Uncovering the Host Range for Maize Pathogen Magnaporthiopsis maydis
Abstract
:1. Introduction
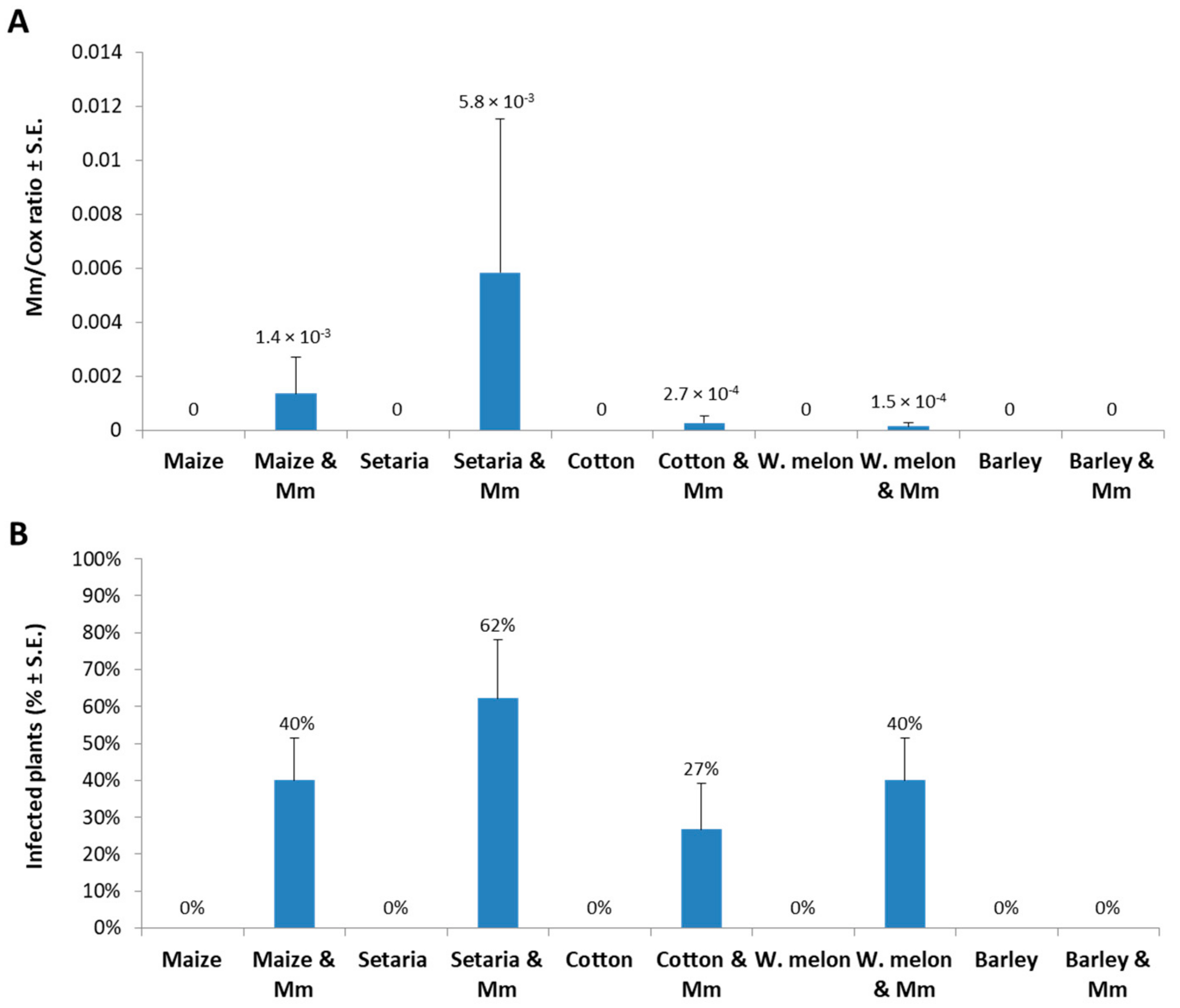
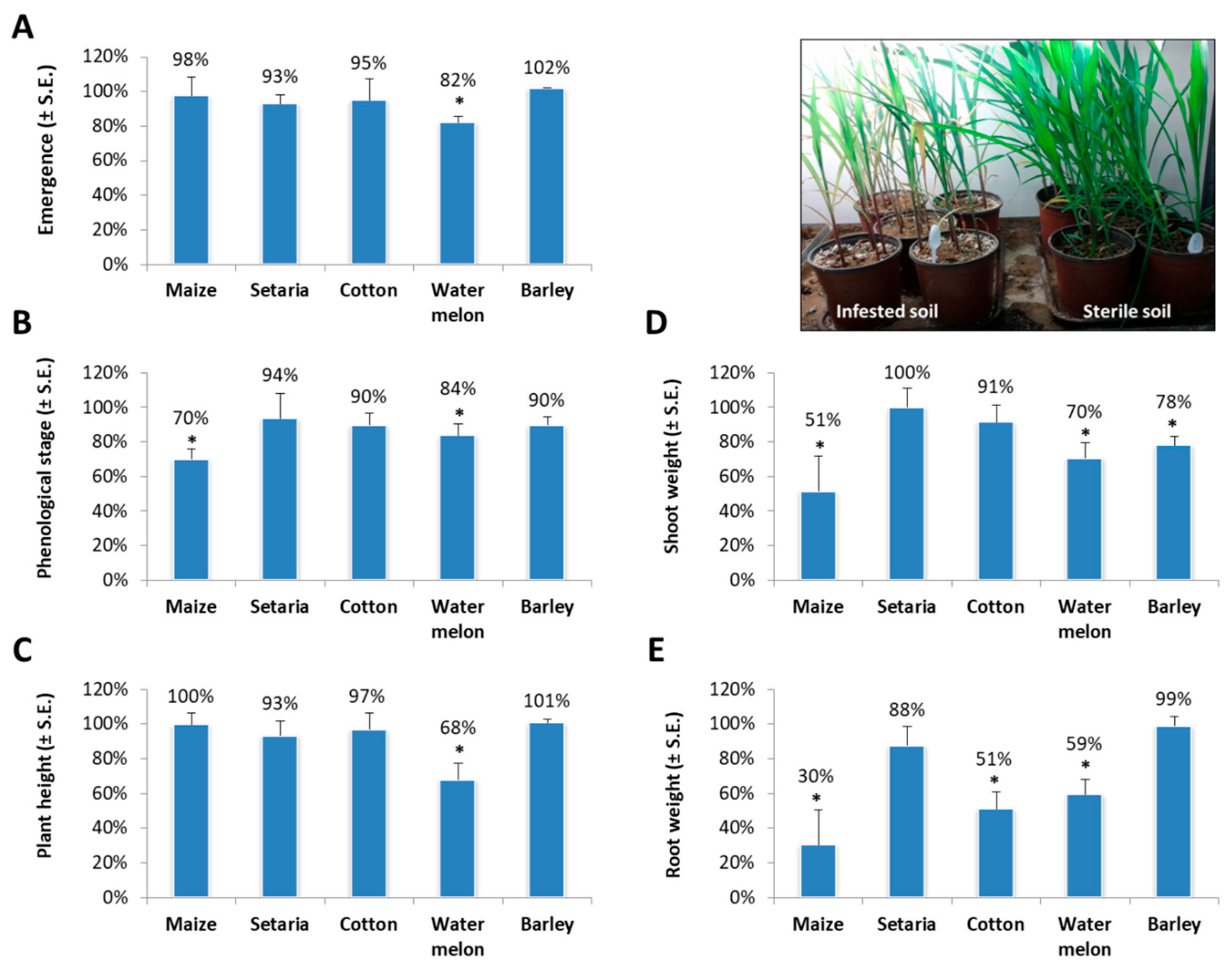
2. Results
3. Discussion
4. Materials and Methods
4.1. Field Survey
4.2. Fungal Isolates and Growth Conditions
4.3. Growth Chamber Experiment
4.4. Molecular Diagnosis of Magnaporthiopsis maydis
4.5. Statistical Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klaubauf, S.; Tharreau, D.; Fournier, E.; Groenewald, J.Z.; Crous, P.W.; de Vries, R.P.; Lebrun, M.H. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Stud. Mycol. 2014, 79, 85–120. [Google Scholar] [CrossRef] [PubMed]
- Gams, W. Phialophora and some similar morphologically little-differentiated anamorphs of divergent ascomycetes. Stud. Mycol. 2000, 45, 187–200. [Google Scholar]
- Sabet, K.A.; Samra, A.S.; Hingorani, M.K.; Mansour, I.M. Stalk and root rots of maize in the United Arab Republic. FAO Plant Prot. Bull. 1961, 9, 121–125. [Google Scholar]
- Payak, M.M.; Lal, S.; Lilaramani, J.; Renfro, B.L. Cephalosporium maydis—A new threat to maize in India. Indian Phytopathol. 1970, 23, 562–569. [Google Scholar]
- Pecsi, S.; Nemeth, L. Appearance of Cephalosporium maydis Samra Sabet and Hingorani in Hungary; Mededelingen Faculteit Landbouwkundige En Toegepaste Biologische Wetenschappen, Universiteit Gent: Gent, Belgium, 1998; Volume 63, pp. 873–877. [Google Scholar]
- Bergstrom, G.; Leslie, F.J.; Huber, D.; Lipps, P.; Warren, H.; Esker, P.; Grau, C.; Botratynski, T.; Bulluck, R.; Floyd, J.; et al. Recovery Plan for Late Wilt of Corn Caused by Harpophora Maydis Syn. Cephalosporium Maydis; National Plant Disease Recovery System (NPDRS): Washington, DC, USA, 2008; p. 24. [Google Scholar]
- Molinero-Ruiz, M.L.; Melero-Vara, J.M.; Mateos, A. Cephalosporium maydis, the cause of late wilt in maize, a pathogen new to Portugal and Spain. Plant Dis. 2011, 94, 379. [Google Scholar] [CrossRef] [PubMed]
