Population Genomic Approaches for Weed Science
Abstract
1. Introduction
2. Developing Weed Genome Sequences as a Fundamental Tool
2.1. What Is a Draft Genome?
2.2. Preparing and Assessing Plant Material
2.3. DNA Extraction
2.4. Sequencing Strategies
2.5. Data Assessment, Correction and Filtering
2.6. Assembly and Assessment
2.7. Polishing
2.8. Scaffolding
2.9. Gene Prediction and Annotation
2.10. Examples: Three Recently Sequenced Weed Genomes
3. Current Application: What Are Agricultural Weeds and Where Do They Come From?
3.1. Detecting the Signatures of Demographic Change and Selection on the Genome
3.2. Example: Convergent Adaptation to Glyphosate in Common Waterhemp
4. Current Application: What Genes Underlie Herbicide Resistance?
4.1. Five Superfamilies of Suspects
4.1.1. Cytochrome P450 Monooxygenases
4.1.2. Glutathione S-Transferases
4.1.3. ATP-Binding Cassette Transporters
4.1.4. MFS Transporters
4.1.5. Glycosyltransferases
4.2. A Role for Genomic Approaches
4.3. Example: Glyphosate NTSR in Morning Glory
5. Future Application: Can We Genetically Alter Weed Population to Make Them Easier to Control?
5.1. The Potential for Manipulation of Weed Populations
5.2. Additional Technical Challenges
5.3. Evolutionary Consequences and the Need for Integration with Other Management Strategies
5.4. Example: Gene Drive in Malaria Vector Mosquitos
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-W.; Harkess, A. A guide to sequence your favorite plant genomes. Appl. Plant Sci. 2018, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dominguez Del Angel, V.; Hjerde, E.; Sterck, L.; Capella-Gutierrez, S.; Notredame, C.; Vinnere Pettersson, O.; Amselem, J.; Bouri, L.; Bocs, S.; Klopp, C.; et al. Ten steps to get started in Genome Assembly and Annotation. F1000Research 2018, 7, 148. [Google Scholar] [CrossRef]
- Armstrong, O.; Fiddes, I.T.; Diekhans, M.; Paten, B. Whole-Genome Alignment and Comparative Annotation. Annu. Rev. Anim. Biosci. 2019, 7, 41–64. [Google Scholar] [CrossRef]
- Gillings, M.R.; Paulsen, I.T.; Tetu, S.G. Genomics and the evolution of antibiotic resistance. Ann. N. Y. Acad. Sci. 2017, 1388, 92–107. [Google Scholar] [CrossRef]
- Loman, N.J.; Pallen, M.J. Twenty years of bacterial genome sequencing. Nat. Rev. Microbiol. 2015, 13, 787–794. [Google Scholar] [CrossRef]
- Hatfull, G.F. Bacteriophage genomics. Curr. Opin. Microbiol. 2008, 11, 447–453. [Google Scholar] [CrossRef]
- Holmes, E.C. Viral Evolution in the Genomic Age. PLoS Biol. 2007, 5, e278. [Google Scholar] [CrossRef][Green Version]
- Gudbjartsson, D.F.; Helgason, H.; Gudjonsson, S.A.; Zink, F.; Oddson, A.; Gylfason, A.; Besenbacher, S.; Magnusson, G.; Halldorsson, B.V.; Hjartarson, E.; et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015, 47, 435–444. [Google Scholar] [CrossRef]
- Stranger, B.E.; Nica, A.C.; Forrest, M.S.; Dimas, A.; Bird, C.P.; Beazley, C.; Ingle, C.E.; Dunning, M.; Flicek, P.; Koller, D.; et al. Population genomics of human gene expression. Nat. Genet. 2007, 39, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, D.L.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Collins, F.S.; De La Vega, F.M.; Donnelly, P.; Egholm, M.; et al. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar]
- Li, J.Z.; Absher, D.M.; Tang, H.; Southwick, A.M.; Casto, A.M.; Ramachandran, S.; Cann, H.M.; Barsh, G.S.; Feldman, M.; Cavalli-Sforza, L.L.; et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008, 319, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Ravet, K.; Patterson, E.L.; Krähmer, H.; Hamouzová, K.; Fan, L.; Jasieniuk, M.; Lawton-Rauh, A.; Malone, J.M.; McElroy, J.S.; Merotto, A.; et al. The power and potential of genomics in weed biology and management. Pest Manag. Sci. 2018, 74, 2216–2225. [Google Scholar] [CrossRef] [PubMed]
- Basu, C.; Halfhill, M.D.; Mueller, T.C.; Stewart, C.N. Weed genomics: New tools to understand weed biology. Trends Plant Sci. 2004, 9, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [CrossRef]
- Michael, T.P.; Jackson, S. The First 50 Plant Genomes. Plant Genome 2013, 6, 1–7. [Google Scholar] [CrossRef]
- Veeckman, E.; Ruttink, T.; Vandepoele, K. Are We There Yet? Reliably Estimating the Completeness of Plant Genome Sequences. Plant Cell 2016, 28, 1759–1768. [Google Scholar] [CrossRef]
- Jung, H.; Winefield, C.; Bombarely, A.; Prentis, P.; Waterhouse, P. Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes. Trends Plant Sci. 2019, 8, 1–25. [Google Scholar] [CrossRef]
- Ekblom, R.; Wolf, J.B.W. A field guide to whole-genome sequencing, assembly and annotation. Evol. Appl. 2014, 7, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Wajid, B.; Serpedin, E. Do it yourself guide to genome assembly. Brief. Funct. Genom. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Leitch, I.; Johnston, E.; Pellicer, J.; Hidalgo, O.; Bennett, M. Angiosperm DNA C-Values Database. Available online: https://cvalues.science.kew.org/ (accessed on 28 May 2019).
- Rice, A.; Glick, L.; Abadi, S.; Einhorn, M.; Kopelman, N.M.; Salman-Minkov, A.; Mayzel, J.; Chay, O.; Mayrose, I. The Chromosome Counts Database (CCDB)—A community resource of plant chromosome numbers. New Phytol. 2015, 206, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Greilhuber, J.; Temsch, E.M.; Loureiro, J.C.M. Nuclear DNA Content Measurement. In Flow Cytometry with Plant Cells; Doležel, J., Greilhuber, J., Suda, J., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; pp. 67–101. ISBN 9783527314874. [Google Scholar]
- Doležel, J.; Bartoš, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Leitch, I.J.; Bennett, M.D. Genome size and its uses: The impact of flow cytometry. In Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes; Doležel, J., Greilhuber, J., Suda, J., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; pp. 153–176. [Google Scholar]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.W.; Kron, P.; Martin, S.L. flowPloidy: An R package for genome size and ploidy assessment of flow cytometry data. Appl. Plant Sci. 2018, 6, e01164. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Bartoš, J.; Voglmayr, H.; Greilhuber, J. Nuclear DNA content and genome size of trout and human. Cytometry 2003, 51A, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Barow, M.; Meister, A. Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ. 2003, 26, 571–584. [Google Scholar] [CrossRef]
- Barow, M.; Jovtchev, G. Endopolyploidy in Plants and its Analysis by Flow Cytometry. In Flow Cytometry with Plant Cells; Doležel, J., Greilhuber, J., Suda, J., Eds.; WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; pp. 349–372. ISBN 9783527314874. [Google Scholar]
- Doležel, J.; Kubaláková, M.; Suchánková, P.; Kovářová, P.; Bartoš, J.; Šimková, H. Chromosome analysis and sorting. In Flow Cytometry with Plant Cells; Doležel, J., Greilhuber, J., Suda, J., Eds.; WILEY-VCH Verlag GmbH & Co.: Weinheim, Germany, 2007; pp. 373–404. ISBN 9783527314874. [Google Scholar]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef]
- Heap, I. The International Survey of Herbicide Resistant Weeds. Available online: www.weedscience.org (accessed on 4 January 2018).
- United State Department of Agriculture Federal Noxious Weeds. Available online: https://plants.usda.gov/java/noxious (accessed on 25 July 2019).
- Australian Government Weeds of National Significance. Available online: https://www.environment.gov.au/biodiversity/invasive/weeds/weeds/lists/wons.html (accessed on 25 July 2019).
- Weber, E.; Gut, D. A survey of weeds that are increasingly spreading in Europe. In Agronomy for Sustainable Development; Springer Verlag/EDP Sciences/INRA: Berlin/Heidelberg, Germany, 2005; pp. 109–121. [Google Scholar]
- Minister of Agriculture and Agri-Food Canada (AAFC). Weed Seeds Order; Minister of Agriculture and Agri-Food (AAFC), Canada: Ottawa, ON, Canada, 2016.
