Abstract
Insulin resistance is an indication of early stage Type 2 diabetes (T2D). Insulin resistant adipose tissues contain higher levels of insulin than the physiological level, as well as higher amounts of intracellular tumor necrosis factor-α (TNF-α) and other cytokines. However, the mechanism of insulin resistance remains poorly understood. To better understand the roles played by insulin and TNF-α in insulin resistance, we performed proteomic analysis of differentiated 3T3-L1 adipocytes treated with insulin (Ins), TNF-α (TNF), and both (Ins + TNF). Out of the 693 proteins identified, the abundances of 78 proteins were significantly different (p < 0.05). Carnitine parmitoyltransferase-2 (CPT2), acetyl CoA carboxylase 1 (ACCAC-1), ethylmalonyl CoA decarboxylase (ECHD1), and methylmalonyl CoA isomerase (MCEE), enzymes required for fatty acid β-oxidation and respiratory electron transport, and β-glucuronidase, an enzyme responsible for the breakdown of complex carbohydrates, were down-regulated in all the treatment groups, compared to the control group. In contrast, superoxide dismutase 2 (SOD2), protein disulfide isomerase (PDI), and glutathione reductase, which are the proteins responsible for cytoskeletal structure, protein folding, degradation, and oxidative stress responses, were up-regulated. This suggests higher oxidative stress in cells treated with Ins, TNF, or both. We proposed a conceptual metabolic pathway impacted by the treatments and their possible link to insulin resistance or T2D.
1. Introduction
The alarmingly increasing frequency of obesity and non-insulin-dependent (type II) diabetes mellitus (T2D) in the United States poses a serious health crisis in the near future. To address this, we need to better understand the molecular factors and cellular pathways that are responsible for insulin resistance. Insulin is a dipeptide hormone secreted by pancreatic islet β cells, which is essential for maintaining normal blood glucose levels [1]. Insulin resistance is defined as the inability of a cell to maintain glucose homeostasis, respond to the physiological level of insulin [2,3], and is a characteristic condition of the early state of T2D.
Insulin lowers blood glucose levels by regulating glucose metabolism via glycolysis, gluconeogenesis, and glycogenesis pathways [4]. Glucose is converted into pyruvate or lactate via glycolysis by sequential glycolytic enzymes, including bifunctional 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1 (PFKFB1) [5]. Insulin dephosphorylates PFKFB1 to activate kinase activity and promote glycolysis. Gluconeogeneis, on the other hand, generates glucose from pyruvate or lactate, and glycogenesis converts glucose into glycogen granules. Gluconeogenesis is regulated by rate-limiting phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) [6]. Insulin is known to reduce PEPCK and G6Pase expression through serine/threonine kinase AKT signaling pathways resulting in the supression of gluconeogenesis [7,8]. Glycogenesis, under other conditions, is regulated by the level of glycogen synthase (GS) and glycogen phosphorylase (GP) [9]. Insulin promotes glycogenesis by dephosphorylation and activation of GS or dephosphorylation and inactivation of GP via the AKT pathway [10,11]. In addition, glucokinase (GK) also plays a key role for glucose utilization in the liver through glucose phosphorylation [12]. Insulin up-regulates GK expression [13], and its activity is also regulated by the interaction with other regulatory proteins [14]. However, the mechanism(s) by which insulin selectively regulates glycolysis, gluconeogenesis, and glycogenesis are still unclear.
Many factors can cause insulin resistance, including genetic factors in some ethnic populations, environmental factors, and behavioral factors, such as physical inactivity, over-nutrition, and obesity [15]. Obesity is known to be linked with an increased risk of insulin resistance through the release of increased amounts of non-esterified fatty acids, glycerol, hormones, and pro-inflammatory cytokines from adipose tissues [16]. The dysfunction of pancreatic islet β-cells, due to insulin resistance, leads to the failure of the body to control blood glucose levels [17]. Higher amounts of TNF-α and other cytokines have been reported in insulin-resistant adipocytes [16]. Insulin resistant individuals also contain an elevated level of circulating insulin to keep blood glucose levels under control [18]. Studies have shown higher levels of TNF-α and higher expression of TNF-α mRNA in the tissues of obese, insulin-resistant patients with T2D, as well as in the adipose tissues of several rodent models of obesity and T2D [19,20,21]. Thus, TNF-α, a pro-inflammatory cytokine, plays a central role in some types of insulin resistance [22]. Consequently, the conditions for insulin resistance can be modeled by treating cells with insulin or TNF-α [18,23]. Cellular metabolic pathways and mechanisms of TNF-α mediated insulin resistance are unclear, and in many cases, contradictory. For example, TNF-α treatment resulted in decreased expression of insulin-sensitive glucose transporter GLUT4 in cultured cells [24], but normal GLUT4 levels in insulin resistant rodents and human muscles [25,26]. The GLUT4 knockout mice are moderately insulin resistant [27] and TNF knockout mice respond more efficiently to an exogenous dose of insulin and glucose than wild-type [28,29].
In this study, we performed a quantitative proteomic analysis of differentiated 3T3-L1 adipocytes treated with Ins only, with TNF only, and both Ins + TNF, to identify proteins that are differentially expressed, due to the treatments. The 3T3-L1 adipocytes develop insulin resistance in response to insulin, TNF or saturated fatty acids, and transcriptional response to TNF begins within the first 1 h of the treatment, and changes in protein phosphorylation associated with insulin resistance can be detected as early as 6 h [30,31]. We identified 693 proteins of which 643 proteins were common to all experimental conditions and expression of 78 proteins was significantly different in one or more treatments compared to the control. Mitochondrial and cytosolic proteins involved in sequential fatty acid beta oxidation and carbohydrate metabolism were down-regulated, and those involved in folding, translation and oxidative stress responses were up-regulated in response to either, Ins only, TNF only or Ins + TNF treatments.
2. Materials and Methods
2.1. Cell Culture
Murine 3T3-L1 pre-adipocytes, [32] were grown, maintained, and induced, in order to differentiate using a standard protocol [33]. Fully differentiated adipocytes were maintained in DMEM (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% FBS (Sigma-Aldrich) until two days before experimentation, when cells were fed with 10% calf serum (Sigma-Aldrich). Prior to treatments, media was changed to low-glucose (5.5 mM) DMEM (Sigma-Aldrich) and 1% calf serum overnight. Cells were treated with 5 nM TNF-α (R&D Systems) for 8 h (TNF), 30 min with 10 nM insulin either alone (Ins) or followed by TNF treatment (Ins + TNF).
