Abstract
At the end of 2019, a severe public health threat named coronavirus disease (COVID-19) spread rapidly worldwide. After two years, this coronavirus still spreads at a fast rate. Due to its rapid spread, the immediate and rapid diagnosis of COVID-19 is of utmost importance. In the global fight against this virus, chest X-rays are essential in evaluating infected patients. Thus, various technologies that enable rapid detection of COVID-19 can offer high detection accuracy to health professionals to make the right decisions. The latest emerging deep-learning (DL) technology enhances the power of medical imaging tools by providing high-performance classifiers in X-ray detection, and thus various researchers are trying to use it with limited success. Here, we propose a robust, lightweight network where excellent classification results can diagnose COVID-19 by evaluating chest X-rays. The experimental results showed that the modified architecture of the model we propose achieved very high classification performance in terms of accuracy, precision, recall, and f1-score for four classes (COVID-19, normal, viral pneumonia and lung opacity) of 21.165 chest X-ray images, and at the same time meeting real-time constraints, in a low-power embedded system. Finally, our work is the first to propose such an optimized model for a low-power embedded system with increased detection accuracy.
1. Introduction
COVID-19 is a highly contagious and infectious disease that causes severe infection of the lower respiratory system. COVID-19 was officially declared a global pandemic by the World Health Organization (WHO) on 11 March 2020 [1]. The total number of COVID-19 cases worldwide up to this point, which changes almost every minute, is over 328 million, including 5 million deaths and 55 million active patients [2]. The increasing number of deaths from COVID-19 and the rapid rate of disease transmission makes it one of the world’s most significant and most serious public health issues.
The clinical findings of COVID-19 infection are those of bronchopneumonia, where cough, dyspnea, fever and respiratory failure with acute respiratory distress syndrome (ARDS) occur [3,4]. Radiographic imaging is the fastest, cheapest, and most essential diagnostic tool for detecting pneumonia while providing lower patient radiation than computed tomography (CT) and magnetic resonance imaging (MRI) [5,6].
In COVID-19 disease, the lungs are affected resulting in severe viral pneumonia [7,8,9]. Deaths from viral pneumonia due to COVID-19 are constantly increasing day by day. Thus, a valid diagnosis of COVID-19 pneumonia is vital but often a difficult task due to the similar symptoms and radiological findings that occur between patients infected with COVID-19 and patients with viral pneumonia [10,11,12]. This difficulty can be alleviated by enhancing the arsenal of health workers using artificial intelligence in the form of machine learning.
In recent years, artificial intelligence (AI) has been established as a powerful tool in the field of medical services for classification, prediction, diagnosis, detection and segmentation [13,14,15,16,17,18]. In medical imaging, DL in chest X-rays has been shown to play an essential role in the detection of viral pneumonia, bacterial pneumonia and other chest diseases [19,20]. With the rapid spread of the virus and the fight against pandemics, researchers are using state-of-the-art DL techniques to classify COVID-19 X-rays [21,22,23,24]. Therefore, the design and development of DL models for the classification of COVID-19 infected radiographs is an urgent need to address the pandemic to make appropriate clinical decisions and significantly reduce workload [25].
The main goal of this study is to propose a robust, lightweight neural network, enhancing the work in [26], that provides high accuracy of chest X-ray prediction for COVID-19 detection executed on a low-power embedded system. The classic deep CNNs such as VGG16, VGG19 [27] have excellent performance, but they are difficult to deploy on devices with low hardware, due to the amount of model parameters and computational complexity. We use low-power embedded systems to design a low-cost portable point-of-care medical device that would be affordable for any hospital to acquire [28,29].
Many models have been developed and modified, but there is still room for improvement, as our research proves.
The main contributions of the proposed study can be summarized as follows:
- We propose a modified neural network structure of the MobileNetV2 model, to maximize the learning ability for classification of chest X-rays. The modified version of our architecture requires significant less training time than other existing DL architectures due to the small number of network parameters.
- Our design can classify four different categories of chest X-rays (COVID-19, normal, viral pneumonia and lung opacity). The accuracy of our approach is significantly higher than standard architecture and surpasses other state-of-the-art methods.