- Drori, R.; Sharon, A.; Goldberg, D.; Rabinovitz, O.; Levy, M.; Degani, O. Molecular diagnosis for Harpophora maydis, the cause of maize late wilt in Israel. Phytopathol. Mediterr. 2013, 52, 16–29. [Google Scholar]
- Subedi, S. A review on important maize diseases and their management in Nepal. J. Maize Res. Dev. 2015, 1, 28–52. [Google Scholar] [CrossRef]
- Degani, O.; Movshowitz, D.; Dor, S.; Meerson, A.; Goldblat, Y.; Rabinovitz, O. Evaluating azoxystrobin seed coating against maize late wilt disease using a sensitive qPCR-based method. Plant Dis. 2019, 103, 238–248. [Google Scholar] [CrossRef]
- El-Hosary, A.A.A.; EL-Fiki, I.A.I. Diallel cross analysis for earliness, yield, its components and resistance to late wilt in maize. Int. J. Agric. Sci. Res. 2015, 5, 199–210. [Google Scholar]
- Degani, O.; Cernica, G. Diagnosis and control of Harpophora maydis, the cause of late wilt in maize. Adv. Microbiol. 2014, 4, 94–105. [Google Scholar] [CrossRef]
- El-Shafey, H.A.; Claflin, L.E. Late Wilt; APS Press: St. Paul, MN, USA, 1999; pp. 43–44. [Google Scholar]
- Khokhar, M.K.; Hooda, K.S.; Sharma, S.S.; Singh, V. Post flowering stalk rot complex of maize-present status and future prospects. Maydica 2014, 59, 226–242. [Google Scholar]
- Sabet, K.A.; Samra, A.S.; Mansour, I.M. Saprophytic behaviour of Cephalosporium maydis and C. acremonium. Ann. Appl. Biol. 1970, 66, 265–271. [Google Scholar] [CrossRef]
- Sabet, K.; Samra, A.; Mansour, I. Interaction between Fusarium oxysporum F. Vasinfectum and Cephalosporium maydis on cotton and maize. Ann. Appl. Biol. 1966, 58, 93–101. [Google Scholar]
- Sahab, A.F.; Osman, A.R.; Soleman, N.K.; Mikhail, M.S. Studies on root-rot of lupin in Egypt and its control. Egypt J. Phytopathol. 1985, 17, 23–35. [Google Scholar]
- Samra, A.S.; Sabet, K.A.; Hingorani, M.K. Late wilt disease of maize caused by Cephalosporium maydis. Phytopathology 1963, 53, 402–406. [Google Scholar]
