Current Status and Future Prospects in Herbicide Discovery
Abstract
1. Introduction
- The success of GR crops that revolutionized weed management [8].
- The increased cost of R&D programs for production of a single new active ingredient from $184 million in 2000 to nearly $286 million in 2016 [9].
- The increased barriers imposed by toxicological and environmental regulations that must be fulfilled to ensure safety of the products [10].
- The severe attrition of the Agchem industry from more than 100 R&D companies to a few dominating companies [11]. This may in part be due to late stage failures (duPont), expense of liabilities and the depth of intellectual property.
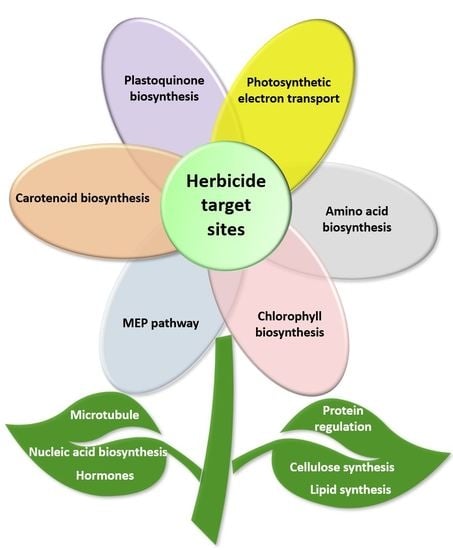
2. Novel Mechanisms of Action
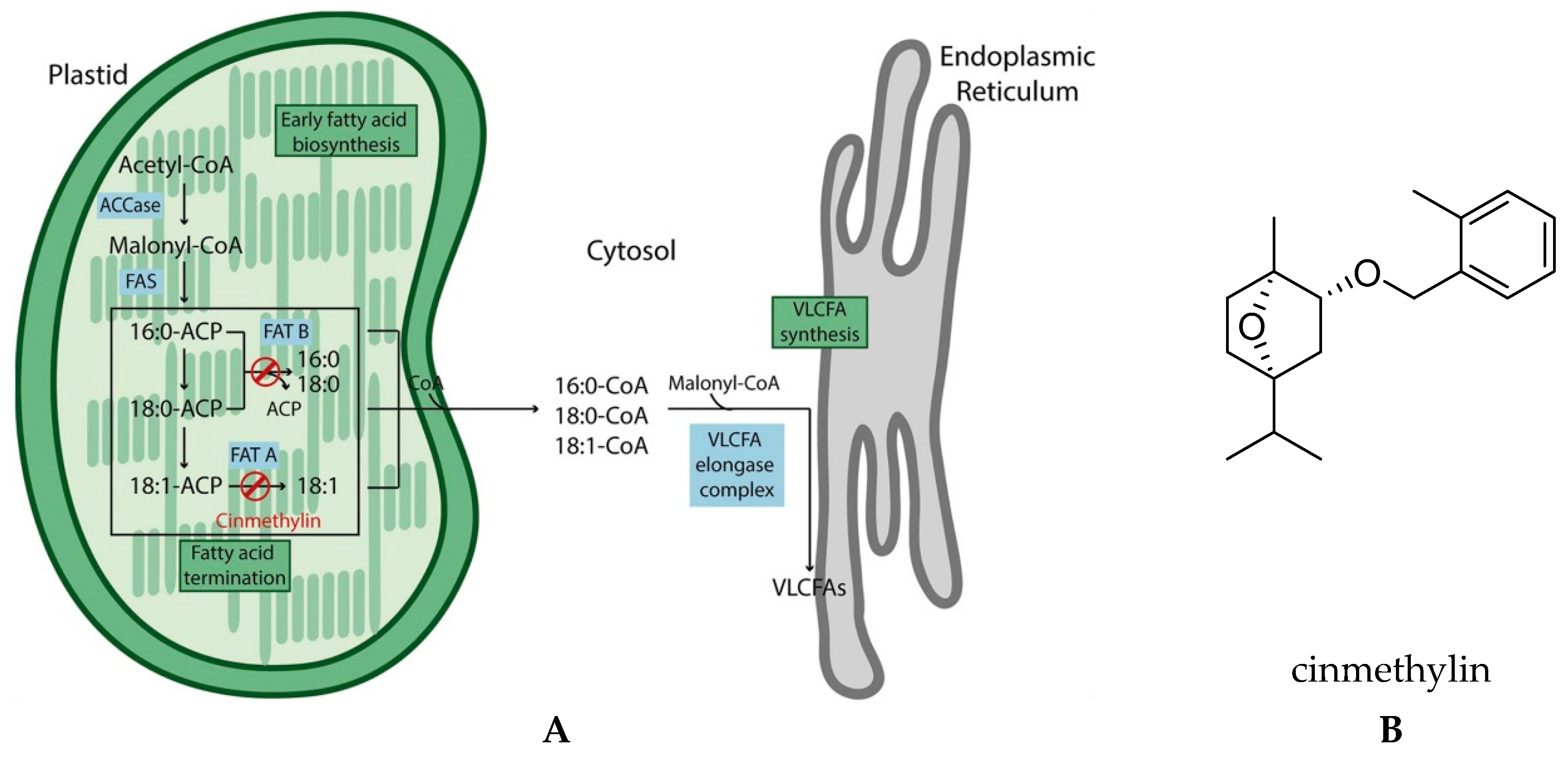
2.1. Lipid Biosynthesis
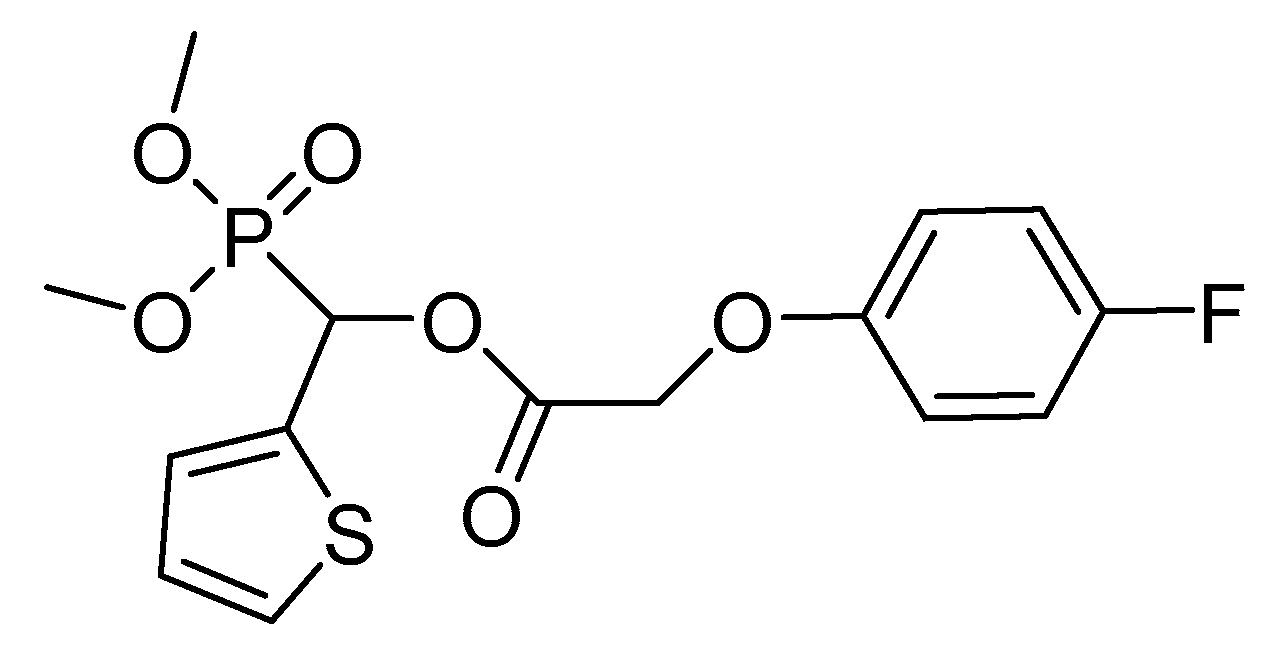
Fatty Acid Thioesterase (FAT)
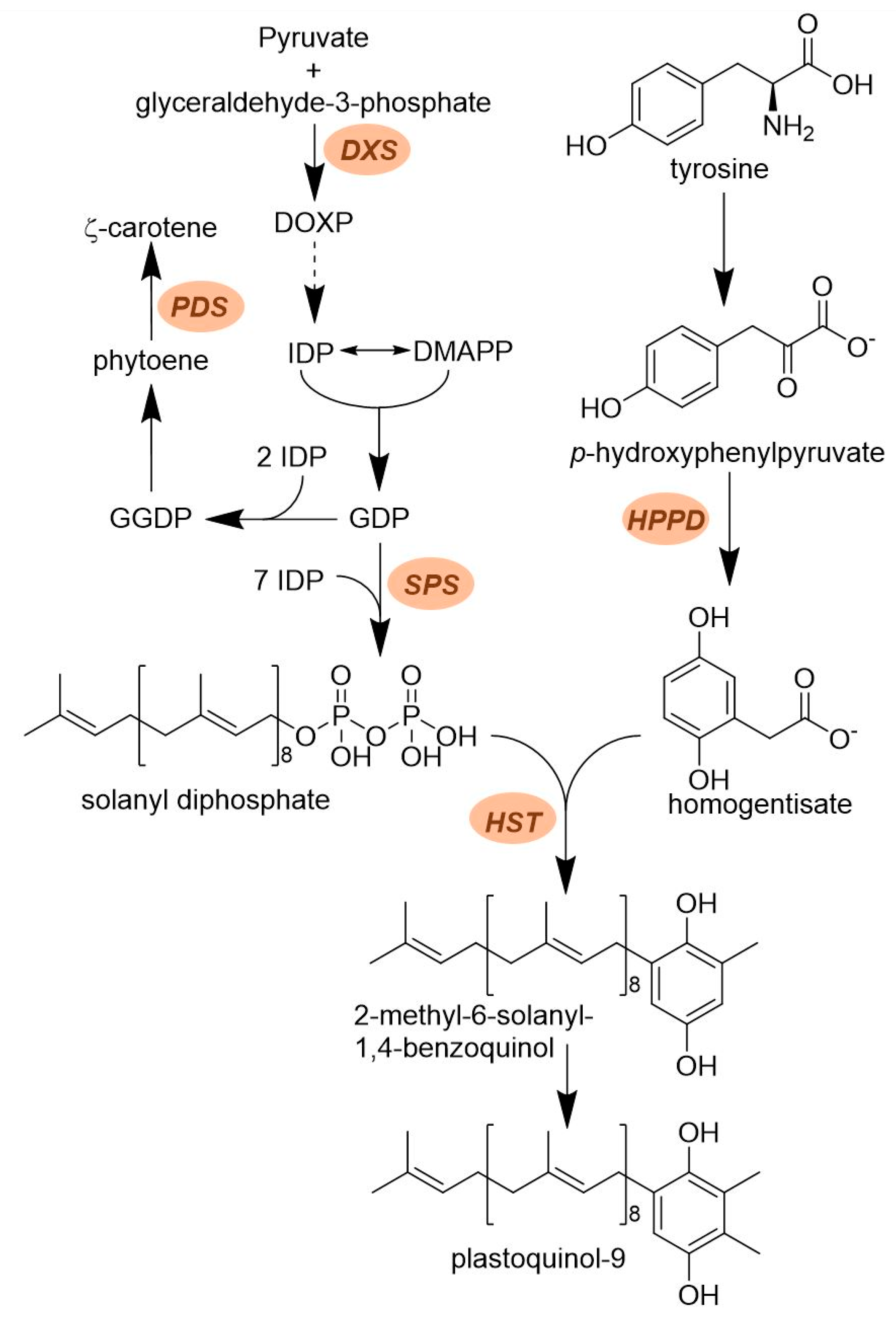
2.2. Plastoquinone Biosynthesis
2.2.1. Solanyl Diphosphate Synthase (SPS)
2.2.2. Homogentisate Solanesyl Transferase (HST)
2.3. Amino Acid Biosynthesis and Protein Regulation
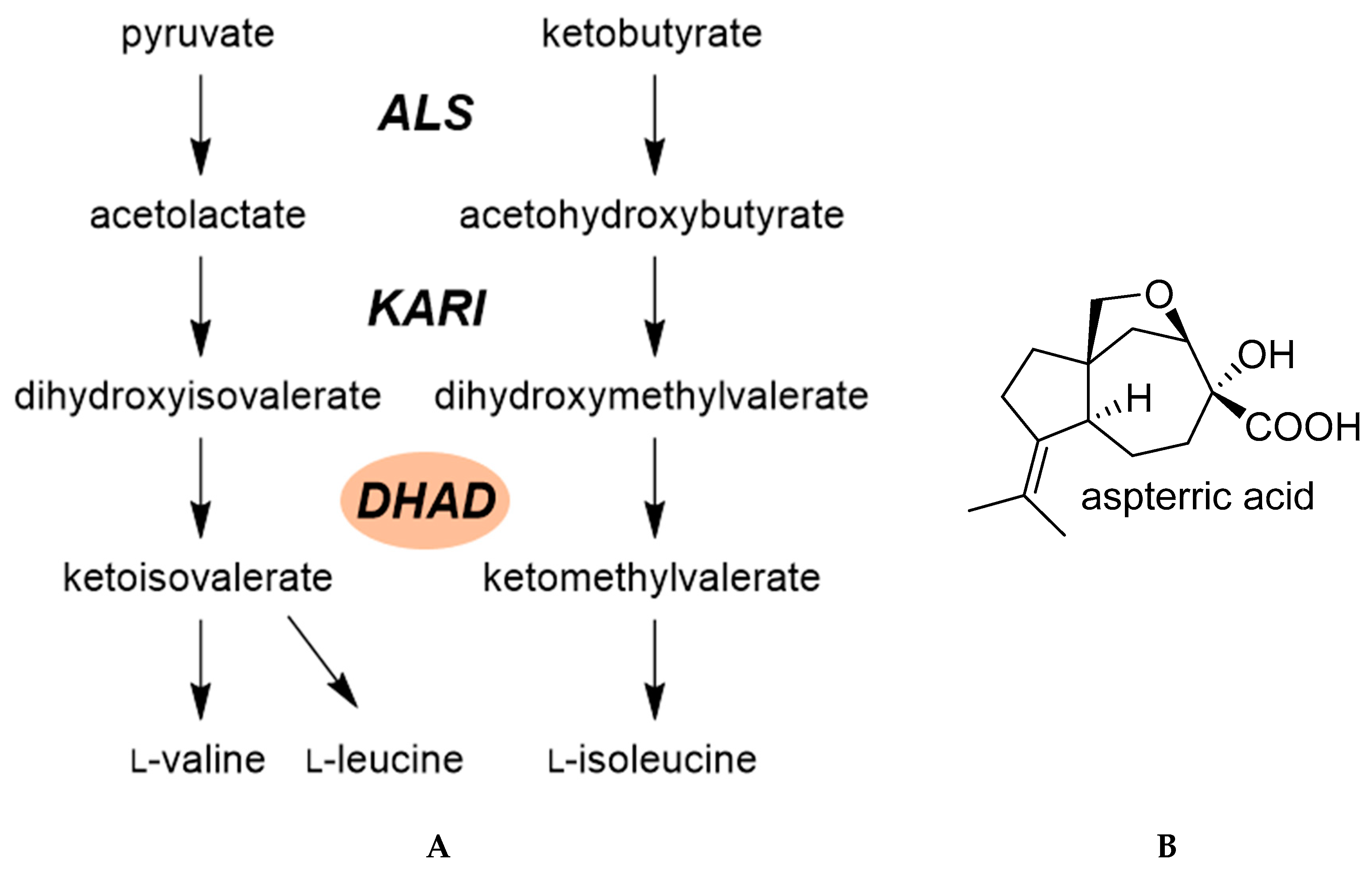
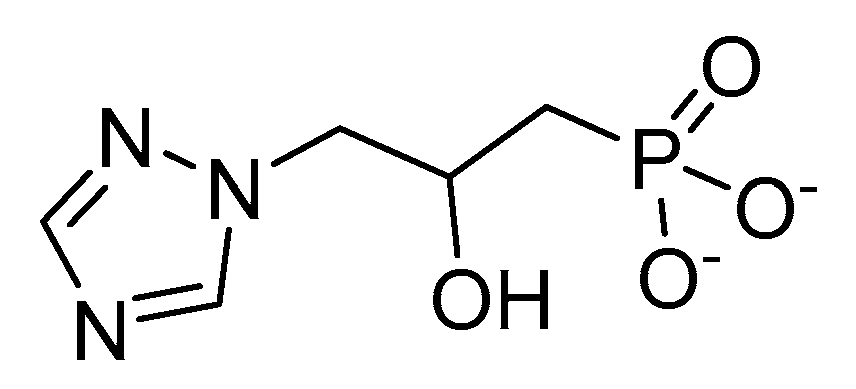
2.3.1. Dihydroxy-Acid Dehydratase (DHAD)
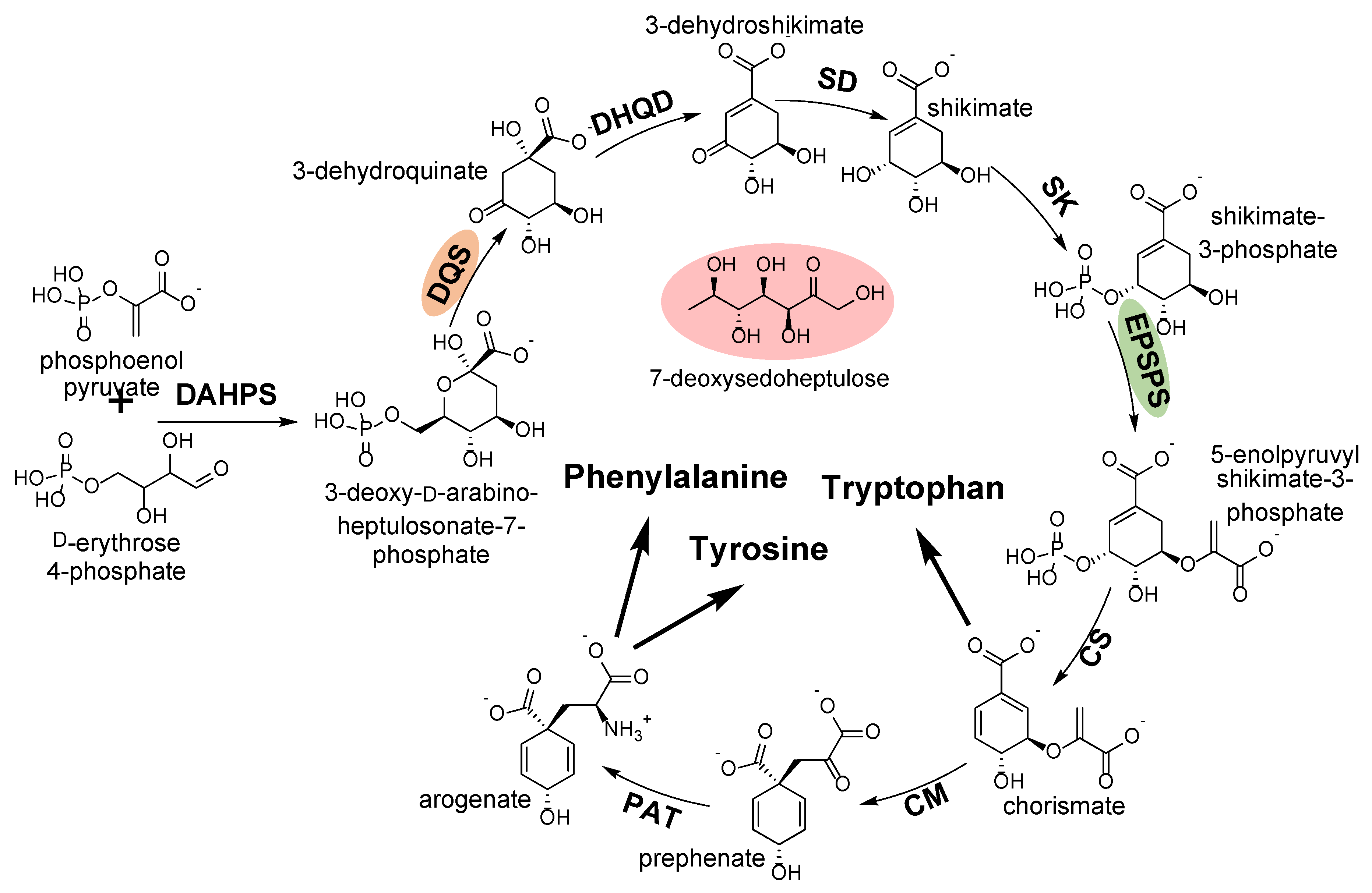
2.3.2. 3-Dehydroquinate Synthase
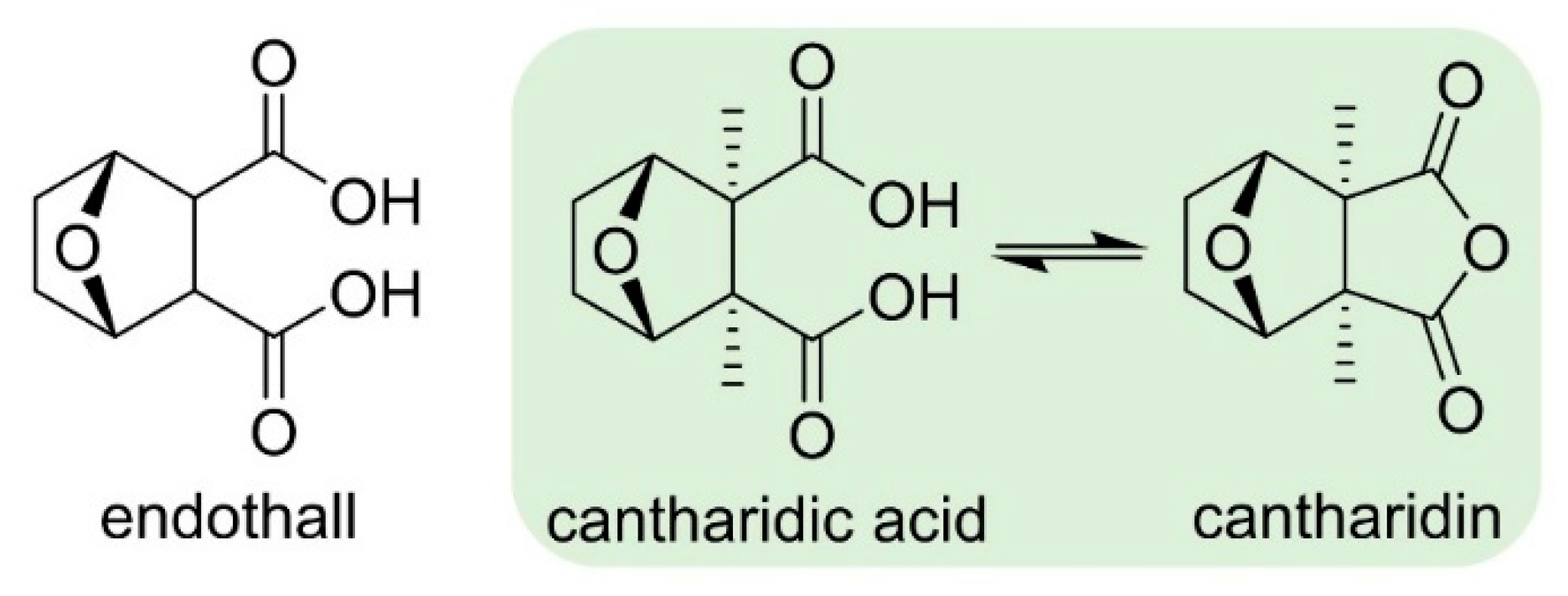
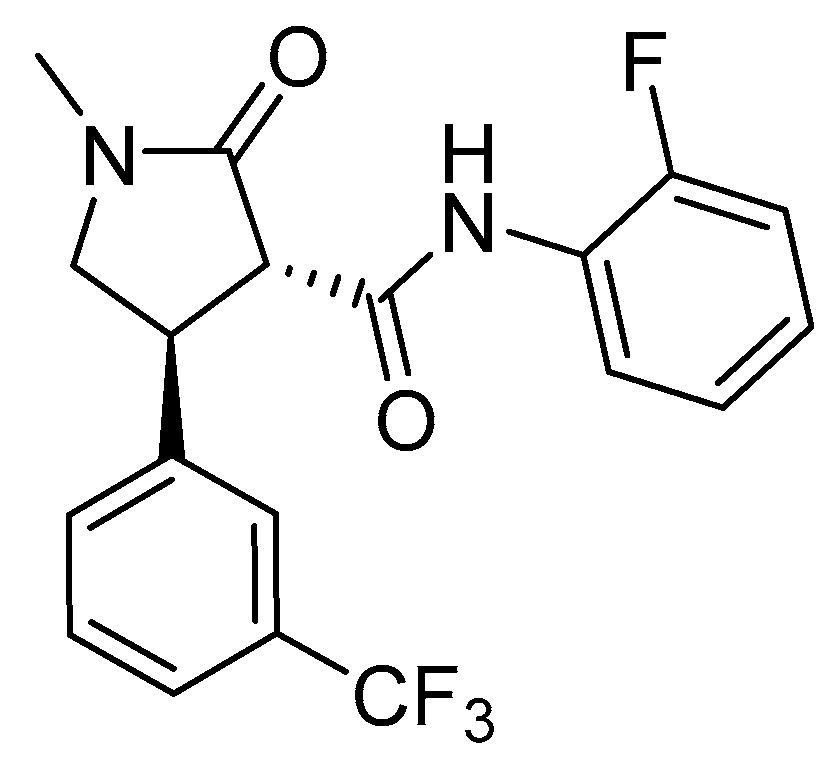
2.3.3. Serine/Threonine Protein Phosphatases (PPs)
2.4. Pyruvate Dehydrogenase Complex (PDHc)
2.5. Imadazoleglycerol Phosphate Dehydratase (IGPD)
2.6. Dihydroorotate Dehydrogenase (DHODH)
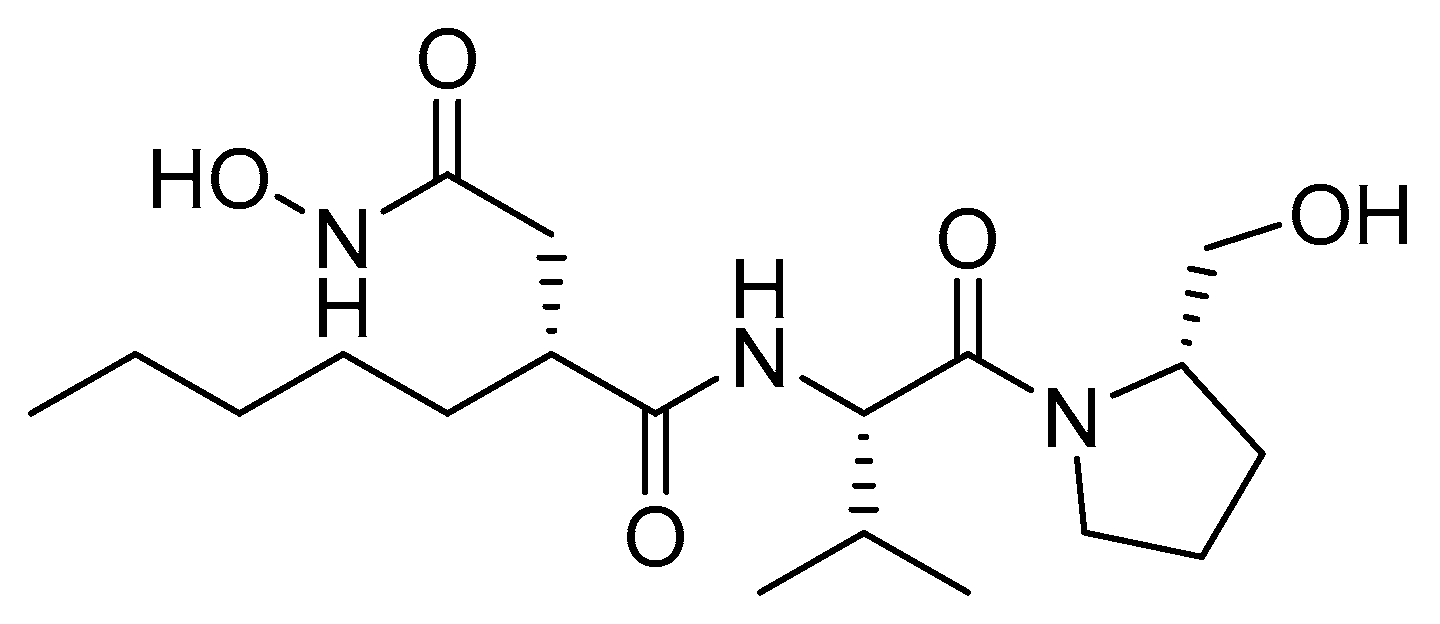
2.7. Peptide Deformylase
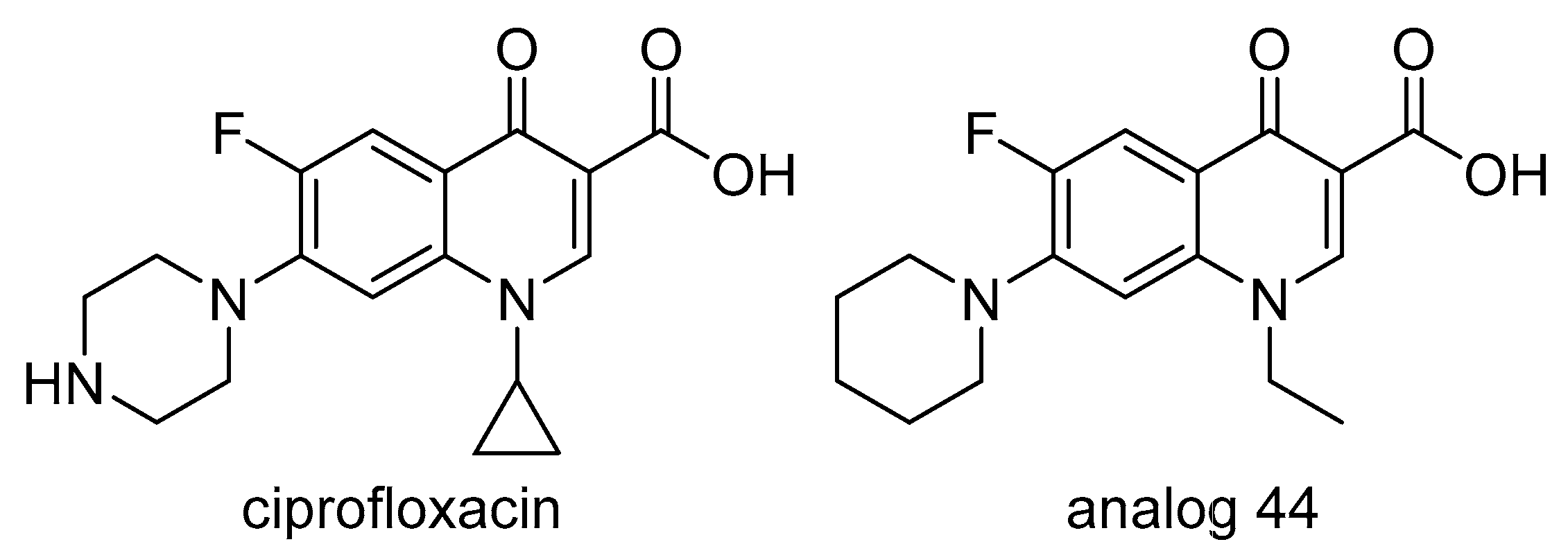
2.8. DNA Gyrase
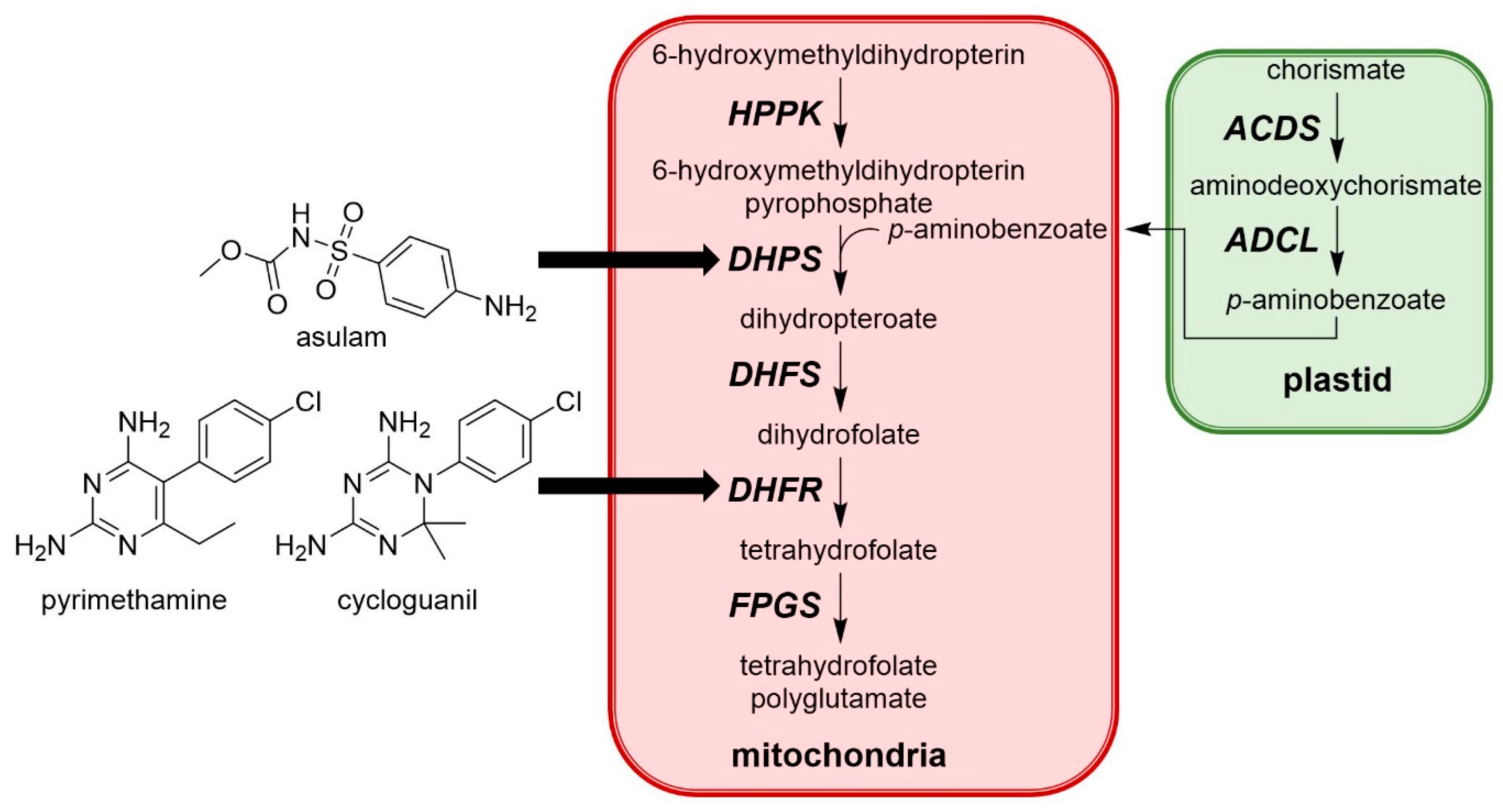
2.9. Dihydrofolate Reductase (DHFR)
3. New Insight on Known Mechanisms of Action
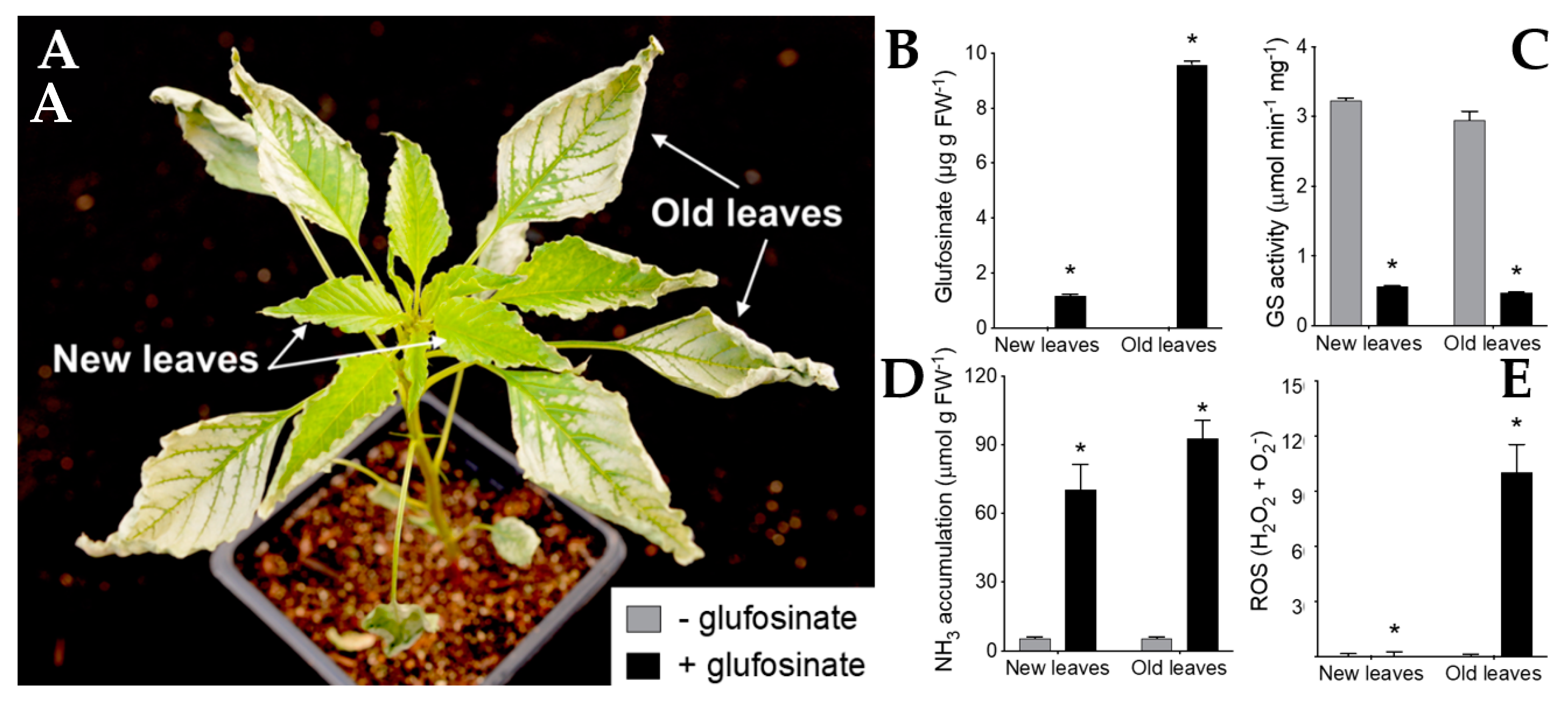
3.1. New Insight on Glufosinate Mechanism of Action
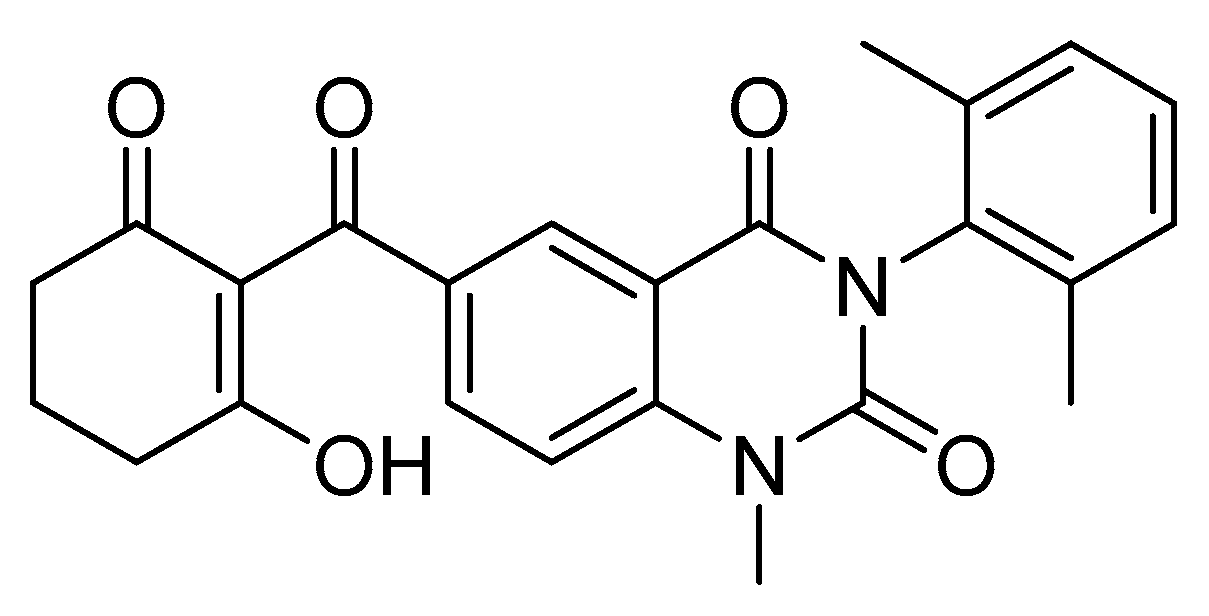
3.2. New Insight on Slow-Binding Properties of HPPD Inhibitors
4. Promising New Chemistry
