- Anderson, L.C.; DeHart, C.J.; Kaiser, N.K.; Fellers, R.T.; Smith, D.F.; Greer, J.B.; LeDuc, R.D.; Blakney, G.T.; Thomas, P.M.; Kelleher, N.L.; et al. Identification and characterization of human proteoforms by top-down LC-21 Tesla FT-ICR mass spectrometry. J. Proteome Res. 2017, 16, 1087–1096. [Google Scholar] [CrossRef]
- Fornelli, L.; Durbin, K.R.; Fellers, R.T.; Early, B.P.; Greer, J.B.; LeDuc, R.D.; Compton, P.D.; Kelleher, N.L. Advancing top-down analysis of the human proteome using a benchtop quadrupole-Orbitrap mass spectrometer. J. Proteome Res. 2017, 16, 609–618. [Google Scholar] [CrossRef]
- Fornelli, L.; Srzentić, K.; Huguet, R.; Mullen, C.; Sharma, S.; Zabrouskov, V.; Fellers, R.T.; Durbin, K.R.; Compton, P.D.; Kelleher, N.L. Accurate sequence analysis of a monoclonal antibody by top-down and middle-down Orbitrap mass spectrometry applying multiple ion activation techniques. Anal. Chem. 2018, 90, 8421–8429. [Google Scholar] [CrossRef]
- McCool, E.N.; Lubeckyj, R.A.; Shen, A.; Chen, D.; Kou, Q.; Liu, X.; Sun, L. Deep top-down proteomics using capillary zone electrophoresis-tandem mass spectrometry: Identification of 5700 proteoforms from the Escherichia coli proteome. Anal. Chem. 2018, 90, 5529–5533. [Google Scholar] [CrossRef] [PubMed]
- McCool, E.N.; Lubeckyj, R.; Shen, X.; Kou, Q.; Liu, X.; Sun, L. Large-scale top-down proteomics using capillary zone electrophoresis tandem mass spectrometry. J. Vis. Exp. 2018, 140, e58644. [Google Scholar] [CrossRef] [PubMed]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Prudova, A.; auf dem Keller, U.; Butler, G.S.; Overall, C.M. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteom. 2010, 9, 894–911. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, J.; Timms, J.F. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS. Methods 2011, 54, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.W.; Tholey, A. Tandem mass tag protein labeling for top-down identification and quantification. Anal. Chem. 2012, 84, 161–170. [Google Scholar] [CrossRef]
- Gygi, S.P.; Rist, B.; Gerber, S.A.; Turecek, F.; Gelb, M.H.; Aebersold, R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999, 17, 994–999. [Google Scholar] [CrossRef]
- Blein-Nicolas, M.; Zivy, M. Thousand and one ways to quantify and compare protein abundances in label-free bottom-up proteomics. Biochim. Biophys. Acta Proteins Proteom. 2015, 1864, 883–895. [Google Scholar] [CrossRef]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 222. [Google Scholar] [CrossRef]
- Aebersold, R.; Bensimon, A.; Collins, B.C.; Ludwig, C.; Sabido, E. Applications and developments in targeted proteomics: From SRM to DIA/SWATH. Proteomics 2016, 16, 2065–2067. [Google Scholar] [CrossRef]
- Autelitano, F.; Loyaux, D.; Roudières, S.; Déon, C.; Guette, F.; Fabre, P.; Ping, Q.; Wang, S.; Auvergne, R.; Badarinarayana, V.; et al. Identification of novel tumor-associated cell surface sialoglycoproteins in human glioblastoma tumors using quantitative proteomics. PLoS ONE 2014, 9, e110316. [Google Scholar] [CrossRef] [PubMed]
- Schelletter, L.; Albaum, S.; Walter, S.; Noll, T.; Hoffrogge, R. Clonal variations in CHO IGF signaling investigated by SILAC-based phosphoproteomics and LFQ-MS. Appl. Microbiol. Biotechnol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next proteomics currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef] [PubMed]
- Kusch, K.; Uecker, M.; Liepold, T.; Möbius, W.; Hoffmann, C.; Neumann, H.; Werner, H.B.; Jahn, O. Partial immunoblotting of 2D-Gels: A novel method to identify post-translationally modified proteins exemplified for the myelin acetylome. Proteomes 2017, 5, 3. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).