- Straub, S.C.K.; Cronn, R.C.; Edwards, C.; Fishbein, M.; Liston, A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol. Evol. 2013, 5, 1872–1885. [Google Scholar] [CrossRef]
- Byrne, S.L.; Erthmann, P.Ø.; Agerbirk, N.; Bak, S.; Hauser, T.P.; Nagy, I.; Paina, C.; Asp, T. The genome sequence of Barbarea vulgaris facilitates the study of ecological biochemistry. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bettgenhaeuser, J.; Corke, F.M.K.; Opanowicz, M.; Green, P.; Hernández-Pinzón, I.; Doonan, J.H.; Moscou, M.J. Natural Variation in Brachypodium Links Vernalization and Flowering Time Loci as Major Flowering Determinants. Plant Physiol. 2017, 173, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Wang, X.; Liu, B.; Wu, J.; Liang, J.; Cui, Y.; Cheng, F.; Wang, X. Brassica rapa Genome 2.0: A Reference Upgrade through Sequence Re-assembly and Gene Re-annotation. Mol. Plant 2017, 10, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011, 12, R102. [Google Scholar] [CrossRef] [PubMed]
- Kasianov, A.S.; Klepikova, A.V.; Kulakovskiy, I.V.; Gerasimov, E.S.; Fedotova, A.V.; Besedina, E.G.; Kondrashov, A.S.; Logacheva, M.D.; Penin, A.A. High-quality genome assembly of Capsella bursa-pastoris reveals asymmetry of regulatory elements at early stages of polyploid genome evolution. Plant J. 2017, 91, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Zhang, H.; Chen, B.; Nie, S.; Liu, H.; Gao, W.; Wang, H.; Gao, Y.; Gu, L. De novo genome assembly of the stress tolerant forest species Casuarina equisetifolia provides insight into secondary growth. Plant J. 2019, 97, 779–794. [Google Scholar] [CrossRef]
- Griesmann, M.; Chang, Y.; Liu, X.; Song, Y.; Haberer, G.; Crook, M.B.; Billault-Penneteau, B.; Lauressergues, D.; Keller, J.; Imanishi, L.; et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 2018, 361, eaat1743. [Google Scholar] [CrossRef]
- Wang, L.; He, F.; Huang, Y.; He, J.; Yang, S.; Zeng, J.; Deng, C.; Jiang, X.; Fang, Y.; Wen, S.; et al. Genome of Wild Mandarin and Domestication History of Mandarin. Mol. Plant 2018, 11, 1024–1037. [Google Scholar] [CrossRef]
- Peng, Y.; Lai, Z.; Lane, T.; Nageswara-Rao, M.; Okada, M.; Jasieniuk, M.; O’Geen, H.; Kim, R.W.; Sammons, R.D.; Rieseberg, L.H.; et al. De novo genome assembly of the economically important weed horseweed using integrated data from multiple sequencing platforms. Plant Physiol. 2014, 166, 1241–1254. [Google Scholar] [CrossRef]
- Sarkar, D.; Mahato, A.K.; Satya, P.; Kundu, A.; Singh, S.; Jayaswal, P.K.; Singh, A.; Bahadur, K.; Pattnaik, S.; Singh, N.; et al. The draft genome of Corchorus olitorius cv. JRO-524 (Navin). Genom. Data 2017, 12, 151–154. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; Gonzalez, V.M.; Henaff, E.; Camara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 1–17. [Google Scholar]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L.; et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Honig, J.A.; Zelzion, E.; Wagner, N.E.; Kubik, C.; Averello, V.; Vaiciunas, J.; Bhattacharya, D.; Bonos, S.A.; Meyer, W.A. Microsatellite identification in perennial ryegrass using next-generation sequencing. Crop Sci. 2017, 57, S-331–S-340. [Google Scholar] [CrossRef]
- Vining, K.J.; Johnson, S.R.; Ahkami, A.; Lange, I.; Parrish, A.N.; Trapp, S.C.; Croteau, R.B.; Straub, S.C.K.; Pandelova, I.; Lange, B.M. Draft Genome Sequence of Mentha longifolia and Development of Resources for Mint Cultivar Improvement. Mol. Plant 2017, 10, 323–339. [Google Scholar] [CrossRef]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W.; et al. The genome of broomcorn millet. Nat. Commun. 2019, 10, 491–500. [Google Scholar] [CrossRef]
- Guo, L.; Guo, L.; Winzer, T.; Yang, X.; Li, Y.; Ning, Z.; He, Z.; Teodor, R.; Lu, Y.; Tim, A.; et al. The opium poppy genome and morphinan production. Science 2018, 362, 343–347. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Wang, X.-R.; Zeng, Q.-Y. De novo assembly of white poplar genome and genetic diversity of white poplar population in Irtysh River basin in China. Sci. China Life Sci. 2019, 62, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Moghe, G.D.; Hufnagel, D.E.; Tang, H.B.; Xiao, Y.L.; Dworkin, I.; Town, C.D.; Conner, J.K.; Shiu, S.H. Consequences of Whole-Genome Triplication as Revealed by Comparative Genomic Analyses of the Wild Radish Raphanus raphanistrum and Three Other Brassicaceae Species. Plant Cell 2014, 26, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Xiaohui, Z.; Zhen, Y.; Shiyong, M.; Yang, Q.; Xinhua, Y.; Xiaohua, C.; Feng, C.; Zhangyan, W.; Yuyan, S.; Yi, J.; et al. A de novo Genome of a Chinese Radish Cultivar. Hortic. Plant J. 2015, 1, 155–164. [Google Scholar]
- Nakamura, N.; Hirakawa, H.; Sato, S.; Otagaki, S.; Matsumoto, S.; Tabata, S.; Tanaka, Y. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses. DNA Res. 2018, 25, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E.; Schmutzer, T.; Barilar, I.; Mascher, M.; Gundlach, H.; Martis, M.M.; Twardziok, S.O.; Hackauf, B.; Gordillo, A.; Wilde, P.; et al. Towards a whole-genome sequence for rye (Secale cereale L.). Plant J. 2017, 89, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Giolai, M.; Paajanen, P.; Verweij, W.; Witek, K.; Jones, J.D.G.; Clark, M.D. Comparative analysis of targeted long read sequencing approaches for characterization of a plant’s immune receptor repertoire. BMC Genom. 2017, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Dorn, K.M.; Fankhauser, J.D.; Wyse, D.L.; Marks, M.D. A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Res. 2015, 22, 121–131. [Google Scholar] [CrossRef]
- Creber, H.M.C.; Davies, M.S.; Francis, D.; Walker, H.D. Variation in DNA C value in natural populations of Dactylis glomerata L. New Phytol. 1994, 128, 555–561. [Google Scholar] [CrossRef]
- Beck, J. Meiotic Chromosome Counting in Flowering Plants Part 1 [Video File]. Available online: www.youtube.com/watch?v=iXqni6knH5A&t (accessed on 30 May 2019).
- Beck, J. Meiotic Chromosome Counting in Flowering Plants Part 2 [Video File]. Available online: www.youtube.com/watch?v=xVV4qBfSQLs&t (accessed on 30 May 2019).
- Kato, A. Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem. 1999, 74, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Lamb, J.C.; Albert, P.S.; Danilova, T.; Han, F.; Gao, Z.; Findley, S.; Birchler, J.A. Chromosome Painting for Plant Biotechnology. In Plant Chromosome Engineering. Methods in Molecular Biology (Methods and Protocols); Birchler, J.A., Ed.; Humana Press: Totowa, NJ, USA, 2011; Volume 701, pp. 67–96. ISBN 9781617379574. [Google Scholar]
- Mandáková, T.; Lysak, M.A. Chromosome Preparation for Cytogenetic Analyses in Arabidopsis. Curr. Protoc. Plant Biol. 2016, 1, 43–51. [Google Scholar]
- Chikhi, R.; Medvedev, P. Informed and automated k-mer size selection for genome assembly. Bioinformatics 2014, 30, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 2010, 11, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; MacAs, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.L.; Smith, T.; James, T.; Shalabi, F.; Kron, P.; Sauder, C.A. An update to the Canadian range and abundance of Camelina spp. (Brassicaceae) east of the Rocky Mountains. Botany 2017, 95, 405–417. [Google Scholar] [CrossRef]
- Roessler, K.; Muyle, A.; Diez, C.M.; Gaut, G.R.J.; Bousios, A.; Stitzer, M.C.; Seymour, D.K.; Doebley, J.F.; Liu, Q.; Gaut, B.S. The Genomics of Selfing in Maize (Zea mays ssp. mays): Catching Purging in the Act. bioRxiv 2019, 594812. [Google Scholar] [CrossRef]
- Palmer, C.E.D.; Keller, W.A. Overview of Haploidy. In Biotechnology in Agriculture and Forestry Haploids in Crop Improvement II Vol.56; Palmer, C.E., Keller, W.A., Kasha, K.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 56, pp. 3–9. [Google Scholar]
- Forster, B.P.; Thomas, W.T.B. Doubled haploids in genetics and plant breeding. In Plant Breeding Reviews Vol. 25; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005; Volume 25, pp. 57–88. ISBN 9780471666936. [Google Scholar]
- Dunwell, J.M. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol. J. 2010, 8, 377–424. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidou, M.; Tai, H.H.; Anglin, N.L.; Ellis, D.; Strömvik, M.V. Current Strategies of Polyploid Plant Genome Sequence Assembly. Front. Plant Sci. 2018, 9, 1660–1675. [Google Scholar] [CrossRef]
- Carter, R.; Bryson, C.T.; Darbyshire, S.J. Preparation and Use of Voucher Specimens for Documenting Research in Weed Science. Weed Technol. 2007, 21, 1101–1108. [Google Scholar] [CrossRef]