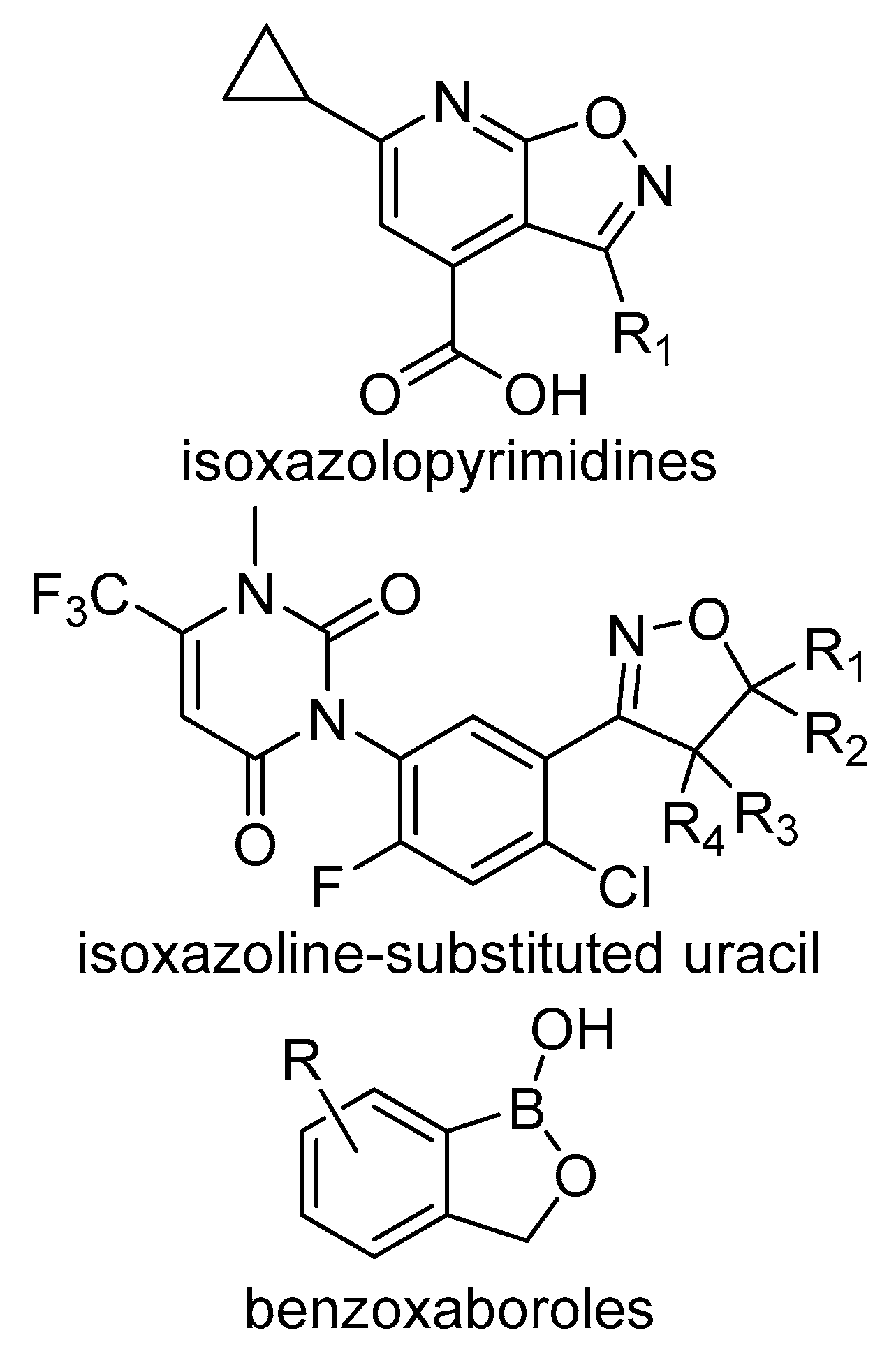
4.1. Isoxazolopyridine Herbicides
4.2. Isoxazoline-Substituted Uracil Herbicides
4.3. Benzoxaboroles Herbicides
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Sterling, T.M.; Hall, J.C. Mechanism of action of natural auxins and the auxinic herbicides. In Herbicide Activity: Toxicology, Biochemistry and Molecular Biology; Roe, R.M., Burton, J.D., Kuhr, R.J., Eds.; IOS Press: Amsterdam, The Netherlands, 1997; pp. 111–141. [Google Scholar]
- Pannell, D.J.; Tillie, P.; Rodríguez-Cerezo, E.; Ervin, D.; Frisvold, G.B. Herbicide resistance: Economic and environmental challenges. AgBioForum 2017, 19, 136–155. [Google Scholar]
- Shaw, D.R. The “Wicked” nature of the herbicide resistance problem. Weed Sci. 2016, 64, 552–558. [Google Scholar] [CrossRef]
- Ervin, D.E.; Breshears, E.H.; Frisvold, G.B.; Hurley, T.; Dentzman, K.E.; Gunsolus, J.L.; Jussaume, R.A.; Owen, M.D.K.; Norsworthy, J.K.; Al Mamun, M.M.; et al. Farmer attitudes toward cooperative approaches to herbicide resistance management: A common pool ecosystem service challenge. Ecol. Econ. 2019, 157, 237–245. [Google Scholar] [CrossRef]
- Davis, A.S.; Frisvold, G.B. Are herbicides a once in a century method of weed control? Pest Manag. Sci. 2017, 73, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Duke, S.O. Why have no new herbicide modes of action appeared in recent years? Pest Manag. Sci. 2012, 68, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E. Is there a natural route to the next generation of herbicides? Outlooks Pest Manag. 2018, 29, 54–57. [Google Scholar] [CrossRef]
- Duke, S.O. The history and current status of glyphosate. Pest Manag. Sci. 2018, 74, 1027–1034. [Google Scholar] [CrossRef]
- McDougall, P. Agrochemical Research and Development: The Cost of New Product Discovery, Development and Registratoin; Pathhead: Midlothian, UK, 2016; p. 43. [Google Scholar]
- Peters, B.; Strek, H.J. Herbicide discovery in light of rapidly spreading resistance and ever-increasing regulatory hurdles. Pest Manag. Sci. 2018, 74, 2211–2215. [Google Scholar] [CrossRef]