2.2. Cell Lysis and Protein Extraction
Prior to harvesting, cells were thoroughly rinsed at 37 °C. Differentiated 3T3-L1 cells were pelleted by centrifugation (Centrifuge 5810 R, Eppendorf, Hauppauge, NY, USA) at 14,000 rpm for 5 min at 4 °C. After washing cell pellets 3× with 200 µL of 20 mM PBS buffer, pH 7.5, the cells were suspended in 150 µL of wash buffer (20 mM PBS buffer, pH 7.5) and transferred to 150 µL size Barocycler microtubes. Freshly prepared serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) was added to each tube to achieve the final concentration of 1 mM, sealed and lysed in the Barocyler (Pressure Biosciences Inc., South Easton, MA, USA) at 35,000 psi for 60 min. The Barocycler was operated at 4 °C, holding 50 s at 35,000 psi followed by 10 s at atmospheric pressure per cycle for a total of 60 cycles or 1 h. Cell lysate was centrifuged at 14,000 rpm for 15 min at 4 °C and proteins in the supernatant were precipitated by adding 4 volume of cold (−20 °C) acetone and incubating overnight at −20 °C. After centrifugation at 14,000 rpm for 15 min at 4 °C, pellets were dried in a vacuum centrifuge (~1 min with heating system on), and dried protein pellets were dissolved in 40 µL of 8 M urea by incubating at room temperature for 1 h with continuous vortexing. The protein concentration was measured using a bicinchoninic acid (BCA) assay using bovine serum albumin (BSA) as a standard (Pierce Chemical Co., Rockford, IL, USA).
2.3. Trypsin/LysC Digestion
Samples containing 50 µg of total protein were transferred to a new tube for reduction, alkylation, and proteolysis as described previously [34,35]. Briefly, proteins were reduced in 10 mM dithiothreitol (DTT), alkylated in 20 mM iodoacetamide (IAA), and digested in the Barocycler using a mass spec grade trypsin and Lys-C mixture from Promega at a 1:25 (w/w) enzyme-to-substrate ratio. Digestions were performed at 50 °C and 20,000 psi for 1 h (60 cycles; 50 s at 20,000 psi and 10 s at atmospheric pressure. The digested peptides were desalted using Pierce C18 spin columns (Pierce Biotechnology, Rockford, IL, USA). Peptides were eluted using 80% acetonitrile (ACN) containing 0.1% formic Acid (FA) and dried in a vacuum centrifuge at room temperature. Dried peptides were re-suspended in 80 µL of the buffer containing 97% water, 3% ACN and 0.1% FA. Peptide concentration was measured again using BCA assay. The volume of each peptide sample was adjusted to 0.2 µg/µL and 5 μL (1 µg of the total peptides) was used for LC-MS/MS analysis.
2.4. Mass Spectrometry Analysis
Samples were analyzed by reverse-phase HPLC-ESI-MS/MS using a Dionex UltiMate 3000 RSLC Nano System (Thermo Fisher Scientific, Waltham, MA, USA), which was coupled to a Q-Exactive™ HF Hybrid Quadrupole-Orbitrap MS (Thermo Fisher Scientific) and a Nanospray Flex™ Ion Source (Thermo Fisher Scientific). Peptides were loaded to the trap column (300 μm ID × 5 mm) packed with 5 μm 100 Å C18 PepMap™ medium and then separated on an Acclaim™ PepMap™ 100 C18 analytical column (75 μm ID × 15 cm) packed with 2 μm 100 Å PepMap C18 medium (Thermo Fisher Scientific).
The peptides were separated using a 120 min gradient method. Mobile phase solvent A was 0.1% FA in water and solvent B was 0.1% FA in 80% ACN. Peptides were loaded to the trap column in 100% buffer A for 5 minutes, and eluted with a linear 80 min gradient of 5 to 30% buffer B and reaching 45% B in 91 min, 100% of B in 93 min at which point the gradient was held for 7 min before reverting back to 5% B at 100 min. The column was equilibrated at 5% of B for 20 min. The samples were loaded at a flow rate of 5 μL/min for 5 min and eluted from the analytical column at a flow rate of 300 nL/min. The mass spectrometer was operated using standard data-dependent mode acquiring MS/MS for the top 20 precursors. The full scan MS spectra were collected in the 400–1600 m/z range with a maximum injection time of 100 milliseconds at a resolution of 120,000 (at 200 m/z). Fragmentation of precursor ions was performed by high-energy C-trap dissociation (HCD) with the normalized collision energy of 27 eV. MS/MS scans were acquired at a resolution of 15,000 (at m/z 200). The dynamic exclusion was set to 15 s to avoid repeated scanning of identical peptides.
2.5. Data Analysis
LC-MS/MS data were searched using MaxQuant software (v. 1.5.3.28) [36,37,38] for protein identification and label free MS1 quantitation. The MS/MS spectra were searched against the Uniprot mouse database (downloaded on 02/15/2018). The minimal length of 6 amino acids was required in the database search. The database search was performed with the precursor mass tolerance set to 10 ppm and MS/MS fragment ions tolerance was set to 20 ppm. Database searches were performed with enzyme specificity for trypsin and Lys-C, allowing up to two missed cleavages. The oxidation of methionine was defined as a variable modification, and carbamidomethylation of cysteine was defined as a fixed modification. The ‘unique plus razor peptides’ were used for peptide quantitation. Razor peptides are those assigned to the protein group with the highest number of peptides identified. The false discovery rate (FDR) of peptides and proteins identification was set at 1%. The results were filtered to remove contaminants and also those identified as reverse hit. Similarly, proteins with LFQ ≠ 0 and MS/MS ≥ 2 in at least two replicates were retained for further analysis.
2.6. Gene Ontology (GO), Statistical, and Cluster Analysis
GO terms for the cellular component and molecular function were mapped using the Panther Gene Ontology Consortium Slim Cellular Component and Biological Processes analysis. Bonferroni analysis was used and only significantly enriched genes were included (p < 0.05).
The statistical analysis of LC-MS/MS search results was conducted using the InfernoRDN software (previously known as DAnTE) [39] that uses an R script for the statistical tests. Differentially expressed proteins were identified by two-way analysis of variance (ANOVA) using protein LFQ values and the proteins with a p-value ≤ 0.05 were considered for further analysis and biological interpretation. Classification for the molecular function category is based on gene ontology (GO).