- Hussing, C.; Kampmann, M.L.; Mogensen, H.S.; Børsting, C.; Morling, N. Comparison of techniques for quantification of next-generation sequencing libraries. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e276–e278. [Google Scholar] [CrossRef]
- Kreiner, J.M.; Giacomini, D.; Bemm, F.; Waithaka, B.; Regalado, J.; Lanz, C.; Hildebrandt, J.; Sikkema, P.H.; Tranel, P.J.; Weigel, D.; et al. Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus. bioRxiv 2018, 1–17. [Google Scholar] [CrossRef]
- Patterson, E.L.; Saski, C.A.; Sloan, D.B.; Tranel, P.J.; Westra, P.; Gaines, T.A. The draft genome of Kochia scoparia and the mechanism of glyphosate resistance via transposon-mediated EPSPS tandem gene duplication. bioRxiv 2019. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid procedure for DNA purification from small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Waterman, M.S. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics 1988, 2, 231–239. [Google Scholar] [CrossRef]
- Soorni, A.; Haak, D.; Zaitlin, D.; Bombarely, A. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genom. 2017, 18, 49–57. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L.; et al. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. bioRxiv 2016, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Laetsch, D.R.; Blaxter, M.L. BlobTools: Interrogation of genome assemblies. F1000Research 2017, 6, 1287. [Google Scholar] [CrossRef]
- Kajitani, R.; Yoshimura, D.; Okuno, M.; Minakuchi, Y.; Kagoshima, H.; Fujiyama, A.; Kubokawa, K.; Kohara, Y.; Toyoda, A.; Itoh, T. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simao, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Alonge, M.; Soyk, S.; Ramakrishnan, S.; Wang, X.; Goodwin, S.; Sedlazeck, F.J.; Lippman, Z.B.; Schatz, M.C. Fast and accurate reference-guided scaffolding of draft genomes. bioRxiv 2019, 519637. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genome Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Vaser, R.; Nagarajan, N.; Sović, I.; Šikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Oddes, S.; Zelig, A.; Kaplan, N. Three invariant Hi-C interaction patterns: Applications to genome assembly. bioRxiv 2018, 142, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Ghurye, J.; Pop, M.; Koren, S.; Bickhart, D.; Chin, C.S. Scaffolding of long read assemblies using long range contact information. BMC Genom. 2017, 18, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, Z.N.; Rhie, A.; Koren, S.; Concepcion, G.T.; Peluso, P.; Munson, K.M.; Hiendleder, S.; Fedrigo, O.; Jarvis, E.D.; Adam, M.; et al. Extended haplotype phasing of de novo genome assemblies with FALCON-Phase. bioRxiv 2018, 1–27. [Google Scholar] [CrossRef]
- Jibran, R.; Dzierzon, H.; Bassil, N.; Bushakra, J.M.; Edger, P.P.; Sullivan, S.; Finn, C.E.; Dossett, M.; Vining, K.J.; Vanburen, R.; et al. Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Hortic. Res. 2018, 5, 8–19. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Jarvis, D.E.; Ramaraj, T.; Lee, R.; Jellen, E.N.; Maughan, P.J. Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol. 2017, 15, 74. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Ronen, G. International Wheat Genome Sequencing Consortium Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [PubMed]
- Deschamps, S.; Zhang, Y.; Llaca, V.; Ye, L.; Sanyal, A.; King, M.; May, G.; Lin, H. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun. 2018, 9, 4844. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Bosi, E.; Donati, B.; Galardini, M.; Brunetti, S.; Sagot, M.-F.; Lió, P.; Crescenzi, P.; Fani, R.; Fondi, M. MeDuSa: A multi-draft based scaffolder. Bioinformatics 2015, 31, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [PubMed]
- Volfovsky, N.; Hass, B.J.; Salzberg, S.L. A clustering method for repeat analysis in DNA sequences. Genome Biol. 2001, 2, 0027.1–0027.11. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liang, C. Generic Repeat Finder: A high-sensitivity tool for genome-wide de novo repeat detection. Plant Physiol. 2019, 180, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H. SINE-scan: An efficient tool to discover short interspersed nuclear elements (SINEs) in large-scale genomic datasets. Bioinformatics 2017, 33, 743–745. [Google Scholar] [PubMed]
- Chen, J.; Hu, Q.; Zhang, Y.; Lu, C.; Kuang, H. P-MITE: A database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 2014, 42, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; He, L.; Lai, J.; Dooner, H.K.; Du, C. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl. Acad. Sci. USA 2014, 111, 10263–10268. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.S.; Law, M.; Holt, C.; Stein, J.C.; Moghe, G.D.; Hufnagel, D.E.; Lei, J.; Achawanantakun, R.; Jiao, D.; Lawrence, C.J.; et al. MAKER-P: A Tool Kit for the Rapid Creation, Management, and Quality Control of Plant Genome Annotations. Plant Physiol. 2014, 164, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Audano, P.A.; Sulovari, A.; Graves-Lindsay, T.A.; Cantsilieris, S.; Sorensen, M.; Welch, A.M.E.; Dougherty, M.L.; Nelson, B.J.; Shah, A.; Dutcher, S.K.; et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell 2019, 176, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Hackl, T.; Hedrich, R.; Schultz, J.; Förster, F. Proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 2014, 30, 3004–3011. [Google Scholar] [CrossRef] [PubMed]
- Gnerre, S.; Maccallum, I.; Przybylski, D.; Ribeiro, F.J.; Burton, J.N.; Walker, B.J.; Sharpe, T.; Hall, G.; Shea, T.P.; Sykes, S.; et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 2011, 108, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- English, A.C.; Richards, S.; Han, Y.; Wang, M.; Vee, V.; Qu, J.; Qin, X.; Muzny, D.M.; Reid, J.G.; Worley, K.C.; et al. Mind the Gap: Upgrading Genomes with Pacific Biosciences RS Long-Read Sequencing Technology. PLoS ONE 2012, 7, e47768. [Google Scholar] [CrossRef] [PubMed]
- Mayela Soto-Jimenez, L.; Estrada, K.; Sanchez-Flores, A. GARM: Genome assembly, reconciliation and merging. Curr. Top. Med. Chem. 2014, 14, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Linthorst, J.; Hulsman, M.; Holstege, H.; Reinders, M. Scalable multi whole-genome alignment using recursive exact matching. bioRxiv 2015, 022715. [Google Scholar] [CrossRef]
- Gao, S.; Bertrand, D.; Chia, B.K.H.; Nagarajan, N. OPERA-LG: Efficient and exact scaffolding of large, repeat-rich eukaryotic genomes with performance guarantees. Genome Biol. 2016, 17, 102. [Google Scholar] [CrossRef]
- Lukashin, A.V.; Borodovsky, M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998, 26, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [PubMed]
- Petit, S.; Boursault, A.; Le Guilloux, M.; Munier-Jolain, N.; Reboud, X. Weeds in agricultural landscapes. A review. Agron. Sustain. Dev. 2011, 31, 309–317. [Google Scholar] [CrossRef]
- De Wet, J.M.J.; Harlan, J.R. Weeds and Domesticates: Evolution in the man-made habitat. Econ. Bot. 1975, 29, 99–107. [Google Scholar] [CrossRef]
- Barrett, S.H. Crop mimicry in weeds. Econ. Bot. 1983, 37, 255–282. [Google Scholar] [CrossRef]
- Harlan, J.R.; de Wet, J.M.J. Some thoughts about weeds. Econ. Bot. 1965, 19, 16–24. [Google Scholar] [CrossRef]
- Tedin, O. Vererbung, Variation Und Syste-Matik in Der Gattung Camelina (German with English Summary). Hereditas 1925, 6, 275–386. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef]
- Neve, P.; Vila-Aiub, M.; Roux, F. Evolutionary-thinking in agricultural weed management. New Phytol. 2009, 184, 783–793. [Google Scholar] [CrossRef]
- Baker, H.G. The evolution of weeds. Annu. Rev. Ecol. Syst. 1974, 5, 1–24. [Google Scholar] [CrossRef]
- Warwick, S.I.; Stewart, C.N., Jr. Crops come from wild plants: How domestication, transgenes, and linkage together shape ferality. In Crop Ferality and Volunteerism; CRC Press: Boca Raton, FL, USA, 2005; pp. 9–30. ISBN 9783540773405. [Google Scholar]