- This is the first study to investigate a large set of chest X-ray images (21.165 chest X-ray images) combined from many other studies that appear in the literature, which include few X-ray samples and mainly concern binary classifications.
- The modified structure of the architecture yields excellent classification results and, in combination with the small size of the network, leads to an attractive model for diagnosing chest X-rays in embedded systems.
2. Related Work
In the global fight against the virus, the rapid and valid assessment of COVID-19 infected patients is a major challenge for the research community. Several studies have been performed to demonstrate the potential of DL in detecting COVID-19 infected chest X-rays in the field of medical imaging.
Kamal et al. [30] evaluated eight pre-trained CNN models (MobileNetV2, MobileNet, VGG19, ResNet50, ResNet50V2, DenseNet121, InceptionV3 and NasNetMobile). The aperture in the preformed plates of 744 images exposes 186 normal, 186 COVID-19, 186 bacterial pneumonia, and 186 viral pneumonia. Each DL model trained 1000 epochs. The results showed that for 4 class classification, the MobileNetV2 model achieved 95.40% accuracy. In [31] the authors proposed a model called Bayesian CNN. The used dataset consisted of 68 COVID-19, 1.583 normal, 2.786 bacterial pneumonia and 1.504 viral pneumonia images. Their CNN model was trained for 25 epochs and for 4 class classification achieved 89.82% accuracy. Khan et al. [32] proposed a model called CoroNet, which is based on the Xception model and consists of 33 million parameters. The used dataset in their tests consisted of 284 COVID-19, 310 normal, 330 bacterial pneumonia and 327 viral pneumonia chest radiography images. Their DL model was trained for 80 epochs and for 4 class classification (COVID-19, bacterial pneumonia, viral pneumonia, normal) achieved an accuracy of 89.60%.
Mahmud et al. [33] suggest a model called CovXNet. The used dataset consisted of 305 COVID-19, 305 normal 305 viral pneumonia and 305 bacterial pneumonia. Their CNN model was trained for 150 epochs and for 4 class classification achieved 90.20% accuracy. Wang et al. [34] proposed a model called COVID Net, which consists of 11.75 million parameters. The dataset they used in their tests consisted of 358 COVID-19, 8.066 normal and 5.538 pneumonia. Their CNN model was trained for 22 epochs and for 3 class classification achieved 93.30% accuracy. Similarly, in [35] the authors proposed a model called DarkCovidNet. In their tests, the dataset they used consisted of 127 COVID-19, 500 normal and 500 pneumonia. Their DL model was trained for 100 epochs and for 3 class classification achieved an accuracy of 87.02%.
Furthermore, in [36] compared five pre-trained CNN models (MobileNetV2, VGG19, Xception, InceptionV3 and InceptionResNetV2). They used two sets of data, the first set consisting of 224 COVID-19, 504 normal, 700 bacterial pneumonia, while the second set consisted of 224 COVID-19, 504 normal, 714 bacterial and viral pneumonia. Each CNN model was trained with the same hyperparameters for 10 epochs. The DL MobileNetV2 architecture in the first set for 3 class classification achieved 92.85% accuracy and in the second dataset achieved 94.72% accuracy. Misra et al. [37] proposed a multi-channel pre-trained ResNet architecture. The dataset they used consisted of 1.579 normal, 4.245 pneumonia and 184 COVID-19 images. Their DL model was trained up to 500 epochs with early stopping criteria. Their model for 3 class classification achieved 93.90% accuracy.
In [38] the authors evaluated the VGG16, VGG19, DenseNet201, InceptionResNetV2, InceptionV3, Resnet50 and MobileNetV2 models. The dataset they used in their tests consisted of 2.780 images of bacterial pneumonia, 231 of COVID-19 and 1.583 normal. Each CNN model was trained with the same hyperparameters for 300 epochs. The InceptionResNetV2 model for 3 class classification achieved 92.18% accuracy, while the MobileNetV2 85.47%. Narinet al. [39] compared five pre-trained CNN models (ResNet50, ResNet101, ResNet152, InceptionV3 and InceptionResNetV2). In their study they applied three different binary classifications with four classes (COVID-19, viral pneumonia, bacterial pneumonia, normal). The used dataset in their tests consisted of 341 COVID-19, 2.800 normal, 2.772 bacterial pneumonia and 1.493 viral pneumonia chest radiography images. Each DL model trained 30 epochs. The results showed that the pre-trained ResNet50 model provided the highest classification performance in all three different binary classifications with an accuracy of over 96%. The ResNet50 model they propose consists of over 25 million parameters.