3. Result and Discussion
3.1. Experimental Overview and LC-MS Reproducibility
Figure 1 shows experimental overview. Cells were treated with either Ins, TNF and both Ins + TNF. LC-MS/MS data were acquired on a Thermo Q-Exactive Orbitrap HF MS using three biological replicates. LC-MS reproducibility was tested running 3 technical replicates. The average coefficient of variation (CV) of peptide intensities was 17.2% and median CV was 15.8% (Figure S1A), indicating good quantitative reproducibility. The correlation co-efficient (r2) of peptides intensity was 0.945 between replicate 1 and 2, and 0.987 between replicate 1 and 3 (Figure S1B). The r2 of protein intensities was 0.997 between replicate 1 and 2, and 0.998 between replicate 1 and 3 (data not shown). Accuracy of LC-MS/MS analysis was also evaluated using 3 biological replicates, and showed strong correlation as shown in Figure 2A for the control samples. These results were consistent for other biological replicates as well.
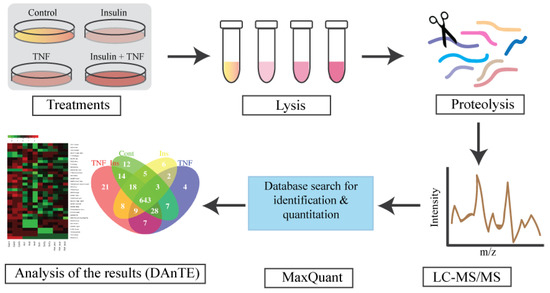
Figure 1.
Experimental design for proteomic analysis. Fully differentiated murine 3T3-L1 adipocytes were treated with TNF-α for 8 h (TNF), insulin for 30 min (Ins), and TNFα for 8 h, followed by insulin 30 min (TNF + Ins). LC-MS/MS data were searches using MaxQuant for protein identification and quantitation. Search results were visualized and plotted using DAnTE [39]. N = 3 biological replicates from each treatment group were analyzed.
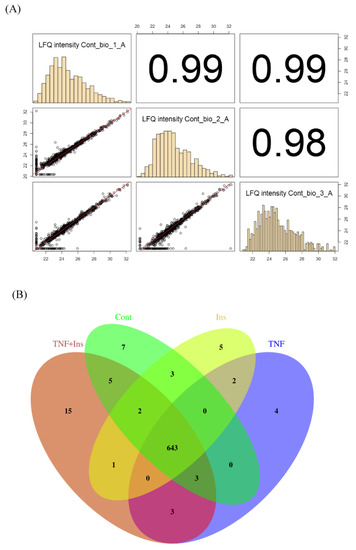
Figure 2.
LC-MS/MS reproducibility. (A). Correlation plots showing reproducibility among biological replicates. (B) Venn diagram showing the overlap of proteins expressed across all treatments.
3.2. Analysis of Differentially Expressed Proteins
We identified a total of 5022 peptides (Supplementary Data Table S1) mapped to 1050 proteins in at least one of the 12 samples (3 replicates for each of the 4 condition). We filtered these proteins to remove those that have no quantifiable MS1 peak intensity, and 0 or 1 MS/MS count. We then only considered proteins that were identified in at least 2 of the 3 biological replicates for downstream statistical analysis. After applying these filtering steps, we have a final list of 693 proteins (Supplementary Data Table S2). Functional classification of these proteins showed that the list was diverse including kinases, transcription and translational factors, transporters, carbohydrate and sugar metabolism, structural proteins, and enzymes (Supplementary Data Table S2). These proteins belong to diverse cellular compartments, including mitochondrion, nucleus, cytosol, extracellular exosomes, integral component of membrane, plasma membrane, and ER (Supplementary Data Table S2). Using a threshold of p value ≤ 0.05, we identified 78 proteins that were significantly different in cells treated with Ins only, TNF only, or both, compared to the control (Table 1). Most quantitative proteomics methods use fold changes to determine differentially expressed proteins, and a 2-fold increase or decrease is the most commonly applied threshold. However, a fold change to determine altered proteins is confounded by biases in protein abundances [40]. For example, a 1.5 or 2-fold increase can be highly stringent for highly abundant proteins, but for low abundance proteins, this level of change might simply represent technical noise. Therefore, rather than relying on fold changes, we used the one-way ANOVA test to determine differentially expressed proteins and used p values of ≤ 0.05 as a cut-off to determine significantly altered protein expression between the treatments. We found that many significantly different proteins across experimental groups have fold changes ≤1.5 (Supplementary Data Table S3). Of the 78 significant proteins, the expression of 18 proteins was at least 2-fold higher and the expression of 28 proteins was at least 2-fold lower in at least one of the 3 treatment groups (Supplementary Data Table S3). Changes in expression of 32 significantly different proteins was less than 2-fold and 24 proteins were uniquely identified in Ins + TNF (Figure 2B and Supplementary Data Table S3). Among the 78 differentially expressed proteins, the expression of 20 proteins was up and 30 proteins was down in all the treatments compared to the control (Table 1). Eight (8) proteins were up and 4 proteins were down in Ins only treatment, 3 proteins were up and 5 proteins were down in TNF only, and 4 proteins were up and 4 proteins were down in Ins + TNF (Table 1). The expression of 7 proteins was detected only in the control, 5 proteins only in Ins, 4 proteins only in TNF, and 15 proteins only in Ins + TNF (Figure 2B). The four proteins elevated in TNF are Sigma non-opioid intracellular receptor 1 (SIGMAR1, OPRS1), 60S ribosomal protein L23 (RPL23), 40S ribosomal protein S28 (RPS28) and ATP dependent RNA helicase (DDX5). Their direct or indirect role or response to TNF is currently unknown. The SIGMAR1 is an endoplasmic reticulum (ER)-resident transmembrane protein which functions in lipid transport from the ER and is involved in a wide variety of disorders, including depression, drug addiction, and pain [41]. The elevated expression of RPL23 and RPS28 might suggest that cells treated with TNF might require a more efficient translational machinery by regulating ribosome biogenesis and global protein synthesis [42]. The increased expression of DDX5 is quite interesting. DDX5 is known to participate in all aspects of RNA metabolism ranging from transcription to translation, RNA decay, and mRNA processing [43]. Its role in cell cycle regulation, tumorigenesis, apoptosis, cancer development, and adipogenesis has been well established [43]. Understanding how elevated expression of DDX5 is linked to TNF treatment will provide new information about TNF-induced cellular outcomes of adipose tissues.

Table 1.
Differentially expressed proteins in cells treated with insulin (Ins), TNF-α (TNF) and insulin + TNF-α (Ins + TNF) compared to the control group. p-values were determined based on the ANOVA test. ‘+’ indicates up-regulation and ‘−’ indicates down-regulation of proteins in the treated cells compared to the control.