- Ellstrand, N.C.; Heredia, S.M.; Leak-Garcia, J.A.; Heraty, J.M.; Burger, J.C.; Yao, L.; Nohzadeh-Malakshah, S.; Ridley, C.E. Crops gone wild: Evolution of weeds and invasives from domesticated ancestors. Evol. Appl. 2010, 3, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Vigueira, C.C.; Olsen, K.M.; Caicedo, A.L. The red queen in the corn: Agricultural weeds as models of rapid adaptive evolution. Heredity 2013, 110, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Royal, C.D.; Novembre, J.; Fullerton, S.M.; Goldstein, D.B.; Long, J.C.; Bamshad, M.J.; Clark, A.G. Inferring Genetic Ancestry: Opportunities, Challenges, and Implications. Am. J. Hum. Genet. 2010, 86, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Kidd, K.K.; Speed, W.C.; Pakstis, A.J.; Furtado, M.R.; Fang, R.; Madbouly, A.; Maiers, M.; Middha, M.; Friedlaender, F.R.; Kidd, J.R. Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci. Int. 2014, 10, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred From Metric Distances Among DNA Haplotypes: Application. Genetics 1992, 131, 479–491. [Google Scholar] [PubMed]
- Pickrell, J.K.; Pritchard, J.K. Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 1993, 47, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Vekemans, X.; Hardy, O.J. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 2004, 13, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Aguillon, S.M.; Fitzpatrick, J.W.; Bowman, R.; Schoech, S.J.; Clark, A.G.; Coop, G.; Chen, N. Deconstructing isolation-by-distance: The genomic consequences of limited dispersal. PLoS Genet. 2017, 13, e1006911. [Google Scholar] [CrossRef] [PubMed]
- Bradburd, G.S.; Coop, G.M.; Ralph, P.L. Inferring continuous and discrete population genetic structure across space. Genetics 2018, 210, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.J.; van Dorp, L.; Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 2018, 9, 3258. [Google Scholar] [CrossRef] [PubMed]
- Gutenkunst, R.N.; Hernandez, R.D.; Williamson, S.H.; Bustamante, C.D. Inferring the Joint Demographic History of Multiple Populations from Multidimensional SNP Frequency Data. PLoS Genet. 2009, 5, e1000695. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Dupanloup, I.; Huerta-Sánchez, E.; Sousa, V.C.; Foll, M. Robust Demographic Inference from Genomic and SNP Data. PLoS Genet. 2013, 9, e1003905. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. Camb. 1974, 23, 23–35. [Google Scholar] [CrossRef]
- Przeworski, M. The Signature of Positive Selection at Randomly Chosen Loci. Genetics 2002, 160, 1179–1189. [Google Scholar] [PubMed]
- Fay, J.C.; Wu, C.-I. Hitchhiking Under Positive Darwinian Selection. Genetics 2000, 155, 1405–1413. [Google Scholar] [PubMed]
- Hermisson, J.; Pennings, P.S. Soft Sweeps: Molecular Population Genetics of Adaptation From Standing Genetic Variation. Genetics 2005, 169, 2335–2352. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef]
- Wright, S. Genetical Structure of Populations. Nature 1950, 247–249. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- Fay, J.C.; Wu, C.-I. Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genomics Hum. Genet. 2003, 4, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Pavlidis, P.; Živković, D.; Stamatakis, A.; Alachiotis, N. SweeD: Likelihood-Based Detection of Selective Sweeps in Thousands of Genomes. Mol. Biol. Evol. 2013, 30, 2224–2234. [Google Scholar] [CrossRef] [PubMed]
- Degiorgio, M.; Huber, C.D.; Hubisz, M.J.; Hellmann, I.; Nielsen, R. SweepFinder2: Increased sensitivity, robustness and flexibility. Bioinformatics 2016, 32, 1895–1897. [Google Scholar] [CrossRef]
- Nielsen, R.; Williamson, S.; Kim, Y.; Hubisz, M.J.; Clark, A.G.; Bustamante, C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005, 15, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.J.; Coop, G. A Population Genetic Signal of Polygenic Adaptation. PLoS Genet. 2014, 10, e1004412. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.J.; Harpak, A.; Sinnott-armstrong, N.; Joergensen, A.M.; Mostafavi, H.; Field, Y.; Boyle, E.A.; Zhang, X.; Racimo, F.; Pritchard, J.K.; et al. Reduced signal for polygenic adaptation of height in UK Biobank. Elife 2019, 8, e39725. [Google Scholar] [CrossRef] [PubMed]
- Mosyakin, S.L.; Robertson, K.R. Amaranthus. In Flora of North America North of Mexico; Flora of North America Editorial Committee: New York, NY, USA; Oxford, UK, 2004. [Google Scholar]
- Délye, C.; Menchari, Y.; Michel, S.; Cadet, E.; Le Corre, V. A new insight into arable weed adaptive evolution: Mutations endowing herbicide resistance also affect germination dynamics and seedling emergence. Ann. Bot. 2013, 111, 681–691. [Google Scholar] [CrossRef]
- Baucom, R.S. Evolutionary and ecological insights from herbicide-resistant weeds: What have we learned about plant adaptation, and what is left to uncover? New Phytol. 2019, 223, 68–82. [Google Scholar] [CrossRef]
- Yuan, J.S.; Tranel, P.J.; Stewart, C.N. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop Prot. 2012, 35, 15–28. [Google Scholar] [CrossRef]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013, 62, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Ghanizadeh, H.; Harrington, K.C. Non-target Site Mechanisms of Resistance to Herbicides. CRC. Crit. Rev. Plant Sci. 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.J.; Zeng, Q.Y. Overexpression of three orthologous glutathione S-transferases from Populus increased salt and drought resistance in Arabidopsis. Biochem. Syst. Ecol. 2019, 83, 57–61. [Google Scholar] [CrossRef]
- Conte, S.S.; Lloyd, A.M. Exploring multiple drug and herbicide resistance in plants-Spotlight on transporter proteins. Plant Sci. 2011, 180, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Cummins, I.; Wortley, D.J.; Sabbadin, F.; He, Z.; Coxon, C.R.; Straker, H.E.; Sellars, J.D.; Knight, K.; Edwards, L.; Hughes, D.; et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA 2013, 110, 5812–5817. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, M.; Lee, K.M.; Chang, S.-M.; Baucom, R.S. Parallel and nonparallel genomic responses contribute to herbicide resistance in Ipomoea purpurea, a common agricultural weed. bioRxiv 2019. [Google Scholar] [CrossRef]
- Salas-Perez, R.A.; Saski, C.A.; Noorai, R.E.; Srivastava, S.K.; Lawton-Rauh, A.L.; Nichols, R.L.; Roma-Burgos, N. RNA-Seq transcriptome analysis of Amaranthus palmeri with differential tolerance to glufosinate herbicide. PLoS ONE 2018, 13, e0195488. [Google Scholar] [CrossRef]
- Bai, S.; Liu, W.; Wang, H.; Zhao, N.; Jia, S.; Zou, N.; Guo, W.; Wang, J. Enhanced herbicide metabolism and metabolic resistance genes identified in tribenuron-methyl resistant Myosoton aquaticum L. J. Agric. Food Chem. 2018, 66, 9850–9857. [Google Scholar] [CrossRef]
- Kreuz, K.; Tommasini, R.; Martinoia, E. Old Enzymes for a New Job Herbicide Detoxification in Plants. Plant Physiol. 1996, 111, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Del Buono, D.; Fordham, M.; Skipsey, M.; Brazier, M.; Dixon, D.P.; Cummins, I. Differential induction of glutathione transferases and glucosyltransferases in wheat, maize and Arabidopsis thaliana by herbicide safeners. Z. Nat. 2005, 60, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A.; Werck-Reichhart, D. Functional Genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R.; Schuler, M.A.; Paquette, S.M.; Werck-Reichhart, D.; Bak, S. Comparative Genomics of Rice and Arabidopsis. Analysis of 727 Cytochrome P450 Genes and Pseudogenes from a Monocot and a Dicot. Plant Physiol. 2004, 135, 756–772. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Provart, N.J.; Werck-Reichhart, D. Functional annotation of the Arabidopsis P450 superfamily based on large-scale co-expression analysis. Biochem. Soc. Trans. 2006, 34, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Hehn, A.; Diderjean, L. Cytochromes P450 for engineering herbicide tolerance. Plant Cell 2004, 5, 116–123. [Google Scholar] [CrossRef]
- Inui, H.; Ueyama, Y.; Shiota, N.; Ohkawa, Y.; Ohkawa, H. Herbicide Metabolism and Cross-Tolerance in Transgenic Potato Plants Expressing Human CYP1A1. Pestic. Biochem. Physiol. 1999, 64, 33–46. [Google Scholar] [CrossRef]
- Kawahigashi, H.; Hirose, S.; Ohkawa, H.; Ohkawa, Y. Herbicide resistance of transgenic rice plants expressing human CYP1A1. Biotechnol. Adv. 2007, 25, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Kawahigashi, H.; Hirose, S.; Ohkawa, H.; Ohkawa, Y. Phytoremediation of the herbicides atrazine and metolachlor by transgenic rice plants expressing human CYP1A1, CYP2B6, and CYP2C19. J. Agric. Food Chem. 2006, 54, 2985–2991. [Google Scholar] [CrossRef]
- Hirose, S.; Kawahigashi, H.; Ozawa, K.; Shiota, N.; Inui, H.; Ohkawa, H.; Ohkawa, Y. Transgenic rice containing human CYP2B6 detoxifies various classes of herbicides. J. Agric. Food Chem. 2005, 53, 3461–3467. [Google Scholar] [CrossRef]
- Herbicide Resistance Action Committee. Available online: https://www.hracglobal.com/ (accessed on 26 January 2019).