Most of the above studies used a limited dataset, which varies and includes minimal samples of COVID-19 X-rays ranging from 100 samples up to 500 using millions of parameters. Studies have shown an urgent need for efficient DL models in large-scale data with fewer parameters that will provide higher accuracy. This work presents a modified version of the MobileNetV2 model for detecting COVID-19 infection from chest X-ray images and focuses on 4 class classifications.
In contrast with all other authors, we propose a well-tuned DL architecture that can be executed on a low-power embedded system in real time (sustainable performance with over 120 fps). Furthermore, we have trained our model using a dataset of over 21 k images. In our study, our goal is for the model to combine small network size a small number of parameters and to show very high performance for the classification of the four most important cases for lung deceases (COVID-19, normal, lung opacity and viral pneumonia) in a large dataset 21.165 chest X-ray images.
3. Materials and Methods
In this section, we will describe the dataset. Then, we will describe the details of the proposed model architecture, the training on the embedded GPU and the performance metrics.
3.1. Dataset Description
In this study, the dataset we used in our experiments is the COVID-19 Radiography Database [40]. This dataset is currently one of the largest public databases with 21.165 chest X-ray images and includes 3.616 COVID-19 positive cases, 10.192 normal, 6.012 lung opacity (Non-COVID lung infection) and 1.345 viral pneumonia images. All the images are in PNG file format, and the resolution is pixels. Figure 1 shows a sample of the COVID-19 Radiography Database (normal, COVID-19, lung opacity and viral pneumonia).
Figure 1.
A sample of the COVID-19 Radiography Database.
3.2. Splitting the Dataset
We randomly split the dataset into training 70% (14.813 images), validation 20% (4.232 images), and testing 10% (2.120 images). The training set consists of 14.813 images, in which there were 2.530 COVID-19 images, 7.134 normal images, 4.208 lung opacity images and 941 viral pneumonia images. The validation set consists of 4.232 images, in which there were 723 COVID-19 images, 2.038 normal images, 1.202 lung opacity images and 269 viral pneumonia images. The test set consists of 2.120 images, in which there were 363 COVID-19 images, 1.020 normal images, 602 lung opacity images and 135 viral pneumonia images. The images used for testing were never used in the training process. The summary of the dataset (training, validation, and test) after the split is presented in Table 1.

Table 1.
The COVID-19 Radiography Database summary used in our research.
3.3. Data Pre-Processing
Most CNNs are trained on image resolution with pixels [41]. Therefore, in all the images of the COVID-19 Radiography Database [40] we changed the size from pixels to pixels. The size change was performed using the Python Imaging Library (PIL). The data augmentation methods such as random rotation, width shift, height shift, horizontal and vertical flip operations were not used because they have many limitations on medical images due to their strict format [42].
3.4. Proposed Modified Model Architecture
Most state-of-the-art CNNs have many millions of parameters to improve accuracy, and thus they are very time consuming during the training process. The proposed model is based on the MobileNetV2 architecture [41]. MobileNetV2 is one of the most popular lightweight network architectures for vision applications (classification, object detection and semantic segmentation). It is a very efficient model that improves the state-of-the-art performance on many visual recognition tasks. The MobileNetV2 is based on an inverted residual and linear bottleneck, significantly decreasing the number of operations and memory needed while retaining high accuracy. Although the classification accuracy of MobileNetV2 is 71% top-1 accuracy such as that of deep CNNs such as VGG16 and VGG19 (up 138 million parameters and 520 MB size) [27], it has the unique advantages of a smaller size network, fewer parameters, fewer operations, high efficiency, low-power and low latency.