Figure 3A shows the heat map of the significant proteins in each group, which showed a 2-fold change in abundances in at least one of the 3 treatments. The PCA analysis was performed to project LFQ based proteome measurements into a two-dimensional data space (Figure 3B). We applied PCA to 78 significant proteins that were quantified in each of the condition. The component 1 of the PCA (PC1) accounts for 42.6% of the variability, and clearly discriminates proteins among different treatment groups. The PC2 accounted for 20.3% of the total variations, and altogether, all 2 components accounted for 62.8% of the total variation. Proteins expressed in control segregated from the others, and proteins expressed in Ins only, TNF only, and Ins + TNF were also clearly discrete from each other. Figure 3B also shows that the distance between replicates within each group is much smaller than the separation between the groups, supporting reproducible LC-MS/MS analysis and peptide quantification, a prerequisite for accurate label-free protein quantitation
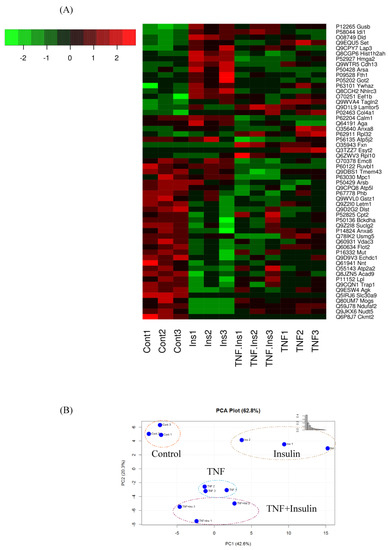
Figure 3.
(A) Heatmap of differentially expressed proteins (p ≤ 0.05) which showed ≥ 2-fold increase or decrease in expression in one or more treatment groups compared to the control group. (B) Principal component analysis, using the LFQ intensity of all significant proteins, displaying the four groups of Control, Ins, TNF, and TNF + Ins.
Figure 4 shows the top 5 GO biological processes and molecular functions of the 78 significantly up- or down-regulated proteins (p ≤ 0.05) in one or more of the 3 treatments compared to the control. For biological processes, down-regulated proteins were enriched for metabolic process, oxidation-reduction process, and TCA cycle in Ins and TNF, and ion transport and regulation and ATPase activity proteins were enriched in Ins + TNF (Figure 4A). With regard to molecular functions, down-regulated proteins were enriched for oxidoreductase activity, electron carrier activity and fatty-acyl-CoA binding activity in Ins only and TNF only treatments, and ATPase activity, voltage gated anion channel and prion activity were enriched in Ins + TNF (Figure 4C). Among the up-regulated proteins, those involved in biological processes, such as platelet degradation, translation, glycolysis, and the response to amino acids were enriched in one or more of the treatments (Figure 4B). For molecular functions, proteins involved in homodimerization, calcium ion binding, structural constituents of ribosomes, and the extracellular matrix were enriched among the up-regulated proteins (Figure 4D). The complete list of all the GO molecular functions, biological processes, and cellular components for all 693 proteins and 78 differentially expressed proteins are shown in the Supplementary Data Table S2 and S3. The analysis of cellular components of 693 proteins by GO annotation showed that ~30% of these proteins were resident in mitochondria (Supplementary Data Table S2). The same analysis showed >50% of the differentially expressed proteins (p ≤ 0.05) resident in mitochondria including mitochondrial matrix and mitochondrial inner membrane (Supplementary Data Table S3), suggesting that relatively higher mitochondrial resident proteins were affected by the treatments. Other major cellular components of differentially expressed proteins included extracellular exosomes, focal adhesion, cytoplasm/cytosol, lysosome, and cell-cell adhesion junction.
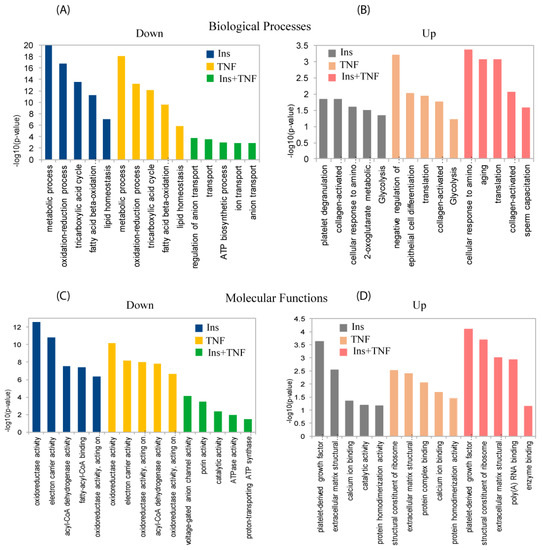
Figure 4.
Gene ontology (GO) analysis for biological processes (upper panels) and molecular functions (lower panels) of differentially expressed proteins. Top five down-regulated (A) and up-regulated (B) biological processes in one or more treatments, compared to the control. Top five down-regulated (C) and up-regulated (D) molecular functions in one or more treatments compared to the control. The complete list of GO biological processes, molecular functions and cellular components of the total and differentially expressed proteins can be found in Supplementary Data Tables S2 and S3, respectively.
3.3. Differentially Expressed Mitochondrial Proteins
Of the 78 proteins (p ≤ 0.05), 39 proteins were resident to mitochondria (Table 1 and Supplementary Data Table S3). The mitochondrial creatine kinase S-type (CKMT2), which provides a spatial and temporal energy buffer to maintain cellular homeostasis, was detected only in the control samples (Supplementary Data Table S2). The amount of carnitine o-palmitoyltransferase 2 (CTP2), a mitochondrial membrane protein, decreased in all three treatments (Table 1). There was a coordinated down-regulation of proteins involved in fatty acid oxidation and tricarboxylic acid (TCA) cycle (Table 1). The expression of acyl-CoA dehydrogenases, including long-chain specific acyl-CoA dehydrogenase (ACADL), short-chain specific acyl-CoA (ACADS), very long-chain specific acyl-CoA dehydrogenase (ACADV), as well as acyl-CoA dehydrogenase family member 9 (ACAD9) and acetyl-CoA carboxylase 1 (ACACA1) were also generally down-regulated in all the treatments (Table 1 and Supplementary Data Table S2). These enzymes work in a stepwise fashion to fatty acid synthesis. Previous studies have shown that mice deficient in ACADL develop hepatic insulin resistance [44], and those deficient in ACADV were protected from high-fat diet-induced obesity and liver and muscle insulin resistance [45]. Inhibition of CPT2 activity is known to inhibit insulin resistance in diet-induced obese mice [46].