- Copping, L.G. The evolution of crop protection companies. Outlooks Pest Manag. 2018, 29, 25–37. [Google Scholar] [CrossRef]
- Gerwick, B.C. Thirty years of herbicide discovery: surveying the past and contemplating the future. In Chapters VII–IX in Agrow Report; Informa: London, UK, 2010; pp. VII–IX. [Google Scholar]
- Gandy, M.N.; Corral, M.G.; Mylne, J.S.; Stubbs, K.A. An interactive database to explore herbicide physicochemical properties. Org. Biomol. Chem. 2015, 13, 5586–5590. [Google Scholar] [CrossRef]
- Dayan, F.E.; Barker, A.; Bough, R.; Ortiz, M.; Takano, H.; Duke, S.O. Herbicide mechanisms of action and resistance. In Comprehensive Biotechnology, 3rd ed.; Grodzinski, B., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 4, in press. [Google Scholar]
- Shaner, D.L. Herbicide Handbook, 10th ed.; Weed Science Society of America: Lawrence, KS, USA, 2014; p. 513. [Google Scholar]
- Beckie, H.J.; Harker, K.N. Our top 10 herbicide-resistant weed management practices. Pest Manag. Sci. 2017, 73, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Haesaert, G.; Van Leeuwen, T.; Holden-Dye, L.; Crossthwaite, A.; Nauen, R. Pesticides modes of action and resistance: A perspective from the 2019 IUPAC congress. Outlooks Pest Manag. 2019, 30, 157–163. [Google Scholar] [CrossRef]
- Campe, R.; Hollenbach, E.; Kämmerer, L.; Hendriks, J.; Höffken, H.W.; Kraus, H.; Lerchl, J.; Mietzner, T.; Tresch, S.; Witschel, M.; et al. A new herbicidal site of action: Cinmethylin binds to acyl-ACP thioesterase and inhibits plant fatty acid biosynthesis. Pestic. Biochem. Physiol. 2018, 148, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Romagni, J.G.; Duke, S.O. Herbicides: Cinmethylin. In Encyclopedia of Agrochemicals; Plimmer, J.R., Gammon, D.W., Ragsdale, N.N., Eds.; John Wiley & Sons: New York, NY, USA, 2003; Volume 2, pp. 754–757. [Google Scholar]
- Norris, S.R.; Barrette, T.R.; DellaPenna, D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 1995, 7, 2139–2149. [Google Scholar] [PubMed]