The weights of the network are initialized with weights from a model pre-trained on ImageNet [43]. The top layer of the MobileNetV2 is discarded. To enhance the learning ability of the model, we added three additional new layers. In contrast with all other authors, we present an enhanced MobileNetV2 with more layers, improving the learning ability. The number and types of layers were determined after enough research into various networks and performing many different executions.
The first layer that we added is a global average pooling to minimize the overfitting and the number of parameters in the model by applying corresponding mathematical computation. Global average pooling generates one feature map for each corresponding category of the classification task in the last fully connected layer. The advantage of global average pooling is more native to the convolution structure between feature maps and categories [44].
After that, a fully connected layer with 512 nodes is added as a second layer with Rectified Linear Unit (ReLU) activation function [45]. The fully connected layer functions similar to a multilayer perceptron. Mathematical computation of ReLU activation function is shown in Equation (1).
A dropout layer is added, as the third layer, with a dropout rate of 20% to prevent network overfitting and divergence [46,47,48]. Dropout is a technique that randomly sets a portion of units, along with their connections, from the fully connected layers to zero during training. Dropout improves the performance of neural networks on supervised learning tasks in computer vision [48].
Finally, a fully connected layer is added as an output layer with 4 nodes, each one for every category, and SoftMax [49] is used as the activation function. This layer is used to predict output images. Mathematical computation of SoftMax activation function is shown in Equation (2).
where represent input data and m the number of classes.
The details of the architecture, parameters and output shape of the proposed model are presented in Table 2. It is important to notice that the global average pooling and the dropout layer consist of 0 parameters, while the fully connected layer with 512 nodes required our model to carry only 655.872 parameters but gave it higher accuracy than the standard architecture (omitting our 3 addition layers). Our model has a total of 2.915.908 parameters and is much lighter than many other studies that appear in the literature, which include more than 25 million parameters. The overall proposed methodology of the model is shown in Figure 2.

Table 2.
Details of the proposed architecture, parameters and output shape.
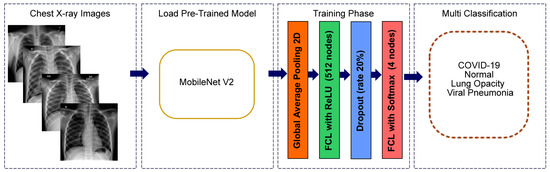
Figure 2.
Modified MobileNetV2 architecture for multiclass classification problem (COVID-19, normal, viral pneumonia and lung opacity).
3.5. Training Strategy
We train our model using a two-step strategy. In the first step, we freeze the internal layers of the network and train only the three new layers (global average pooling layer, fully connected ReLU layer and dropout layer) for 10 epochs using an Adam optimizer [50] ( and ) with a learning rate of 0.001. In this step, the learning is enhanced in the features of our dataset.
In the second step, we are reducing the learning rate of 0.0001 for 20 epochs and training throughout the network to inform all the weights in the network. The batch size of 32 remained constant throughout the training. In both phases of training, the activation function at intermediate layers is ReLU, and at the output layer is SoftMax. Loss function is categorical cross-entropy, and the dropout value is 0.20. The hyperparameters used are presented in Table 3.

Table 3.
The hyperparameters were used for all the experiments on the Jetson AGX Xavier embedded system.
3.6. Training the Model on the Embedded GPU
Training and testing are performed on the Nvidia Jetson AGX Xavier [51] with 512 CUDA cores, an 8 core ARM processor, 32 GB RAM, 32 GB eMMC and various other peripherals. Nvidia Jetson CPU-GPU heterogeneous architecture achieves high-performance computing and where can be quickly programmed to accelerate complex DL tasks [52]. The embedded GPU has shown great acceleration potential and involves low-power, high accuracy and efficiency in point-of-care medical applications [28,29,53,54]. At the same time, they provide local processing, eliminating security and privacy issues where required in biomedical systems [55]. Finally, we need a portable tool to facilitate medical diagnosis with low-weight, low-cost devices with accuracy, speed and power efficiency. The CNN models have trained on the Keras [56] framework.
The system can process up to 115 images per second suitable for real-time processing with approximately 28.5 Watts of maximum power consumption, which is much lower than the typical 1 kWatt for a PC with a GPU that executes very large DL models and cannot reach our highly optimized accuracy.