The expression of cAMP-dependent protein kinase type II (PRKAR2b), enoyl-CoA hydratase domain-containing protein 3 (ECHDC3), 3-hydroxyisobutyryl-CoA hydrolase (HIBCH), methylmalonyl-CoA mutase (MUTA) and glutaryl-CoA dehydrogenase (GCDH) also decreased in Ins and TNF treatments (Table 1, Supplementary Data Table S2). Several of these proteins are involved in the oxidation of unsaturated fatty acid enoyl CoA esters. Relative abundances of TCA cycle enzymes, succinate-CoA ligase (GDP-forming) subunit beta (SUCB2), and fumarate hydratase (FUMH) were also lower in Ins, TNF and both Ins + TNF. Expression of mitochondrial pyruvate dehydrogenase E1 component subunit beta (PDHB) was also reduced in all three treatments (Supplementary Data Table S2). PDHB catalyzes the conversion of pyruvate to acetyl-CoA, thus links the glycolytic pathway to the TCA cycle. The pyruvate dehydrogenase E1 component subunit alpha also showed reduced levels, but the alteration was not significantly different compared to the control.
3.4. Glycolysis and Glycogenesis
Glycolytic enzymes triosephosphate isomerase (TPI1) and phosphoglycerate mutase 1 (PGAM1) were significantly up-regulated in all the treatments, but phosphoglycerate kinase (PGK1) was up-regulated only in Ins and Ins + TNF (Table 1). However, the expression of pyruvate carboxylase (PCX), an enzyme that plays a crucial role in lipogenesis, was not statistically significant (Supplementary Data Table S2). Mitochondrial glycerol-3-phosphate dehydrogenase (GPD2), an integral component of mammalian respiratory chain and glycerophosphate shuttle, was also reduced in all the treatments, as compared to the control. GPD2 is an important enzyme of intermediary metabolism that functions in between glycolysis, oxidative phosphorylation, and fatty acid metabolism [47]. Therefore, its metabolic role involves the regulation of cytosolic glycerol-3-phosphate (G3P) as a metabolite connecting glycolysis, lipogenesis, and oxidative phosphorylation. Insulin is known to reduce PEPC expression through serine/threonine kinase AKT signaling pathways [7,8]. However, in the present study, PEPC expression was not different in all the treatment groups compared to the control. Our data showed the suppression of gluconeogenesis and activation of glycogenesis in Ins, TNF and Ins + TNF, in agreement with previous reports. Mitochondrial serine/threonine-protein phosphatase 2A (PGAM5) was up-regulated under TNF treated cells (Table 1). PGAM5 displays phosphatase activity for serine/threonine residues to dephosphorylate and activate MAP3K5 kinase. This protein is known as a central mediator for programmed necrosis induced by TNF-α or by reactive oxygen species [48]. In contrast, mitochondrial inorganic pyrophosphatase 2 (PPA2) was significantly up regulated only in TNF. PPA2 hydrolyzes inorganic pyrophosphate, which is essential for correct regulation of mitochondrial membrane potential and mitochondrial membrane organization and function [49,50]. It is possible that increased mitochondrial stress and decreased mitochondrial function may cause disruption in mitochondrial potential and membrane organization, leading to increased expression of PPA2.
The enzymes CKTM2, Slc30a9, NUDT5, NDUFA12, and NDUFC2 were down-regulated in all treated cells compared to the control (Table 1, Supplementary Data Table S2). The cytochrome b-c1 complex subunit 1 (UQCRC1) and cytochrome c oxidase subunit (NDUFA4) were down-regulated in all the treatments, but cytochrome c oxidase assembly factor 3 homolog (COA3) was up-regulated in Ins treated adipocytes. Similarly, β-glucuronidase (GUSB), lipid transport sigma-non-opioid intracellular receptor 1 (SIGMAR1, OPRS1) and cytoplasmic aspartate aminotransferase (GOT1) were up-regulated in all the treatments, compared to the control group.
3.5. Transport, Translation, and Oxidative Stress Responses
We searched for proteins with well-established roles in structure and function of cytoskeleton, protein transport, translation, and oxidative stress responses. In contrast to the control group, the expression of collagen α-1 (I) chain (COL1a1) and collagen α-1 (IV) chain (COL4a1), which support and strengthen many tissues in the body, such as cartilage, bone, and tendon, was higher in all treatments (Table 1 and also Supplementary Data Table S2). The expression of 14-3-3 zeta/delta (YWHAZ) also increased in Ins, TNF, as well as Ins + TNF treatments. 14-3-3 proteins are dimeric phosphoserine-binding proteins that participate in multiple signal transduction pathways and regulate several aspects of cellular biochemistry including diabetic cardiomyopathy [51]. The Zinc transporter 9 (Slc30a9), involved in maintaining zinc concentration was detected only in the control (Supplementary Data Table S2). Sarcoplasmic/endoplasmic reticulum (SR) calcium ATPase 2 (ATP2a2) expression was reduced in Ins and Ins + TNF but increased in TNF only treatment (Table 1). ATP2a2 is a Ca2+ ATPase that transfers Ca2+ from the cytosol to the lumen of the SR at the expense of ATP hydrolysis during muscle relaxation [52]. The expression of calmodulin-like protein 3 (CALL3) increased in all the treated cells in comparison to the control group.
Insulin resistance and T2D can lead to oxidative stress by elevated reactive oxygen species (ROS) generation [53]. The ROS subsequently degrade mitochondrial proteins, and key TCA cycle enzymes are particularly sensitive to such damage [54]. From our data, several distinct responses of proteins to the treatments may reflect increased ROS in mitochondria. For example, mitochondrial dihydrolipoyl dehydrogenase (DLDH) is known to serve as a source of ROS [55], and its expression increased significantly in all the treated cells (Table 1). The expression of Cu-Zn superoxide dismutase (SOD1) was not statistically significant, but generally showed higher levels in all the treatments (Figure 5, Supplementary Data Table S2). Mitochondrial glutathione reductase (GSR), which may play a key role in scavenging ROS was up-regulated in Ins + TNF (Figure 5, Supplementary Data Table S2). However, thioredixin (TRX), which also maintains cell redox homeostasis, decreased in Ins + TNF, indicating distinct responses of proteins involved in maintaining cellular redox homeostasis. Frataxin (FRDA) is a mitochondrial iron-sulfur-cluster assembly protein and affects oxidative energy flux [56]. From our data, FRDA expression was observed only in cells treated either, with TNF only or Ins + TNF. (Supplementary Data Table S2). This may indicate the deterioration of adipocytes due to TNF treatment. It has been shown that disruption of FRDA caused T2D, due to impaired insulin secretion and increased oxidative stress in pancreatic β cells of mice [56]. Our proteomic analysis provided further evidence that FRDA plays a role in promoting cellular defense against ROS. Our experiment may provide new insight into the deterioration of β cell function observed in different subtypes of diabetes in humans.