- Robineau, T.; Batard, Y.; Nedelkina, S.; Cabello-Hurtado, F.; LeRet, M.; Sorokine, O.; Didierjean, L.; Werck-Reichhart, D. The Chemically Inducible Plant Cytochrome P450 CYP76B1 Actively Metabolizes Phenylureas and Other Xenobiotics. Plant Physiol. 2002, 118, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Siminszky, B.; Corbin, F.T.; Ward, E.R.; Fleischmann, T.J.; Dewey, R.E. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc. Natl. Acad. Sci. USA 1999, 96, 1750–1755. [Google Scholar] [CrossRef] [PubMed]
- Höfer, R.; Boachon, B.; Renault, H.; Gavira, C.; Miesch, L.; Iglesias, J.; Ginglinger, J.-F.; Allouche, L.; Miesch, M.; Grec, S.; et al. Dual Function of the Cytochrome P450 CYP76 Family from Arabidopsis thaliana in the Metabolism of Monoterpenols and Phenylurea Herbicides. Plant Physiol. 2014, 166, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Khanom, S.; Jang, J.; Lee, O.R. Overexpression of ginseng cytochrome P450 CYP736A12 alters plant growth and confers phenylurea herbicide tolerance in Arabidopsis. J. Ginseng Res. 2019. [Google Scholar] [CrossRef]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 Are Associated with Resistance to Two Acetolactate Synthase Inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Iwakami, S.; Yamaguchi, T.; Uchino, A.; Sunohara, Y.; Matsumoto, H. Role of CYP81A cytochrome P450s in clomazone metabolism in Echinochloa phyllopogon. Plant Sci. 2019, 283, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, J.; Shen, J.; Xu, Y.; Liu, H.; Deng, W.; Li, X.; Zheng, M. Metabolic Resistance to Acetolactate Synthase Inhibiting Herbicide Tribenuron-Methyl in Descurainia sophia L. Mediated by Cytochrome P450 Enzymes. J. Agric. Food Chem. 2018, 66, 4319–4327. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.C.; Gaines, T.A.; Dayan, F.E.; Patterson, E.L.; Jhala, A.J.; Knezevic, S.Z. Reversing resistance to tembotrione in an Amaranthus tuberculatus (var. rudis) population from Nebraska, USA with cytochrome P450 inhibitors. Pest Manag. Sci. 2018, 74, 2296–2305. [Google Scholar] [CrossRef]
- Hidayat, I.; Preston, C. Cross-resistance to imazethapyr in a fluazifop-P-butyl-resistant population of Digitaria sanguinalis. Pestic. Biochem. Physiol. 2001, 71, 190–195. [Google Scholar] [CrossRef]
- Marrs, K.A. The Functions and Regulation of Glutathione S-Transferases in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef]
- Stavridou, E.; Voulgari, G.; Bosmali, I.; Chronopoulou, E.G.; Lo Cicero, L.; Lo Piero, A.R.; Labrou, N.Ε.; Tsaftaris, A.; Nianiou-Obeidat, I.; Madesis, P. Plant Adaptation to Stress Conditions: The Case of Glutathione S-Transferases (GSTs). In Biotic and Abiotic Stress Tolerance in Plants; Vats, S., Ed.; Springer Nature Singapore Pte Ltd.: Berlin/Heidelberg, Germany, 2018; pp. 173–202. ISBN 978-981-10-9028-8. [Google Scholar]
- Labrou, N.E.; Papageorgiou, A.C.; Pavli, O.; Flemetakis, E. Plant GSTome: Structure and functional role in xenome network and plant stress response. Curr. Opin. Biotechnol. 2015, 32, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hou, M.; Cao, L.; Xia, Y.; Shen, Z.; Hu, Z. Glutathione S-transferases modulate Cu tolerance in Oryza sativa. Environ. Exp. Bot. 2018, 155, 313–320. [Google Scholar] [CrossRef]
- Li, Z.-K.; Chen, B.; Li, X.-X.; Wang, J.-P.; Zhang, Y.; Wang, X.-F.; Yan, Y.-Y.; Ke, H.F.; Yang, J.; Wu, J.-H.; et al. A newly identified cluster of glutathione S-transferase genes provides Verticillium wilt resistance in cotton. Plant J. 2019, 98, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Protein family review Plant glutathione transferases. Genome Biol. 2002, 3, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dixon, D.; Cole, D.J.; Edwards, R. Characterisation of multiple glutathione transferases containing the GST I subunit with activities toward herbicide substrates in maize (Zea mays). Pestic. Sci. 1997, 50, 72–82. [Google Scholar] [CrossRef]
- Grove, G.; Zarlengo, R.P.; Timmerman, K.P.; Li, N.Q.; Tam, M.F.; Tu, C.-P.D. Characterization and heterospecific expression of cDNA clones of genes in the maize GSH S-transferase multigene family. Nucleic Acids Res. 1988, 16, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Karavangeli, M.; Labrou, N.E.; Clonis, Y.D.; Tsaftaris, A. Development of transgenic tobacco plants overexpressing maize glutathione S-transferase I for chloroacetanilide herbicides phytoremediation. Biomol. Eng. 2005, 22, 121–128. [Google Scholar] [CrossRef]
- Milligan, A.S.; Daly, A.; Parry, M.A.J.; Lazzeri, P.A.; Jepson, I. The expression of a maize glutathione S-transferase gene in transgenic wheat confers herbicide tolerance, both in planta and in vitro. Mol. Breed. 2001, 7, 301–315. [Google Scholar] [CrossRef]
- Benekos, K.; Kissoudis, C.; Nianiou-Obeidat, I.; Labrou, N.; Madesis, P.; Kalamaki, M.; Makris, A.; Tsaftaris, A. Overexpression of a specific soybean GmGSTU4 isoenzyme improves diphenyl ether and chloroacetanilide herbicide tolerance of transgenic tobacco plants. J. Biotechnol. 2010, 150, 195–201. [Google Scholar] [CrossRef]
- Cummins, I.; Cole, D.J.; Edwards, R. A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J. 1999, 18, 285–292. [Google Scholar] [CrossRef]
- Petit, C.; Duhieu, B.; Boucansaud, K.; Délye, C. Complex genetic control of non-target-site-based resistance to herbicides inhibiting acetyl-coenzyme A carboxylase and acetolactate-synthase in Alopecurus myosuroides Huds. Plant Sci. 2010, 178, 501–509. [Google Scholar] [CrossRef]
- Wright, A.A.; Rodriguez-Carres, M.; Sasidharan, R.; Koski, L.; Peterson, D.G.; Nandula, V.K.; Ray, J.D.; Bond, J.A.; Shaw, D.R. Multiple Herbicide–Resistant Junglerice (Echinochloa colona): Identification of Genes Potentially Involved in Resistance through Differential Gene Expression Analysis. Weed Sci. 2018, 66, 347–354. [Google Scholar] [CrossRef]
- Nakka, S.; Godar, A.S.; Thompson, C.R.; Peterson, D.E.; Jugulam, M. Rapid detoxification via glutathione S-transferase (GST) conjugation confers a high level of atrazine resistance in Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 2017, 73, 2236–2243. [Google Scholar] [CrossRef] [PubMed]
- Dücker, R.; Zöllner, P.; Lümmen, P.; Ries, S.; Collavo, A.; Beffa, R. Glutathione transferase plays a major role in flufenacet resistance of ryegrass (Lolium spp.) field populations. Pest Manag. Sci. 2019. [Google Scholar] [CrossRef]
- Balabanova, D.; Remans, T.; Vassilev, A.; Cuypers, A.; Vangronsveld, J. Possible involvement of glutathione S-transferases in imazamox detoxification in an imidazolinone-resistant sunflower hybrid. J. Plant Physiol. 2018, 221, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Draicchio, F.; Bull, H.; Herzig, P.; Maurer, A.; Pillen, K.; Thomas, W.T.B.; Flavell, A.J. Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J. Exp. Bot. 2018, 69, 3811–3822. [Google Scholar] [CrossRef] [PubMed]
- Krajewski, M.P.; Kanawati, B.; Fekete, A.; Kowalski, N.; Schmitt-Kopplin, P.; Grill, E. Analysis of Arabidopsis glutathione-transferases in yeast. Phytochemistry 2013, 91, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L. Plant ABC transporters. Biochim. Biophys. Acta Biomembr. 2000, 1465, 79–103. [Google Scholar] [CrossRef]
- Hwang, J.U.; Song, W.Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S.; Lee, E.; Khare, D.; Kim, K.; et al. Plant ABC Transporters Enable Many Unique Aspects of a Terrestrial Plant’s Lifestyle. Mol. Plant 2016, 9, 338–355. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Fernández, R.; Davies, T.G.E.; Coleman, J.O.D.; Rea, P.A. The Arabidopsis thaliana ABC Protein Superfamily, a Complete Inventory. J. Biol. Chem. 2001, 276, 30231–30244. [Google Scholar] [CrossRef] [PubMed]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of atp binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef] [PubMed]
- Busi, R.; Goggin, D.E.; Heap, I.M.; Horak, M.J.; Jugulam, M.; Masters, R.A.; Napier, R.M.; Riar, D.S.; Satchivi, N.M.; Torra, J.; et al. Weed resistance to synthetic auxin herbicides. Pest Manag. Sci. 2018, 74, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Abercrombie, L.L.G.; Yuan, J.S.; Riggins, C.W.; Sammons, R.D.; Tranel, P.J.; Stewart, C.N., Jr. Characterization of the horseweed (Conyza canadensis) transcriptome using GS-FLX 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Manag. Sci. 2010, 66, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Windsor, B.; Roux, S.J.; Lloyd, A. Multiherbicide tolerance conferred by AtPgp1 and apyrase overexpression in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Won, S.H.; Son, D.; Lee, B.H. Paraquat resistance of transgenic tobacco plants over-expressing the Ochrobactrum anthropi pqrA gene. Biotechnol. Lett. 2004, 26, 1391–1396. [Google Scholar] [CrossRef] [PubMed]
- Quistgaard, E.M.; Löw, C.; Guettou, F.; Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Nat. Rev. Mol. Cell Biol. 2016, 17, 123–132. [Google Scholar] [CrossRef]
- Ward, J.M. Identification of novel families of membrane proteins from the model plant Arabidopsis thaliana. Bioinformatics 2001, 17, 560–563. [Google Scholar] [CrossRef]
- Teixeira, M.C.; Duque, P.; Sá-Correia, I. Environmental genomics: Mechanistic insights into toxicity of and resistance to the herbicide 2,4-D. Trends Biotechnol. 2007, 25, 363–370. [Google Scholar] [CrossRef]
- Cabrito, T.R.; Teixeira, M.C.; Duarte, A.A.; Duque, P.; Sá-Correia, I. Heterologous expression of a Tpo1 homolog from Arabidopsis thaliana confers resistance to the herbicide 2,4-D and other chemical stresses in yeast. Appl. Microbiol. Biotechnol. 2009, 84, 927–936. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef]
- Pflugmacher, S.; Schröder, P.; Sandermann Jr, H. Taxonomic distribution of plant glutathione S-transferases acting on xenobiotics. Phytochemistry 2000, 54, 267–273. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Offen, W.A.; Gershater, M.C.; Revett, T.J.; Lim, E.-K.; Bowles, D.J.; Davies, G.J.; Edwards, R. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 20238–20243. [Google Scholar] [CrossRef] [PubMed]
- Loutre, C.; Dixon, D.P.; Brazier, M.; Slater, M.; Cole, D.J.; Edwards, R. Isolation of a glucosyltransferase from Arabidopsis thaliana active in the metabolism of the persistent pollutant 3,4-dichloroaniline. Plant J. 2003, 34, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, A.; Sandermann, H. Plant biochemistry of xenobiotics: Isolation and characterization of a soybean O-glucosyltransferase of DDT metabolism. Arch. Biochem. Biophys. 1994, 314, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Brazier-Hicks, M.; Edwards, R. Functional importance of the family 1 glucosyltransferase UGT72B1 in the metabolism of xenobiotics in Arabidopsis thaliana. Plant J. 2005, 42, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Meβner, B.; Thulke, O.; Schäffner, A.R. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 2003, 217, 138–146. [Google Scholar]
- Cha, J.-Y.; Lee, D.-Y.; Ali, I.; Jeong, S.Y.; Shin, B.; Ji, H.; Kim, J.S.; Kim, M.-G.; Kim, W.-Y. Arabidopsis GIGANTEA negatively regulates chloroplast biogenesis and resistance to herbicide butafenacil. Plant Cell Rep. 2019, 38, 793–801. [Google Scholar] [CrossRef]
- Etter, P.D.; Bassham, S.; Hohenlohe, P.A.; Johnson, E.; Cresko, W.A. SNP Discovery and genotyping for evolutionary genetics using RAD Sequencing. In Molecular Methods for Evolutionary Genetics; Orgogozo, V., Rockman, M.V., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; Volume 772, pp. 1–9. ISBN 978-1-61779-227-4. [Google Scholar]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Catchen, J.M.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Rochette, N.C.; Catchen, J.M. Deriving genotypes from RAD-seq short-read data using Stacks. Nat. Protoc. 2017, 12, 2640–2659. [Google Scholar] [CrossRef]
- Paris, J.R.; Stevens, J.R.; Catchen, J.M. Lost in parameter space: A road map for stacks. Methods Ecol. Evol. 2017, 8, 1360–1373. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- R Core Team, R. A Language and Environment for Statistical Computing; R Core Team R: Vienna, Austria, 2017. [Google Scholar]
- Paradis, E.; Gosselin, T.; Goudet, J.; Jombart, T.; Schliep, K. Linking genomics and population genetics with R. Mol. Ecol. Resour. 2017, 17, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J.; Jombart, T. Hierfstat: Estimation and Tests of Hierarchical F-Statistics; CRAN, 2015; p. 58. Available online: https://cran.r-project.org/web/packages/hierfstat/index.html (accessed on 30 May 2019).