3.7. Model Evaluation Metrics on the Test Dataset
Four metrics (accuracy, precision, recall and f1-score) [57] were used to evaluate the performance of the standard MobileNetV2 and the modified MobileNetV2 architectures. The calculation types of the metrics are shown in Equations (3)–(6), where TP, FN, FP, TN represent the number of true positives, false negatives, false positives and true negatives.
4. Experimental Results
All executions were performed on the Jetson AGX Xavier embedded system. The total training times of standard MobileNetV2 were 1 h 38 min 22 s, and for the modified MobileNetV2, it was 1 h 40 min 52 s. Table 4 shows the performance comparison of the standard MobileNetV2 and the modified MobileNetV2. The total accuracy is 95.80% in the modified MobileNetV2, and the standard MobileNetV2 is 90.47%. The support is the number of occurrences of each class in the testing dataset, as shown in the classification report.

Table 4.
Comparison of classification report of standard MobileNetV2 and modified MobileNetV2 for 4 classes (COVID-19, normal, viral pneumonia and lung opacity) classification.
In Figure 3 and Figure 4 we show the training accuracy/loss and validation accuracy/loss for standard MobileNetV2 and modified MobileNetV2, respectively. Modified MobileNetV2 architecture achieved 95.80% testing accuracy, while MobileNetV2 standard 90.47% for 4 classes of the public COVID-19 Radiography Database. It is clear that the performance of our modified MobileNetV2 model is much better than the standard MobileNetV2. Additionally, the modified MobileNetV2 model is more stable, while the standard MobileNetV2 shows more oscillation. The confusion matrix of the standard MobileNetV2 and proposed method is shown in Figure 5 and Figure 6. It is evident that our modified MobileNetV2 achieves much better results, classifying correctly many more images than the standard MobileNetV2.
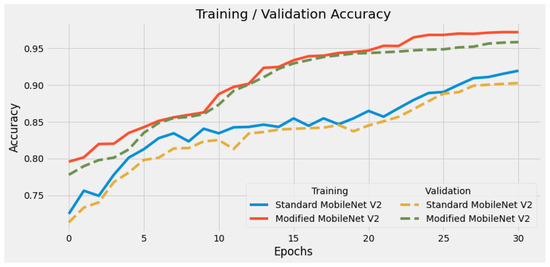
Figure 3.
Comparison of training accuracy of standard MobileNetV2 and modified MobileNetV2. Our modified DL model presents a stable high accuracy due to its minimum loss per epoch.
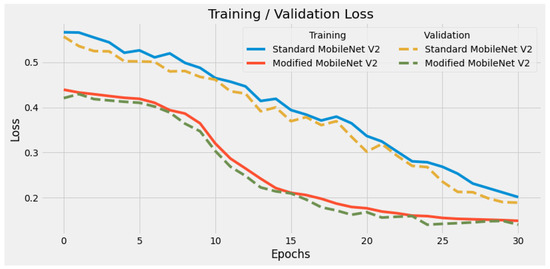
Figure 4.
Comparison of training loss of standard MobileNetV2 and modified MobileNetV2. Our modified DL model has much lower loss in every epoch, illustrating its high optimized structure.
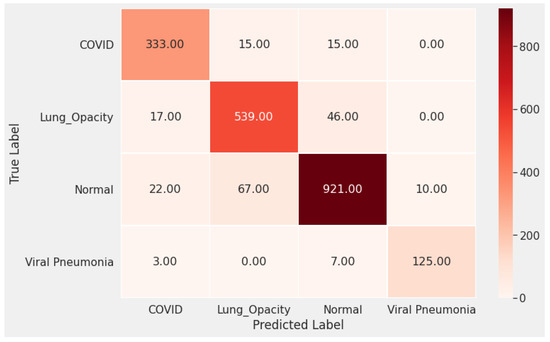
Figure 5.
The confusion matrix of the standard MobileNetV2 shows many false negative and false positive classifications, especially for the COVID-19 cases.