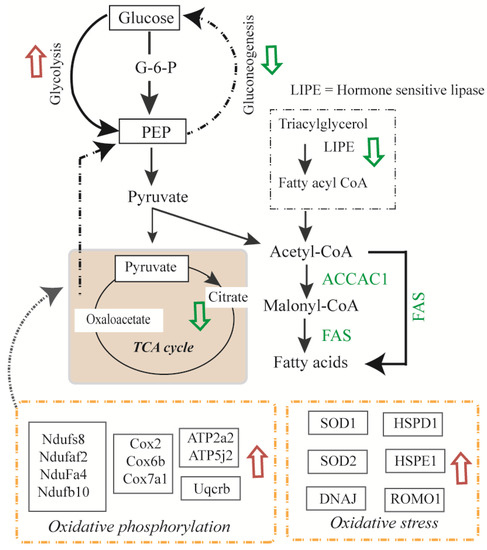
Figure 5.
Overview of the metabolic pathway changes in murine 3T3-L1 adipocytes due to insulin or TNF-α treatments. Pathways that were up-regulated are shown by red arrows and pathways that were down-regulated are shown by green arrows. Proteins involved in pathways of glycolysis, glycogenolysis, oxidative phosphorylation and oxidative stress responses were up-regulated and protein involved in glycogenesis, gluconeogenesis, and TCA cycle, were down-regulated. LIPE; hormone sensitive lipase.
Mitochondrial cytochrome c oxidase, carbohydrate metabolism (β-glucuronidase), lipid transport (sigma-non-opioid intracellular receptor 1) and NADH dehydrogenase [ubiquinone] 1 alpha sub-complex assembly factor 2 (NDUFAF2) were inhibited in all the treatments groups compared to the control (Figure 3A, Table 1). USMG5 (associated protein in insulin sensitive tissues) was expressed in all except Ins + TNF. Proteins involved in vesicle transport (coatomer subunit epsilon), translation (60S and 40S ribosomal proteins), respiration (cytochrome c oxidase), carbohydrate metabolism (β-glucuronidase), lipid transport (sigma-non-opioid intracellular receptor 1), and amino acid biosynthesis were also down-regulated due to treatment with Ins, TNF, or Ins + TNF. Many of these proteins are known to exist as complexes endogenously, so future studies for the determination of their complex composition and changes due to treatment can provide new information about their structure and function.
4. Summary
We showed that the decreased expression of mitochondrial and cytosolic proteins involved in fatty acid beta-oxidation and carbohydrate metabolism, and increased abundances of proteins involved in folding, degradation, and stress responses. The expression of glycolytic enzymes TPI1 and PGAM1 increased in all treatments, and PGK1 increased in Ins only and Ins + TNF (Table 1). PCX, an enzyme involved in gluconeogenesis and lipogenesis showed lower expression in all the treatment groups, compared to the control (Supplementary Data Table S2). GS showed increased expression, but GPD2, an important enzyme of intermediary metabolism that functions in between glycolysis, oxidative phosphorylation, and fatty acid metabolism [47], was reduced in all the treatments (Supplementary Data Table S2). The majority of TCA cycle enzymes were particularly sensitive to the treatments. The abundance of mitochondrial DLDH, which is known to serve as a source of ROS [55], increased significantly in all the treated cells. These results showed a potential relationships among various pathways that may converge to create insulin resistance and T2D (Figure 5). What remains to be explored is how these differentially expressed proteins are organized into functional complexes and understanding how their complex composition and localization changes as a function of the Ins or TNF treatments. Additionally, the changes in post-translational modifications of these proteins due to Ins or TNF treatments will help to better understand their molecular functions and biological processes. All the raw LC-MS/MS data associated with these experiments have been deposited in the Mass Spectrometry Interactive Virtual Environment (http://massive.ucsd.edu) with the ID MSV000081520.
Supplementary Materials
The following are available online at https://www.mdpi.com/2227-7382/7/4/35/s1, Figure S1. LC-MS reproducibility. (A) Percentage CV (coefficient of variation), and (B) correlation coefficients of peptide intensities in 3 technical replicates, Table S1. A list of all non-redundant peptides and mapped proteins, charged state and intensity across samples, Table S2. A list of all proteins with gene ontology (GO) analysis, their abundances (log2 LFQ intensities), and ANOVA and t-test analyses, Table S3. Proteins with p value ≤ 0.05.
Author Contributions
Conceptualization, U.K.A.; Data curation, H.C., V.H. and U.K.A.; Formal analysis, H.C., K.P.B., A.V., V.H., T.J.P.S., R.W.G. and U.K.A.; Funding acquisition, U.K.A.; Investigation, H.C., R.W.G. and U.K.A.; Methodology, H.C., V.H. and U.K.A.; Project administration, U.K.A.; Resources, T.G.S., R.W.G. and U.K.A.; Software, K.P.B. and T.J.P.S.; Visualization, K.P.B.; Writing – original draft, U.K.A., Writing – review & editing, H.C., A.V., V.H., R.W.G., T.G.S. and U.K.A.
Funding
This research was supported in part by #UL1 TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award to Uma K. Aryal. LC-MS experiments were performed at the Purdue Proteomics Facility. The Q-Exactive Orbitrap HF mass spectrometer used in this study was purchased through the grant provided by the Purdue Executive Vice Present for Research and Partnership (EVPRP).