- Pembleton, L.W.; Cogan, N.O.I.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction v2.6.1. PeerJ 2018, 2, e281. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: Phylogenetic tools for comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 1403. [Google Scholar] [CrossRef] [PubMed]
- Küpper, A.; Manmathan, H.K.; Giacomini, D.; Patterson, E.L.; Mccloskey, W.B.; Gaines, T.A. Population Genetic Structure in Glyphosate-Resistant and -Susceptible Palmer Amaranth (Amaranthus palmeri) Populations Using Genotyping-by-sequencing (GBS). Front. Plant Sci. 2018, 9, 29. [Google Scholar] [CrossRef]
- Foll, M.; Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef]
- Günther, T.; Coop, G. Robust identification of local adaptation from allele frequencies. Genetics 2013, 195, 205–220. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Hill, C.M.; Wu, S.; Ruan, J.; Ma, Z. (Sam) DBG2OLC: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 2016, 6, 1–9. [Google Scholar]
- Hoshino, A.; Jayakumar, V.; Nitasaka, E.; Toyoda, A.; Noguchi, H.; Itoh, T.; Shin, T.; Minakuchi, Y.; Koda, Y.; Nagano, A.J.; et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA –Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Neve, P. Gene drive systems: Do they have a place in agricultural weed management? Pest Manag. Sci. 2018, 74, 2671–2679. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Malik, H.S. The gene drive bubble: New realities. PLOS Genet. 2017, 13, e1006850. [Google Scholar] [CrossRef] [PubMed]
- Committee on Gene Drive Research in Non-Human Organisms: Recommendations for Responsible Conduct; Board on Life Sciences; Division on Earth and Life Studies; National Academies of Sciences, Engineering, and Medicine. Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values; The National Academies Press: Washington, DC, USA, 2016; ISBN 9780309437875. [Google Scholar]
- Courtier-Orgogozo, V.; Morizot, B.; Boëte, C. Agricultural pest control with CRISPR-based gene drive: Time for public debate. EMBO Rep. 2017, 18, 878–880. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Smidler, A.L.; Catteruccia, F.; Church, G.M. Concerning RNA-guided gene drives for the alteration of wild populations. Elife 2014, e03401. [Google Scholar] [CrossRef]
- Champer, J.; Buchman, A.; Akbari, O.S. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 2016, 17, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.; Fauser, F.; Puchta, H. DNA recombination in somatic plant cells: Mechanisms and evolutionary consequences. Chromosom. Res. 2014, 22, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.K.; Puchta, H. CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Rep. 2019, 38, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Klompe, S.E.; Vo, P.L.H.; Halpin-Healy, T.S.; Sternberg, S.H. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 2019, 571, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Que, Q.; Chilton, M.D.M.; Elumalai, S.; Zhong, H.; Dong, S.; Shi, L. Repurposing macromolecule delivery tools for plant genetic modification in the era of precision genome engineering. Methods Mol. Biol. 2019, 1864, 3–18. [Google Scholar] [PubMed]
- Grunwald, H.A.; Gantz, V.M.; Poplawski, G.; Xu, X.-R.S.; Bier, E.; Cooper, K.L. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 2019, 566, 105–109. [Google Scholar] [CrossRef]
- Charpentier, M.; Khedher, A.H.; Menoret, S.; Brion, A.; Lamribet, K.; Dardillac, E.; Boix, C.; Perrouault, L.; Tesson, L.; Geny, S.; et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef]
- Kyrou, K.; Hammond, A.M.; Galizi, R.; Kranjc, N.; Burt, A.; Beaghton, A.K.; Nolan, T.; Crisanti, A. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 2018, 36, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Webber, B.L.; Raghu, S.; Edwards, O.R. Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc. Natl. Acad. Sci. USA 2015, 112, 10565–10567. [Google Scholar] [CrossRef] [PubMed]
- Baltzegar, J.; Cavin Barnes, J.; Elsensohn, J.E.; Gutzmann, N.; Jones, M.S.; King, S.; Sudweeks, J. Anticipating complexity in the deployment of gene drive insects in agriculture. J. Responsible Innov. 2018, 5, S81–S97. [Google Scholar] [CrossRef]
- Oye, K.A.; Esvelt, K.; Appleton, E.; Catteruccia, F.; Church, G.; Kuiken, T.; Lightfoot, S.B.-Y.; McNamara, J.; Smidler, A.; Collins, J.P. Regulating gene drives. Science 2014, 345, 626–628. [Google Scholar] [CrossRef] [PubMed]