- Liu, M.; Lu, S. Plastoquinone and ubiquinone in plants: Biosynthesis, physiological function and metabolic engineering. Front. Plant Sci. 2016, 7, 1898. [Google Scholar] [CrossRef] [PubMed]
- Ohara, K.; Sasaki, K.; Yazaki, K. Two solanesyl diphosphate synthases with different subcellular localizations and their respective physiological roles in Oryza sativa. J. Experiment. Bot. 2010, 61, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Sadre, R.; Frentzen, M.; Saeed, M.; Hawkes, T. Catalytic reactions of the homogentisate prenyl transferase involved in plastoquinone-9 biosynthesis. J. Biol. Chem. 2010, 285, 18191–18198. [Google Scholar] [CrossRef] [PubMed]
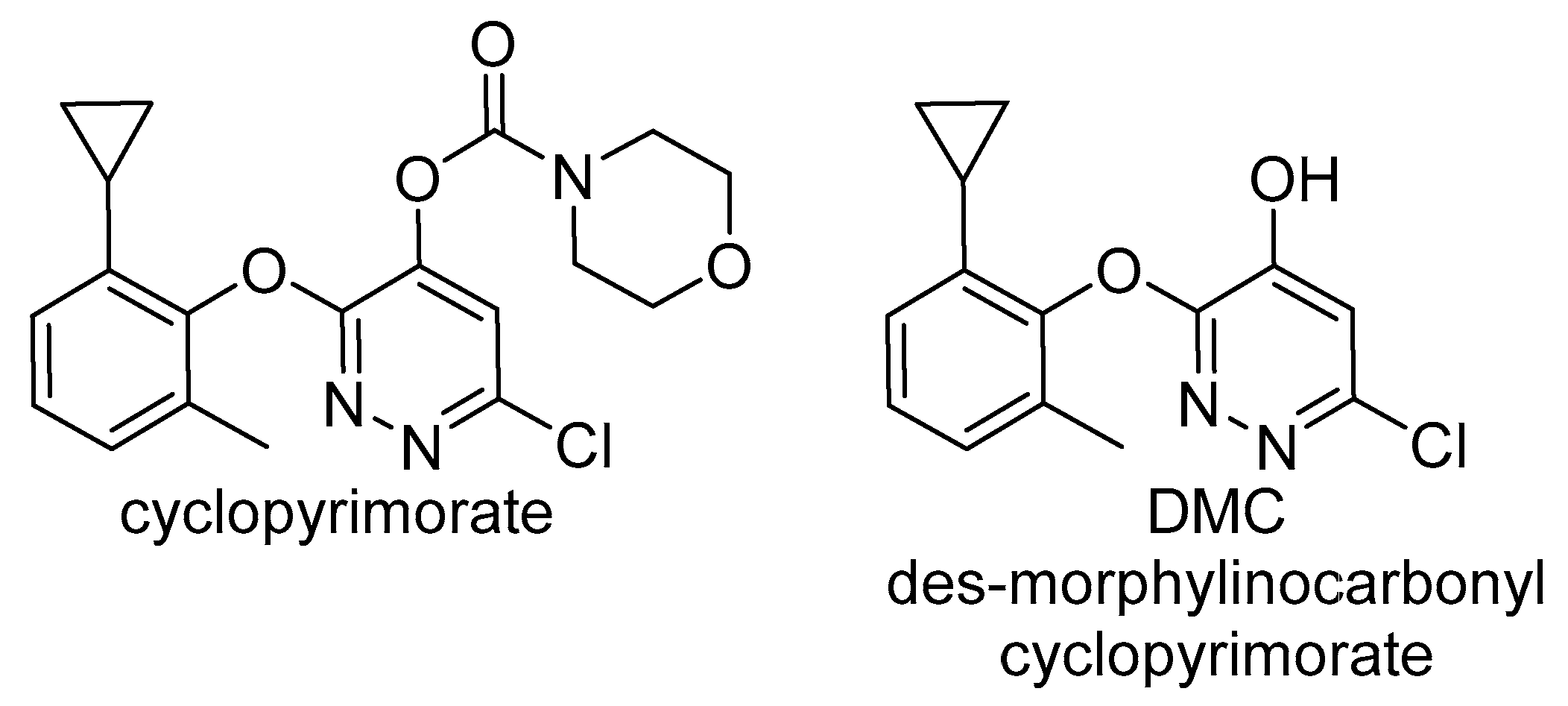
- Shino, M.; Hamada, T.; Shigematsu, Y.; Hirase, K.; Banba, S. Action mechanism of bleaching herbicide cyclopyrimorate, a novel homogentisate solanesyltransferase inhibitor. J. Pestic. Sci. 2018, 43, 233–239. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S.E.; Tang, Y. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Duke, S.O.; Stidham, M.A.; Dayan, F.E. A novel genomic approach to herbicide and herbicide mode of action discovery. Pest Manag. Sci. 2019, 75, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 2013, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Brilisauer, K.; Rapp, J.; Rath, P.; Schöllhorn, A.; Bleul, L.; Weiß, E.; Stahl, M.; Grond, S.; Forchhammer, K. Cyanobacterial antimetabolite 7-deoxy-sedoheptulose blocks the shikimate pathway to inhibit the growth of prototrophic organisms. Nat. Commun. 2019, 10, 545. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Owens, D.K.; Corniani, N.; Silva, F.M.L.; Watson, S.B.; Howell, J.L.; Shaner, D.L. Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci. 2015, 63, 23–63. [Google Scholar] [CrossRef]
- Johnson, L.N. The regulation of protein phosphorylation. Biochem. Soc. Trans. 2009, 37, 627–641. [Google Scholar] [CrossRef] [PubMed]