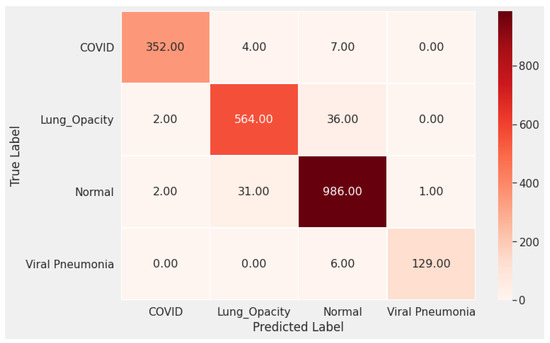
Figure 6.
The confusion matrix of the proposed method (Modified MobileNetV2) shows that our optimized model predicts correctly in most cases much better than the standard DL model.
Comparison with Other Approaches
Comparing the performance of the proposed architecture with other existing methods is not possible because the dataset is different. Most studies are mainly concerned with binary classification, while there is a relatively limited number of studies concerning multiple classes.
However, in Table 5, we summarize the overall accuracy and architecture performance compared to our modified model from other existing models. Specifically, we only look at architectures used to classify 3 or 4 classes, but most have a small number of COVID-19 samples. It is evident that our model has much higher accuracy than every other research, backed up by the biggest dataset of X-rays examined.

Table 5.
Comparison of the proposed model with other existing models for COVID-19 classification using chest X-ray images for 3 or 4 class classification.
As shown in Table 5, the proposed dataset presents the largest number of COVID-19 cases. We introduce a well-tuned DL technique that can be executed on a low-power embedded system in real time with promising results. We propose three additional new layers (global average pooling, fully connected layer with 512 nodes and dropout) and achieve high performance, small network size and a small number of parameters. We achieve the highest accuracy compared to the literature review with an accuracy of 95.80% for multiclass classification.
5. Conclusions and Future Work
COVID-19 has spread rapidly worldwide and is a severe public health problem. Thus, an early and valid prognosis of COVID-19 infected patients is vital to preventing the spread of the disease. This study proposes a lightweight neural network that provides high prediction accuracy from chest X-rays. The results of this study seem promising, but they must be further improved as more and more COVID-19 chest X-ray data become available. In fact, the model benefits of the numerosity of the examined X-ray dataset in reaching much higher accuracy than every other compared research. Our architecture approach achieves 95.80% accuracy, the highest up to this date compared with similar research, and processes up to 115 images per second with approximately 28.5 Watts of maximum power consumption on the Nvidia Jetson AGX Xavier. Furthermore, our work is the first to propose an optimized model for a low-power embedded system with increased detection accuracy. In a later study, we will test the performance of the proposed approach on different modern CNN models with an even larger number of COVID-19 and the number of classes chest X-rays in the dataset.
Author Contributions
Conceptualization, A.S., T.S. and D.T.; Formal analysis, A.S. and D.T.; Investigation, T.S. and A.S.; Methodology, T.S., D.T. and A.S.; Validation, A.S. and T.S.; Project administration, A.S. and M.D.; Supervision, M.D.; Writing—original draft, T.S., D.T. and A.S.; Writing—review and editing, A.S. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database/activity (accessed on 25 May 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial Intelligence |
| ARDS | Acute Respiratory Distress Syndrome |
| ARM | Architecture Reference Manual |
| CNN | Convolutional Neural Network |
| CPU | Central Processing Unit |
| CUDA | Compute Unified Device Architecture |
| DL | Deep Learning |
| eMMC | Embedded MultiMedia Card |
| FN | False Negative |
| FP | False Positive |
| GPU | Graphics Processing Unit |
| RAM | Random access memory |
| ReLU | Rectified Linear Unit |
| PC | Personal Computer |
| PIL | Python Imaging Library |
| PNG | Portable Network Graphics |
| TN | True Negative |
| TP | True Positive |
| WHO | World Health Organization |
References
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/ (accessed on 5 January 2022).
- Worldometer. COVID-19 Coronavirus Pandemic. Available online: https://www.worldometers.info/coronavirus/ (accessed on 15 January 2022).