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
| ER | Endoplasmic Reticulum |
| LC-MS/MS | liquid chromatography tandem MS |
| DMEM | Dulbecco’s modified eagle medium |
| FBS | Fetal bovine serum |
| TNFα | Tumor necrosis Factor-alpha |
| LFQ | Label free quantitation |
| PMSF | Phenylmethylsulfonyl fluoride |
| GO | Gene Ontology |
| HPLC | High Pressure Liquid Chromatography |
| DTT | Dithiothreitol |
| IAA | Iodoacetamide |
| BCA | Bicinchonic Acid |
| CAN | Acetonitril |
| FA | Formic acid |
| HCD | High Energy C-trap Dissociation |
| HF | High Filed |
| ANOVA | Analysis of Variance |
| TCA | Tricarboxylic acid |
| T2D | Type 2 diabetes |
| ROS | Reactive oxygen species |
| kDa | Kilodalton |
| CV | coefficient of variation |
References
- Wilcox, G. Insulin and insulin resistance. Clin. Biochem. Rev. 2005, 26, 19–39. [Google Scholar] [PubMed]
- Moller, D.E.; Flier, J.S. Insulin resistance—Mechanisms, syndromes, and implications. N. Engl. J. Med. 1991, 325, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.M.; Cleary, P.A.; Backlund, J.Y.; Genuth, S.M.; Lachin, J.M.; Orchard, T.J.; Raskin, P.; Zinman, B.; Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Study Research Group. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N. Engl. J. Med. 2005, 353, 2643–2653. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, R.; Kubota, H.; Yugi, K.; Toyoshima, Y.; Komori, Y.; Soga, T.; Kuroda, S. The selective control of glycolysis, gluconeogenesis and glycogenesis by temporal insulin patterns. Mol. Syst. Biol. 2013, 9, 664. [Google Scholar] [CrossRef] [PubMed]
- Okar, D.A.; Manzano, A.; Navarro-Sabate, A.; Riera, L.; Bartrons, R.; Lange, A.J. PFK-2/FBPase-2: Maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem. Sci. 2001, 26, 30–35. [Google Scholar] [CrossRef]
- Yabaluri, N.; Bashyam, M.D. Hormonal regulation of gluconeogenic gene transcription in the liver. J. Biosci. 2010, 35, 473–484. [Google Scholar] [CrossRef]
- Nakae, J.; Park, B.C.; Accili, D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J. Biol. Chem. 1999, 274, 15982–15985. [Google Scholar] [CrossRef]
- Sutherland, C.L.; Heath, A.W.; Pelech, S.L.; Young, P.R.; Gold, M.R. Differential activation of the ERK, JNK, and p38 mitogen-activated protein kinases by CD40 and the B cell antigen receptor. J. Immunol. 1996, 157, 3381–3390. [Google Scholar]
- Bollen, M.; Keppens, S.; Stalmans, W. Specific features of glycogen metabolism in the liver. Biochem. J. 1998, 336, 19–31. [Google Scholar] [CrossRef]
- Aiston, S.; Hampson, L.J.; Arden, C.; Iynedjian, P.B.; Agius, L. The role of protein kinase B/Akt in insulin-induced inactivation of phosphorylase in rat hepatocytes. Diabetologia 2006, 49, 174–182. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Pandey, S.K. Potential mechanism(s) involved in the regulation of glycogen synthesis by insulin. Mol. Cell Biochem. 1998, 182, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Ferre, T.; Pujol, A.; Riu, E.; Bosch, F.; Valera, A. Correction of diabetic alterations by glucokinase. Proc. Natl. Acad. Sci. USA 1996, 93, 7225–7230. [Google Scholar] [CrossRef] [PubMed]
- Ribaux, P.G.; Iynedjian, P.B. Analysis of the role of protein kinase B (cAKT) in insulin-dependent induction of glucokinase and sterol regulatory element-binding protein 1 (SREBP1) mRNAs in hepatocytes. Biochem. J. 2003, 376, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Van Schaftingen, E.; Detheux, M.; Veiga da Cunha, M. Short-term control of glucokinase activity: Role of a regulatory protein. FASEB J. 1994, 8, 414–419. [Google Scholar] [CrossRef]
- Dendup, T.; Feng, X.; Clingan, S.; Astell-Burt, T. Environmental Risk Factors for Developing Type 2 Diabetes Mellitus: A Systematic Review. Int. J. Environ. Res. Public Health 2018, 15, 78. [Google Scholar] [CrossRef]
- Stephens, J.M.; Lee, J.; Pilch, P.F. Tumor necrosis factor-alpha-induced insulin resistance in 3T3-L1 adipocytes is accompanied by a loss of insulin receptor substrate-1 and GLUT4 expression without a loss of insulin receptor-mediated signal transduction. J. Biol. Chem. 1997, 272, 971–976. [Google Scholar] [CrossRef]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef]
- Chen, Y.L.; Qiao, Y.C.; Xu, Y.; Ling, W.; Pan, Y.H.; Huang, Y.C.; Geng, L.J.; Zhao, H.L.; Zhang, X.X. Serum TNF-alpha concentrations in type 2 diabetes mellitus patients and diabetic nephropathy patients: A systematic review and meta-analysis. Immunol. Lett. 2017, 186, 52–58. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Shargill, N.S.; Spiegelman, B.M. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef]
- Kern, P.A.; Saghizadeh, M.; Ong, J.M.; Bosch, R.J.; Deem, R.; Simsolo, R.B. The expression of tumor necrosis factor in human adipose tissue. Regulation by obesity, weight loss, and relationship to lipoprotein lipase. J. Clin. Investig. 1995, 95, 2111–2119. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Investig. 1995, 95, 2409–2415. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Song, P.; Jang, J.H.; Kim, D.K.; Choi, S.; Kim, J.; Ghim, J.; Kim, D.; Park, S.; Lee, H.; et al. Proteomic analysis of tumor necrosis factor-alpha (TNF-alpha)-induced L6 myotube secretome reveals novel TNF-alpha-dependent myokines in diabetic skeletal muscle. J. Proteome Res. 2011, 10, 5315–5325. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J.; Williams, M.G.; Frost, S.C. Development of insulin resistance in 3T3-L1 adipocytes. J. Biol. Chem. 1997, 272, 7759–7764. [Google Scholar] [CrossRef] [PubMed]
- Stephens, J.M.; Pekala, P.H. Transcriptional repression of the GLUT4 and C/EBP genes in 3T3-L1 adipocytes by tumor necrosis factor-alpha. J. Biol. Chem. 1991, 266, 21839–21845. [Google Scholar] [PubMed]