- DeLong, A. Switching the flip: Protein phosphatase roles in signaling pathways. Curr. Opin. Plant Biol. 2006, 9, 470–477. [Google Scholar] [CrossRef]
- Uhrig, R.G.; Labandera, A.M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef]
- Bajsa, J.; Pan, Z.; Dayan, F.E.; Owens, D.K.; Duke, S.O. Validation of serine-threonine protein phosphatase as the herbicide target site of endothall. Pestic. Biochem. Physiol. 2012, 102, 38–44. [Google Scholar] [CrossRef]
- Ortiz, M.F.; Nissen, S.J.; Gray, C.J. Endothall behavior in Myriophyllum spicatum and Hydrilla verticillata. Pest Manag. Sci. 2019, in press. [Google Scholar] [CrossRef]
- Jordan, F.; Nemeria, N.; Guo, F.; Baburina, I.; Gao, Y.; Kahyaoglu, A.; Li, H.; Wang, J.; Yi, J.; Guest, J.R.; et al. Regulation of thiamin diphosphate-dependent 2-oxo acid decarboxylases by substrate and thiamin diphosphate.Mg(II) – evidence for tertiary and quaternary interactions. Biochim. Biophys. Acta - Prot. Struct. Mol. Enzymol. 1998, 1385, 287–306. [Google Scholar] [CrossRef]
- Peng, H.; Wang, T.; Xie, P.; Chen, T.; He, H.W.; Wan, J. Molecular docking and three-dimensional quantitative structure−activity relationship studies on the binding modes of herbicidal 1-(substituted phenoxyacetoxy)alkylphosphonates to the E1 component of pyruvate dehydrogenase. J. Agric. Food Chem. 2007, 55, 1871–1880. [Google Scholar] [CrossRef]
- He, H.-W.; Peng, H.; Wang, T.; Wang, C.; Yuan, J.L.; Chen, T.; He, J.; Tan, X. α-(Substituted-phenoxyacetoxy)-α-heterocyclylmethylphosphonates: Synthesis, herbicidal activity, inhibition on pyruvate dehydrogenase complex (PDHc), and application as postemergent herbicide against broadleaf weeds. J. Agric. Food Chem. 2013, 61, 2479–2488. [Google Scholar] [CrossRef] [PubMed]
- Gohda, K.; Kimura, Y.; Mori, I.; Ohta, D.; Kikuchi, T. Theoretical evidence of the existence of a diazafulvene intermediate in the reaction pathway of imidazoleglycerol phosphate dehydratase: Design of a novel and potent heterocycle structure for the inhibitor on the basis of the electronic structure-activity relationship study. Biochim. Biophys. Acta — Prot. Struct. Mol. Enzymol. 1998, 1385, 107–114. [Google Scholar] [CrossRef]