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Puderbach, M.; Eichinger, M.; Haeselbarth, J.; Ley, S.; Kopp-Schneider, A.; Tuengerthal, S.; Schmaehl, A.; Fink, C.; Plathow, C.; Wiebel, M.; et al. Assessment of morphological MRI for pulmonary changes in cystic fibrosis (CF) patients: Comparison to thin-section CT and chest X-ray. Investig. Radiol. 2007, 42, 715–724. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Tiwari, P.; Kumar, S.; Gupta, D.; Khanna, A.; Rodrigues, J.J. Identifying pneumonia in chest X-rays: A deep learning approach. Measurement 2019, 145, 511–518. [Google Scholar] [CrossRef]
- Bai, H.X.; Hsieh, B.; Xiong, Z.; Halsey, K.; Choi, J.W.; Tran, T.M.L.; Pan, I.; Shi, L.B.; Wang, D.C.; Mei, J.; et al. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology 2020, 296, E46–E54. [Google Scholar] [CrossRef]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [Green Version]
- Toussie, D.; Voutsinas, N.; Finkelstein, M.; Cedillo, M.A.; Manna, S.; Maron, S.Z.; Jacobi, A.; Chung, M.; Bernheim, A.; Eber, C.; et al. Clinical and chest radiography features determine patient outcomes in young and middle-aged adults with COVID-19. Radiology 2020, 297, E197–E206. [Google Scholar] [CrossRef]
- Gattinoni, L.; Chiumello, D.; Caironi, P.; Busana, M.; Romitti, F.; Brazzi, L.; Camporota, L. COVID-19 pneumonia: Different respiratory treatments for different phenotypes? Intensive Care Med. 2020, 46, 1099–1102. [Google Scholar] [CrossRef]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef] [Green Version]
- Hurt, B.; Kligerman, S.; Hsiao, A. Deep learning localization of pneumonia: 2019 coronavirus (COVID-19) outbreak. J. Thorac. Imaging 2020, 35, W87–W89. [Google Scholar] [CrossRef]
- Longoni, C.; Bonezzi, A.; Morewedge, C.K. Resistance to medical artificial intelligence. J. Consum. Res. 2019, 46, 629–650. [Google Scholar] [CrossRef]
- Sanida, T.; Varlamis, I. Application of Affinity Analysis Techniques on Diagnosis and Prescription Data. In Proceedings of the 2017 IEEE 30th International Symposium on Computer-Based Medical Systems (CBMS), Thessaloniki, Greece, 22–24 June 2017; pp. 403–408. [Google Scholar] [CrossRef]
- Sanchez-Reyes, L.M.; Rodriguez-Resendiz, J.; Salazar-Colores, S.; Avecilla-Ramírez, G.N.; Pérez-Soto, G.I. A High-accuracy mathematical morphology and multilayer perceptron-based approach for melanoma detection. Appl. Sci. 2020, 10, 1098. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Perez, D.; Rodríguez-Reséndiz, J.; Gómez-Loenzo, R.A. A study of computing zero crossing methods and an improved proposal for EMG signals. IEEE Access 2020, 8, 8783–8790. [Google Scholar] [CrossRef]
- Ortiz-Echeverri, C.J.; Salazar-Colores, S.; Rodríguez-Reséndiz, J.; Gómez-Loenzo, R.A. A new approach for motor imagery classification based on sorted blind source separation, continuous wavelet transform, and convolutional neural network. Sensors 2019, 19, 4541. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Reyes, L.M.; Rodríguez-Reséndiz, J.; Avecilla-Ramírez, G.N.; García-Gomar, M.L.; Robles-Ocampo, J.B. Impact of eeg parameters detecting dementia diseases: A systematic review. IEEE Access 2021, 9, 78060–78074. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020, 14, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Bullock, J.; Luccioni, A.; Pham, K.H.; Lam, C.S.N.; Luengo-Oroz, M. Mapping the landscape of artificial intelligence applications against COVID-19. J. Artif. Intell. Res. 2020, 69, 807–845. [Google Scholar] [CrossRef]
- Tayarani-N, M.H. Applications of artificial intelligence in battling against COVID-19: A literature review. Chaos Solitons Fractals 2020, 142, 110338. [Google Scholar] [CrossRef]
- Yang, W.; Sirajuddin, A.; Zhang, X.; Liu, G.; Teng, Z.; Zhao, S.; Lu, M. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19). Eur. Radiol. 2020, 30, 4874–4882. [Google Scholar] [CrossRef] [Green Version]