- Unckless, R.L.; Messer, P.W.; Connallon, T.; Clark, A.G. Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 2015, 201, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Unckless, R.L.; Clark, A.G.; Messer, P.W. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 2017, 205, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Champer, J.; Reeves, R.; Oh, S.Y.; Liu, C.; Liu, J.; Clark, A.G.; Messer, P.W. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 2017, 13, e1006796. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.; Olejarz, J.; Esvelt, K.M.; Church, G.M.; Nowak, M.A. Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 2017, 3, e1601964. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Hawkes, N.J.; McCarroll, L.; Ranson, H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. Biol. 2004, 34, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Ranson, H.; Lissenden, N. Insecticide Resistance in African Anopheles Mosquitoes: A Worsening Situation that Needs Urgent Action to Maintain Malaria Control. Trends Parasitol. 2016, 32, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Cohuet, A.; Harris, C.; Robert, V.; Fontenille, D. Evolutionary forces on Anopheles: What makes a malaria vector? Trends Parasitol. 2010, 26, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Ranson, H.; Magill, A.; Kolaczinski, J.; Fornadel, C.; Gimnig, J.; Coetzee, M.; Simard, F.; Roch, D.K.; Hinzoumbe, C.K.; et al. Averting a malaria disaster: Will insecticide resistance derail malaria control? Lancet 2016, 387, 1785–1788. [Google Scholar] [CrossRef]
- Namountougou, M.; Simard, F.; Baldet, T.; Diabaté, A.; Ouédraogo, J.B.; Martin, T.; Dabiré, R.K. Multiple Insecticide Resistance in Anopheles gambiae s.l. Populations from Burkina Faso, West Africa. PLoS ONE 2012, 7, e48412. [Google Scholar] [CrossRef] [PubMed]
- Edi, C.V.A.; Koudou, B.G.; Jones, C.M.; Weetman, D.; Ranson, H. Multiple-Insecticide Resistance in Anopheles gambiae Mosquitoes, Southern Côte d’Ivoire. Emerg. Infect. Dis. 2012, 18, 1508–1511. [Google Scholar] [CrossRef] [PubMed]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Gribble, M.; Baker, D.; Marois, E.; Russell, S.; et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef]
- Holt, R.A.; Broder, S.; Subramanian, G.M.; Halpern, A.L.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, J.M.C.; et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. Hijacking evolution. Nature 2019, 571, 160–162. [Google Scholar] [CrossRef]
- Collins, C.M.; Bonds, J.A.S.; Quinlan, M.M.; Mumford, J.D. Effects of the removal or reduction in density of the malaria mosquito, Anopheles gambiae s.l., on interacting predators and competitors in local ecosystems. Med. Vet. Entomol. 2019, 33, 1–15. [Google Scholar] [CrossRef]
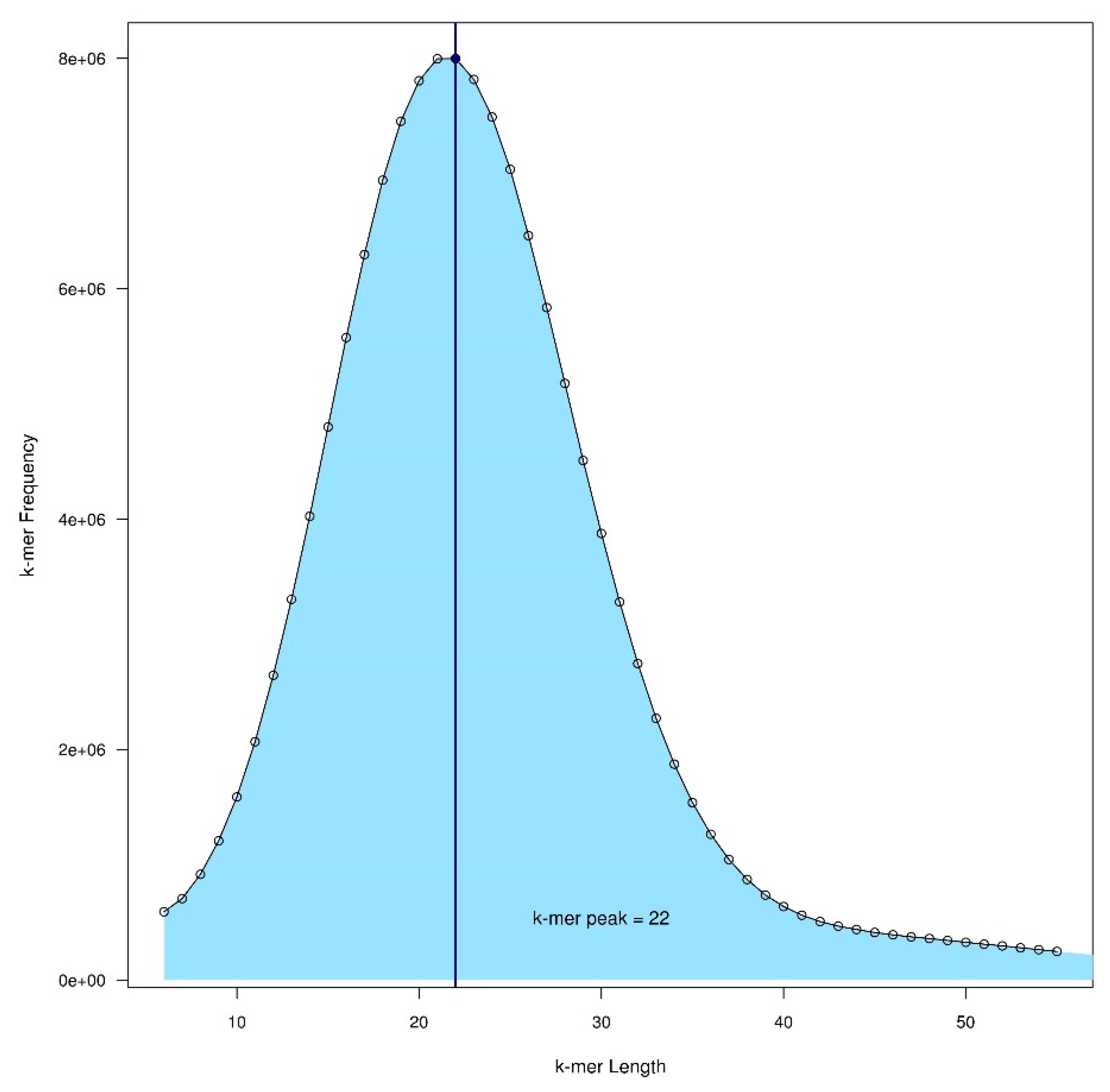
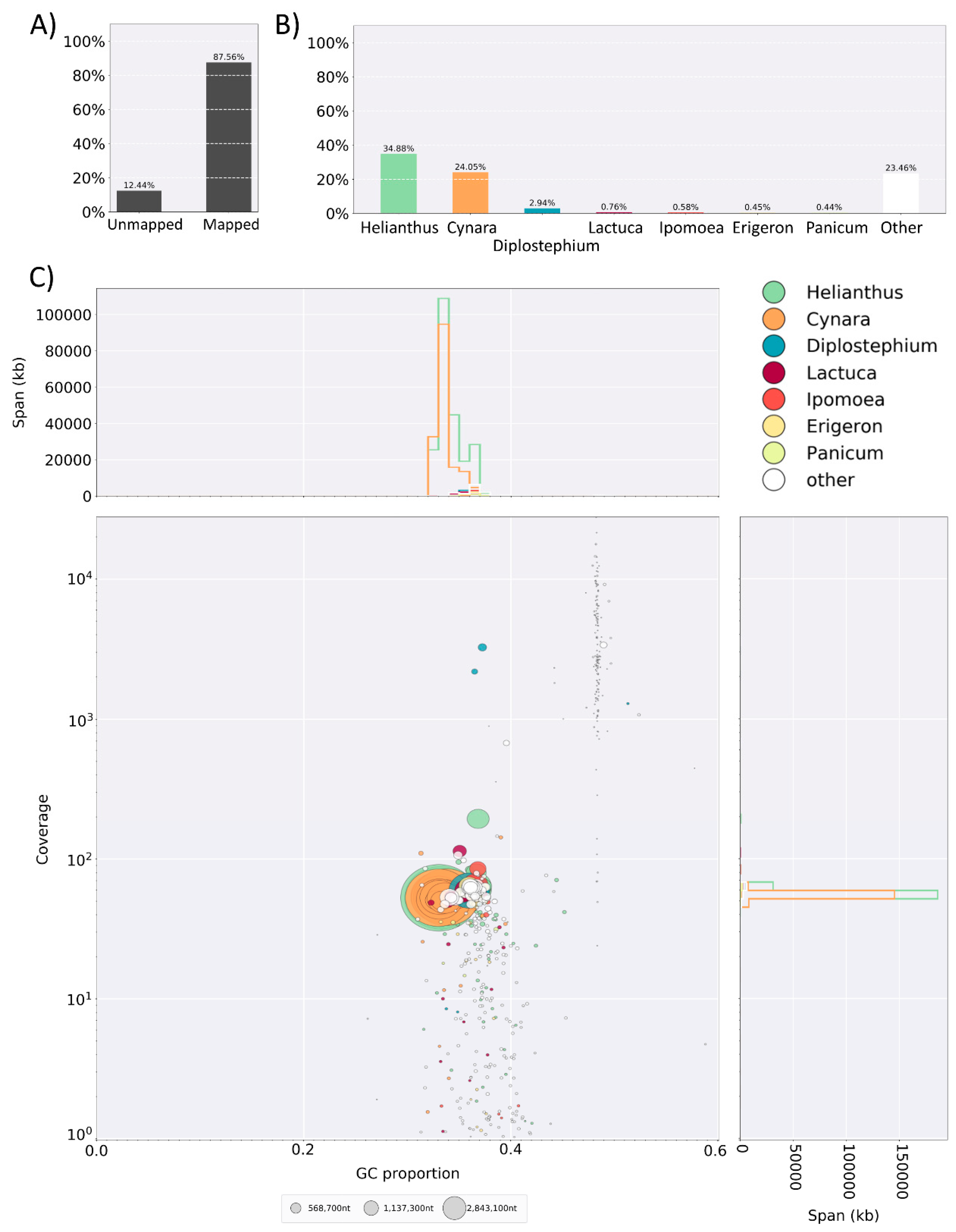
| Common Name | Latin Name | Year | Level of Assembly | No. of Contigs | Est. Genome Size (Mbp) | Assembled Size (Mbp) | N50 | NG50 | BUSCOS (Percentage of 2121 Genes) | Coverage | Sequencing Technology | Assembly Method | Reference or Lead Submitting Author | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | S | D | F | M | |||||||||||||
| Milkweed | Asclepias syriaca | 2017 | Scaffold | 221,885 | 411 1 | 237 | 2555 | NA2 | 76 | 75 | 1 | 13 | 11 | 80.4 | Illumina | Platanus SCUBAT | [40] |
| Winter Cress | Barbarea vulgaris | 2016 | Scaffold | 7810 | 270 | 167 | 56,351 | 19,454 | 95 | 93 | 2 | 3 | 3 | 66.5 | Illumina | Celera | [41] |
| Japanese Barberry | Berberis thunbergii | 2018 | Contig | 11,815 | 1515 1 | 2241 | 397,058 | 654,137 | 88 | 30 | 57 | 3 | 9 | 104.8 | PacBio | FALCON-Unzip | R. Bartaula |
| False Brome | Brachypodium distachyon | 2018 | Chromosome | 11 | 355 | 271 | 59,130,575 | 59,130,575 | 80 | 76 | 4 | 6 | 15 | 9.4 | ABI 3739 | ARACHNE | [42] |
| Bird Rape | Brassica rapa | 2017 | Scaffold | 70,673 | 485 | 386 | 3,737,062 | 2,395,810 | 98 | 80 | 18 | 1 | 1 | 212 | Illumina PacBio | SOAPdenovo | [43] |
| Hemp | Cannabis sativa | 2018 | Chromosome | 6653 3 | 820 | 892 | 60,968,100 | 62,039,859 | 88 | 72 | 16 | 4 | 7 | 79 | PacBio | FALCON | [44] |
| Shepherd’s Purse | Capsella bursa-pastoris | 2017 | Scaffold | 8186 | 391 1 | 268 | 627,605 | 320,701 | 96 | 13 | 83 | 2 | 3 | 40 | Illumina | Newbler Platanus | [45] |
| Horsetail Sheoak | Casuarina equisetifolia subsp. incana | 2018 | Scaffold | 2936 | 340 1 | 301 | 1,020,118 | 894,734 | 97 | 93 | 4 | 1 | 2 | 546.9 | Illumina PacBio | SOAPdenovo2 FALCON DISCOVAR | [46] |
| Swamp Oak | Cauarina glauca | 2018 | Scaffold | 39,787 | 340 | 283 | 964,272 | 627,004 | 97 | 93 | 5 | 1 | 2 | 890 | Illumina | SOAPdenovo | [47] |
| Mandarin Orange | Citrus reticulata | 2018 | Scaffold | 67,725 | 460 | 344 | 1,376,405 | 577,147 | 98 | 96 | 2 | 1 | 1 | 200 | Illumina | Platanus | [48] |
| Horseweed | Conyza canadensis | 2014 | Contig | 20,075 | 335 | 326 | 20,748 | 20,226 | 66 | 44 | 22 | 10 | 24 | 350 | Roche 454 Illumina PacBio | Newbler SOAPdenovo CLC NGS Cell | [49] |
| Jute Mallow | Corchorus olitorius | 2017 | Contig | 52,373 | 450 | 377 | 16,573 | 13,050 | 93 | 90 | 3 | 3 | 4 | 47.7 | Illumina | Newbler | [50] |
| Muskmelon | Cucumis melo | 2012 | Scaffold | 10,823 | 450 | 375 | 4,428,067 | 3,741,400 | 94 | 92 | 2 | 2 | 4 | 13.5 | Roche 454 Illumina | Newbler | [51] |
| Globe artichoke | Cynara cardunculus | 2018 | Chromosome | 8283 3 | 1084 | 725 | 25,947,084 | 173,700 | 96 | 90 | 6 | 2 | 2 | 80 | Illumina | AllPaths | [52] |
| Orchardgrass | Dactylis glomerata | 2018 | Scaffold | 1,072,009 | 3327 4 | 840 | 3242 | NA2 | 76 | 72 | 4 | 8 | 16 | 50 | Illumina | SOAPdenovo | J. Li |
| Carrot | Daucus carota subsp. sativus | 2016 | Chromosome | 4826 3 | 473 | 422 | 36,610,139 | 36,610,139 | 94 | 88 | 6 | 2 | 5 | 186 | Roche 454 Illumina Sanger | SOAPdenovo GapCloser | [53] |