- Glynn, S.E.; Baker, P.J.; Sedelnikova, S.E.; Davies, C.L.; Eadsforth, T.C.; Levy, C.W.; Rodgers, H.F.; Blackburn, G.M.; Hawkes, T.R.; Viner, R.; et al. Structure and mechanism of imidazoleglycerol-phosphate dehydratase. Structure 2005, 13, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.M. Substituted propyl phosphonic acid derivatives and their use as herbicides. EP Patent 78613, 11 May 1983. [Google Scholar]
- Bisson, C.; Britton, K.L.; Sedelnikova, S.E.; Rodgers, H.F.; Eadsforth, T.C.; Viner, R.C.; Hawkes, T.R.; Baker, P.J.; Rice, D.W. Crystal structures reveal that the reaction mechanism of imidazoleglycerol-phosphate dehydratase is controlled by switching Mn(II) coordination. Structure 2015, 23, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, A.; Knecht, W.; Piskur, J.; Löffler, M. Plant dihydroorotate dehydrogenase differs significantly in substrate specificity and inhibition from the animal enzymes. FEBS Lett. 2002, 529, 346–350. [Google Scholar] [CrossRef]
- Chen, D.Z.; Patel, D.V.; Hackbarth, C.J.; Wang, W.; Dreyer, G.; Young, D.C.; Margolis, P.S.; Wu, C.; Ni, Z.J.; Trias, J.; et al. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 2000, 39, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Duke, S.O. Natural compounds as next generation herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef] [PubMed]
- Fernández-San Millán, A.; Obregón, P.; Veramendi, J. Over-expression of peptide deformylase in chloroplasts confers actinonin resistance, but is not a suitable selective marker system for plastid transformation. Transgenic Res. 2011, 20, 613–624. [Google Scholar] [CrossRef]
- Hou, C.X.; Dirk, L.M.A.; Goodman, J.P.; Williams, M.A. Metabolism of the peptide deformylase inhibitor actinonin in tobacco. Weed Sci. 2006, 54, 246–254. [Google Scholar] [CrossRef]