- Pedersen, O.; Bak, J.F.; Andersen, P.H.; Lund, S.; Moller, D.E.; Flier, J.S.; Kahn, B.B. Evidence against altered expression of GLUT1 or GLUT4 in skeletal muscle of patients with obesity or NIDDM. Diabetes 1990, 39, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Garvey, W.T.; Maianu, L.; Hancock, J.A.; Golichowski, A.M.; Baron, A. Gene expression of GLUT4 in skeletal muscle from insulin-resistant patients with obesity, IGT, GDM, and NIDDM. Diabetes 1992, 41, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Kitamura, Y.; Kahn, C.R.; Accili, D. Mouse models of insulin resistance. Physiol. Rev. 2004, 84, 623–647. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. The role of TNFalpha and TNF receptors in obesity and insulin resistance. J. Intern. Med. 1999, 245, 621–625. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Mechanisms of TNF-alpha-induced insulin resistance. Exp. Clin. Endocrinol. Diabetes 1999, 107, 119–125. [Google Scholar] [CrossRef]
- Ruan, H.; Hacohen, N.; Golub, T.R.; Van Parijs, L.; Lodish, H.F. Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: Nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes 2002, 51, 1319–1336. [Google Scholar] [CrossRef]
- Ruan, H.; Miles, P.D.; Ladd, C.M.; Ross, K.; Golub, T.R.; Olefsky, J.M.; Lodish, H.F. Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-alpha: Implications for insulin resistance. Diabetes 2002, 51, 3176–3188. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Kehinde, O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 1975, 5, 19–27. [Google Scholar] [CrossRef]
- Richard, A.J.; Fuller, S.; Fedorcenco, V.; Beyl, R.; Burris, T.P.; Mynatt, R.; Ribnicky, D.M.; Stephens, J.M. Artemisia scoparia enhances adipocyte development and endocrine function in vitro and enhances insulin action in vivo. PLoS ONE 2014, 9, e98897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mittal, L.; Aryal, U.K.; Camarillo, I.G.; Ferreira, R.M.; Sundararajan, R. Quantitative proteomic analysis of enhanced cellular effects of electrochemotherapy with Cisplatin in triple-negative breast cancer cells. Sci. Rep. 2019, 9, 13916. [Google Scholar] [CrossRef]
- Sooreshjani, M.A.; Gursoy, U.K.; Aryal, U.K.; Sintim, H.O. Proteomic analysis of RAW macrophages treated with cGAMP or c-di-GMP reveals differentially activated cellular pathways. RSC Adv. 2018, 8, 36840–36851. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Polpitiya, A.D.; Qian, W.J.; Jaitly, N.; Petyuk, V.A.; Adkins, J.N.; Camp, D.G.; Anderson, G.A.; Smith, R.D. DAnTE: A statistical tool for quantitative analysis of -omics data. Bioinformatics 2008, 24, 1556–1558. [Google Scholar] [CrossRef]
- Deng, W.J.; Nie, S.; Dai, J.; Wu, J.R.; Zeng, R. Proteome, phosphoproteome, and hydroxyproteome of liver mitochondria in diabetic rats at early pathogenic stages. Mol. Cell Proteom. 2010, 9, 100–116. [Google Scholar] [CrossRef]
- Cobos, E.J.; Entrena, J.M.; Nieto, F.R.; Cendan, C.M.; Del Pozo, E. Pharmacology and therapeutic potential of sigma (1) receptor ligands. Curr. Neuropharmacol. 2008, 6, 344–366. [Google Scholar] [CrossRef] [PubMed]
- Silvera, D.; Formenti, S.C.; Schneider, R.J. Translational control in cancer. Nat. Rev. Cancer 2010, 10, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Chen, G.; Jia, H.; He, X.; Jing, Z. DDX5 RNA Helicases: Emerging Roles in Viral Infection. Int. J. Mol. Sci. 2018, 19, 1122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, Z.X.; Choi, C.S.; Tian, L.; Kibbey, R.; Dong, J.; Cline, G.W.; Wood, P.A.; Shulman, G.I. Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc. Natl. Acad. Sci. USA 2007, 104, 17075–17080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Christianson, J.; Liu, Z.X.; Tian, L.; Choi, C.S.; Neschen, S.; Dong, J.; Wood, P.A.; Shulman, G.I. Resistance to high-fat diet-induced obesity and insulin resistance in mice with very long-chain acyl-CoA dehydrogenase deficiency. Cell Metab. 2010, 11, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Keung, W.; Ussher, J.R.; Jaswal, J.S.; Raubenheimer, M.; Lam, V.H.; Wagg, C.S.; Lopaschuk, G.D. Inhibition of carnitine palmitoyltransferase-1 activity alleviates insulin resistance in diet-induced obese mice. Diabetes 2013, 62, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Mracek, T.; Drahota, Z.; Houstek, J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta 2013, 1827, 401–410. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, H.; Chen, S.; Du, F.; Wang, X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012, 148, 228–243. [Google Scholar] [CrossRef]
- Guimier, A.; Gordon, C.T.; Godard, F.; Ravenscroft, G.; Oufadem, M.; Vasnier, C.; Rambaud, C.; Nitschke, P.; Bole-Feysot, C.; Masson, C.; et al. Biallelic PPA2 Mutations Cause Sudden Unexpected Cardiac Arrest in Infancy. Am. J. Hum. Genet. 2016, 99, 666–673. [Google Scholar] [CrossRef]
- Kennedy, H.; Haack, T.B.; Hartill, V.; Matakovic, L.; Baumgartner, E.R.; Potter, H.; Mackay, R.; Alston, C.L.; O’Sullivan, S.; McFarland, R.; et al. Sudden Cardiac Death Due to Deficiency of the Mitochondrial Inorganic Pyrophosphatase PPA2. Am. J. Hum. Genet. 2016, 99, 674–682. [Google Scholar] [CrossRef]
- Watanabe, K.; Thandavarayan, R.A.; Gurusamy, N.; Zhang, S.; Muslin, A.J.; Suzuki, K.; Tachikawa, H.; Kodama, M.; Aizawa, Y. Role of 14-3-3 protein and oxidative stress in diabetic cardiomyopathy. Acta Physiol. Hung. 2009, 96, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Vandecaetsbeek, I.; Vangheluwe, P.; Raeymaekers, L.; Wuytack, F.; Vanoevelen, J. The Ca2+ pumps of the endoplasmic reticulum and Golgi apparatus. Cold Spring Harb. Perspect. Biol. 2011, 3, a004184. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Cruzat, V.F.; Keane, K.N.; Carlessi, R.; de Bittencourt, P.I. Molecular mechanisms of ROS production and oxidative stress in diabetes. Biochem. J. 2016, 473, 4527–4550. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Eubel, H.; Millar, A.H. Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol. Cell Proteom. 2010, 9, 2125–2139. [Google Scholar] [CrossRef] [PubMed]
- Tahara, E.B.; Barros, M.H.; Oliveira, G.A.; Netto, L.E.; Kowaltowski, A.J. Dihydrolipoyl dehydrogenase as a source of reactive oxygen species inhibited by caloric restriction and involved in Saccharomyces cerevisiae aging. FASEB J. 2007, 21, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Ristow, M.; Pfister, M.F.; Yee, A.J.; Schubert, M.; Michael, L.; Zhang, C.Y.; Ueki, K.; Michael, M.D.; Lowell, B.B.; Kahn, C.R. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc. Natl. Acad. Sci. USA 2000, 97, 12239–12243. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).