| Guinea yam | Dioscorea rotundata | 2017 | Chromosome | 21 | 694 1 | 457 | 25,272,979 | NA2 | 83 | 78 | 5 | 3 | 14 | 100 | Illumina | Allpaths-LG SSPACE Premium | S. Natsume |
| Barnyardgrass | Echinochloa crus-galli | 2017 | Scaffold | 4113 | 1400 | 486 | 705,200 | NA2 | 89 | 26 | 63 | 2 | 10 | 170 | Illumina PacBio | SOAPdenovo2 CANU | [54] |
| Paterson’s curse | Echium plantagineum | 2019 | Chromosome | 1091 3 | 333 1 | 349 | 1,429,328 | 1,517,519 | 96 | 46 | 50 | 1 | 3 | 115 | Illumina PacBio | MECAT LACHESIS | C.-Y. Tang |
| Common sunflower | Helianthus annuus | 2017 | Chromosome | 1528 3 | 3600 | 3028 | 178,899,001 | 174,509,413 | 89 | 80 | 9 | 3 | 8 | 100 | PacBio | PBcR | [55] |
| Littlebell | Ipomoea triloba | 2018 | Chromosome | 16 | 496 1 | 462 | 29,809,665 | 28,894,297 | 97 | 89 | 7 | 1 | 2 | 290 | Illumina PacBio | SOAPdenovo2 SSPACE PBJelly, Pilon | [56] |
| Perennial Ryegrass | Lolium perenne | 2016 | Scaffold | 666,180 | 2621 1 | 481 | 1361 | NA2 | 31 | 29 | 2 | 22 | 47 | 5 | Illumina | CLC Genomic Workbench | [57] |
| Horsemint | Mentha longifolia | 2016 | Scaffold | 190,876 | 400 | 353 | 3915 | 3044 | 58 | 52 | 5 | 20 | 22 | 33 | Illumina PacBio | MaSuRCA | [58] |
| Amur silver grass | Miscanthus sacchariflorus | 2018 | Chromosome | 105,321 3 | 2513 1 | 2075 | 37,709 | 24,189 | 49 | 41 | 8 | 17 | 33 | 60 | Illumina | ABySS SOAPdenovo2 | J. De Vega |
| Longstamen Rice | Oryza longistaminata | 2014 | Scaffold | 9688 | 782 1 | 362 | 30,401,905 | NA2 | 86 | 80 | 6 | 4 | 10 | 52.5 | Illumina | SOAPdenovo2 | C. Brian |
| Red Rice | Oryza punctata | 2014 | Chromosome | 12 | 586 1 | 394 | 31,244,610 | 28,494,620 | 81 | 74 | 7 | 6 | 13 | 130 | Roche 454 Illumina | AllPaths | R. A. Wing |
| Brownbeard Rice | Oryza rufipogon | 2015 | Scaffold | 3818 | 450 1 | 339 | 27,785,585 | 26,200,591 | 83 | 76 | 6 | 5 | 13 | 120 | Q. Zhao | ||
| Rice | Oryza sativa | 2019 | Chromosome | 367 3 | 489 1 | 415 | 28,085,715 | 26,003,091 | 88 | 81 | 6 | 3 | 9 | 148 | PacBio | CANU | L. Wang |
| Broomcorn Millet | Panicum miliaceum | 2018 | Chromosome | 466 | 923 | 848 | 48,259,421 | 45,112,342 | 83 | 25 | 58 | 4 | 13 | 160 | Illumina PacBio | CANU | [59] |
| Opium Poppy | Papaver somniferum | 2018 | Chromosome | 34,381 3 | 2870 | 2716 | 204,470,928 | 180,516,484 | 95 | 29 | 65 | 1 | 4 | 239 | Illumina PacBio ONT | DeNovoMAGIC FALCON | [60] |
| White Poplar | Populus alba | 2019 | Contig | 6087 | 508 1 | 707 | 248,703 | 390,844 | 95 | 52 | 43 | 1 | 3 | 130 | Illumina PacBio | SMARTdenovo | [61] |
| Algarrobo blanco | Prosopis alba | 2019 | Contig | 4454 | 391 1 | 500 | 237,044 | 357,710 | 70 | 49 | 21 | 3 | 27 | 30 | PacBio | CANU | W. Kong |
| Wild Radish | Raphanus raphistrum | 2014 | Contig | 64,732 | 515 | 254 | 10,333 | NA2 | 95 | 82 | 12 | 3 | 2 | 47 | Roche 454 Illumina | ABySS, Newbler, Celera Assembler, Minimus2 | [62] |
| Radish | Raphanus sativa | 2017 | Chromosome | 44,239 3 | 573 | 383 | 35,166,889 | 26,198,371 | 96 | 82 | 14 | 2 | 1 | 225 | Illumina | SOAPdenovo2 | [63] |
| Japanese Rose | Rosa multiflora | 2017 | Scaffold | 83,189 | 711 | 740 | 90,830 | 95,085 | 91 | 66 | 25 | 4 | 5 | 327 | Illumina | SOAPdenovo2 GapCloser | [64] |
| Wild Sugarcane | Saccharum spontaneum | 2018 | Chromosome | 15,303 3 | 1565 1 | 3133 | 91,359,291 | 109,189,819 | 78 | 20 | 58 | 5 | 17 | 90 | Illumina PacBio | CANU HiC | [65] |
| Rye | Secale cerale | 2017 | Scaffold | 1,581,707 | 7900 | 1685 | 2200 | NA2 | 66 | 62 | 4 | 13 | 21 | 50 | Illumina | CLC Assembly Cell, CarmA | [66] |
| Green Foxtail | Setaria viridis | 2019 | Chromosome | 14 | 782 1 | 396 | 46,702,114 | 35,460,007 | 81 | 75 | 6 | 6 | 13 | 118 | PacBio | MECAT | P. Huang |
| White Campion | Silene latifolia | 2018 | Scaffold | 319,506 | 2640 1 | 1185 | 11,019 | NA2 | 68 | 66 | 4 | 13 | 18 | 40 | PacBio | SOAPdenovo2, CLC, PBJelly, SSPACE | [67] |
| Milk Thistle | Silybum marianum | 2016 | Contig | 258,575 | 792 1 | 1478 | 6967 | NA2 | 38 | 33 | 6 | 8 | 54 | 96 | Illumina PacBio | Celera Assembler | Y. Lv |
| Sorghum | Sorghum bicolor | 2017 | Chromosome | 869 3 | 730 | 709 | 68,658,214 | 68,658,214 | 86 | 80 | 5 | 4 | 10 | 8 | Illumina Sanger | ARACHNE | [68] |
| Stinkweed | Thlapsi arvense | 2015 | Scaffold | 6768 | 539 | 343 | 140,815 | NA2 | 98 | 97 | 2 | 1 | 1 | 80 | Illumina PacBio | CLC NGS Cell | [69] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, S.L.; Parent, J.-S.; Laforest, M.; Page, E.; Kreiner, J.M.; James, T. Population Genomic Approaches for Weed Science. Plants 2019, 8, 354. https://doi.org/10.3390/plants8090354
Martin SL, Parent J-S, Laforest M, Page E, Kreiner JM, James T. Population Genomic Approaches for Weed Science. Plants. 2019; 8(9):354. https://doi.org/10.3390/plants8090354
Chicago/Turabian StyleMartin, Sara L., Jean-Sebastien Parent, Martin Laforest, Eric Page, Julia M. Kreiner, and Tracey James. 2019. "Population Genomic Approaches for Weed Science" Plants 8, no. 9: 354. https://doi.org/10.3390/plants8090354
APA StyleMartin, S. L., Parent, J.-S., Laforest, M., Page, E., Kreiner, J. M., & James, T. (2019). Population Genomic Approaches for Weed Science. Plants, 8(9), 354. https://doi.org/10.3390/plants8090354