- Wall, M.K.; Mitchenall, L.A.; Maxwell, A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 2004, 101, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Evans-Roberts, K.M.; Mitchenall, L.A.; Wall, M.K.; Leroux, J.; Mylne, J.S.; Maxwell, A. DNA gyrase is the target for the quinolone drug ciprofloxacin in Arabidopsis thaliana. J. Biol. Chem. 2016, 291, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.D.; Waraich, N.F.; Debowski, A.W.; Corral, M.G.; Maxwell, A.; Mylne, J.S.; Stubbs, K.A. Developing ciprofloxacin analogues against plant DNA gyrase: A novel herbicide mode of action. Chem. Commun. 2018, 54, 1869–1872. [Google Scholar] [CrossRef] [PubMed]
- Veerasekaran, P.; Kirkwood, R.C.; Parnell, E.W. Studies of the mechanism of action of asulam in plants. Part II: Effect of asulam on the biosynthesis of folic acid. Pestic. Sci. 1981, 12, 330–338. [Google Scholar] [CrossRef]
- Corral, M.G.; Haywood, J.; Stehl, L.H.; Stubbs, K.A.; Murcha, M.W.; Mylne, J.S. Targeting plant DIHYDROFOLATE REDUCTASE with antifolates and mechanisms for genetic resistance. Plant J. 2018, 95, 727–742. [Google Scholar] [CrossRef] [PubMed]
- Takano, H.K.; Beffa, R.; Preston, C.; Westra, P.; Dayan, F.E. Reactive oxygen species trigger the fast action of glufosinate. Planta 2019, 249, 1837–1849. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.K.; Whitfield, A.C.; Gowans, L.A.; Auton, T.R.; Provan, W.M.; Lock, E.A.; Smith, L.L. Inhibition of 4-hydroxyphenylpyruvate dioxygenase by 2-(2-nitro-4-trifluoromethylbenzoyl)-cyclohexane-1,3-dione and 2-(2-chloro-4-methanesulfonylbenzoyl)-cyclohexane-1,3-dione. Toxicol. Appl. Pharmacol. 1995, 133, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.W.; Lin, H.Y.; Cao, R.J.; Ming, Z.Z.; Chen, T.; Hao, G.F.; Yang, W.C.; Yang, G.F. Design, synthesis and herbicidal activity of novel quinazoline-2,4-diones as 4-hydroxyphenylpyruvate dioxygenase inhibitors. Pest Manag. Sci. 2015, 71, 1122–1132. [Google Scholar] [CrossRef]
- Adamczyk-Woźniak, A.; Borys, K.M.; Sporzyński, A. Recent developments in the chemistry and biological applications of benzoxaboroles. Chem. Rev. 2015, 115, 5224–5247. [Google Scholar] [CrossRef]