- Greenspan, H.; Van Ginneken, B.; Summers, R.M. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Sanida, T.; Tsiktsiris, D.; Sideris, A.; Dasygenis, M. A Heterogeneous Lightweight Network for Plant Disease Classification. In Proceedings of the 2021 10th International Conference on Modern Circuits and Systems Technologies (MOCAST), Thessaloniki, Greece, 5–7 July 2021; pp. 1–4. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Paluru, N.; Dayal, A.; Jenssen, H.B.; Sakinis, T.; Cenkeramaddi, L.R.; Prakash, J.; Yalavarthy, P.K. Anam-Net: Anamorphic depth embedding-based lightweight CNN for segmentation of anomalies in COVID-19 chest CT images. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 932–946. [Google Scholar] [CrossRef]
- An, L.; Peng, K.; Yang, X.; Huang, P.; Luo, Y.; Feng, P.; Wei, B. E-TBNet: Light Deep Neural Network for Automatic Detection of Tuberculosis with X-ray DR Imaging. Sensors 2022, 22, 821. [Google Scholar] [CrossRef]
- Kamal, K.; Yin, Z.; Wu, M.; Wu, Z. Evaluation of deep learning-based approaches for COVID-19 classification based on chest X-ray images. Signal Image Video Process. 2021, 15, 959–966. [Google Scholar]
- Ghoshal, B.; Tucker, A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv 2020, arXiv:2003.10769. [Google Scholar]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Mahmud, T.; Rahman, M.A.; Fattah, S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misra, S.; Jeon, S.; Lee, S.; Managuli, R.; Jang, I.S.; Kim, C. Multi-channel transfer learning of chest x-ray images for screening of COVID-19. Electronics 2020, 9, 1388. [Google Scholar] [CrossRef]
- El Asnaoui, K.; Chawki, Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J. Biomol. Struct. Dyn. 2020, 39, 3615–3626. [Google Scholar] [CrossRef] [PubMed]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021, 24, 1207–1220. [Google Scholar] [CrossRef]
- Kaggle. COVID-19 Radiography Dataset. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database/activity (accessed on 25 May 2021).
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Hinton, G.E. Rectified linear units improve restricted boltzmann machines vinod nair. Citeseer 2010, 7, 1–8. [Google Scholar]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580. [Google Scholar]
- Hawkins, D.M. The problem of overfitting. J. Chem. Inf. Comput. Sci. 2004, 44, 1–12. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Nvidia Developer. Jetson AGX Xavier Developer Kit. Available online: https://developer.nvidia.com/embedded/jetson-agx-xavier-developer-kit (accessed on 30 May 2021).
- Mittal, S.; Vetter, J.S. A survey of CPU-GPU heterogeneous computing techniques. ACM Comput. Surv. (CSUR) 2015, 47, 1–35. [Google Scholar] [CrossRef]
- Ardiyanto, I.; Nugroho, H.A.; Buana, R.L.B. Deep learning-based diabetic retinopathy assessment on embedded system. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 1760–1763. [Google Scholar]
- Page, A.; Shea, C.; Mohsenin, T. Wearable seizure detection using convolutional neural networks with transfer learning. In Proceedings of the 2016 IEEE International Symposium on Circuits and Systems (ISCAS), Montreal, QC, Canada, 22–25 May 2016; pp. 1086–1089. [Google Scholar]
- Attaran, N.; Puranik, A.; Brooks, J.; Mohsenin, T. Embedded low-power processor for personalized stress detection. IEEE Trans. Circuits Syst. II Express Briefs 2018, 65, 2032–2036. [Google Scholar] [CrossRef]
- Géron, A. Hands-on Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems; O’Reilly Media: Sebastopol, CA, USA, 2019. [Google Scholar]
- Hossin, M.; Sulaiman, M. A review on evaluation metrics for data classification evaluations. Int. J. Data Min. Knowl. Manag. Process. 2015, 5, 1. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