| Group | Type | Mechanism/Target |
|---|---|---|
| Biochemical pathways and physiological processes involved with photosynthesis | Light reaction | Photosystem II |
| Photosystem I | ||
| Carotenoid | Deoxyxylulose-5-phosphate synthase | |
| Phytoene desaturase | ||
| Plastoquinone | p-Hydroxyphenylpyruvate dioxygenase | |
| Homogentisate solanesyltransferase1 | ||
| Solanyl diphosphate synthase1 | ||
| Chlorophylls | Protoporphyrinogen oxidase | |
| Uncouplers | Oxidative (photo)phosphorylation | |
| Formation of biological building blocks or their assembly into macromolecules | Amino acids | 5-enolpyruvylshikimate-3-phosphate (EPSP) Synthase |
| Acetolactate synthase | ||
| Glutamine synthetase | ||
| Dihydroxy-acid dehydratase1 | ||
| Lipids | Acetyl-CoA carboxylase | |
| Fatty acid thioesterase1 | ||
| Very long chain fatty acid elongases | ||
| Cell walls | Cellulose synthase and others | |
| Microtubule assembly | α- and/or β-Tubulin | |
| Microtubule organization | Microtubule organizing centers | |
| Folates | Dihydropteroate synthetase | |
| Dihydrofolate reductase1 | ||
| Nucleic acids | DNA gyrase1 | |
| Dihydroorotate dehydrogenase1 | ||
| Other processes | Protein synthesis | Peptide deformylase |
| Protein regulation | Serine-threonine protein phosphatases1 | |
| Hormone | Synthetic auxins | |
| Auxin-transport inhibition |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dayan, F.E. Current Status and Future Prospects in Herbicide Discovery. Plants 2019, 8, 341. https://doi.org/10.3390/plants8090341
Dayan FE. Current Status and Future Prospects in Herbicide Discovery. Plants. 2019; 8(9):341. https://doi.org/10.3390/plants8090341
Chicago/Turabian StyleDayan, Franck E. 2019. "Current Status and Future Prospects in Herbicide Discovery" Plants 8, no. 9: 341. https://doi.org/10.3390/plants8090341
APA StyleDayan, F. E. (2019). Current Status and Future Prospects in Herbicide Discovery. Plants, 8(9), 341. https://doi.org/10.3390/plants8090341