- Degani, O.; Dor, S.; Movshowitz, D.; Fraidman, E.; Rabinowitz, O.; Graph, S. Effective chemical protection against the maize late wilt causal agent, Harpophora maydis, in the field. PLoS ONE 2018, 13, e0208353. [Google Scholar] [CrossRef]
- Huang, P.; Feldman, M.; Schroder, S.; Bahri, B.A.; Diao, X.; Zhi, H.; Estep, M.; Baxter, I.; Devos, K.M.; Kellogg, E.A. Population genetics of Setaria Viridis, a new model system. Mol. Ecol. 2014, 23, 4912–4925. [Google Scholar] [CrossRef]
- Degani, O.; Goldblat, Y. Ambient stresses regulate the development of the maize late wilt causing agent, Harpophora maydis. Agric. Sci. 2014, 5, 571–582. [Google Scholar]
- Degani, O.; Dor, S.; Movshovitz, D.; Rabinovitz, O. Methods for studying Magnaporthiopsis maydis, the maize late wilt causal agent. Agronomy 2019, 9, 181. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [Green Version]
- Weller, S.; Elphinstone, J.; Smith, N.; Boonham, N.; Stead, D. Detection of Ralstonia solanacearumstrains with a quantitative, multiplex, real-time, fluorogenic PCR (taqman) assay. Appl. Environ. Microbiol. 2000, 66, 2853–2858. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, N.C. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hartung, J.S.; Levy, L. Quantitative real-time PCR for detection and identification of candidatus liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 2006, 66, 104–115. [Google Scholar] [CrossRef] [PubMed]
| Crop | Location | Date | Growing Stage and Health | Sample Size (Plants) | qPCR M. maydis Detection |
|---|---|---|---|---|---|
| Cotton | Yavne | 28/07/2016 | End session-healthy | 5 | - |
| 28/07/2016 | End session-diseased | 5 | - | ||
| 20/08/2017 | End session-diseased | 22 | 6 | ||
| Garlic | Hulata | 05/04/2016 | End session-healthy | 10 | - |
| Pea | Manara | 05/04/2016 | End session-crop residues | 10 | - |
| Hulata | 05/04/2016 | End session-healthy | 10 | - | |
| Amir | 21/06/2016 | End session-healthy | 20 | - | |
| Wheat | Manara | 05/04/2016 | End session-post harvest | 20 | - |
| Tomato (Niva) | Amir | 18/05/2016 | End session-healthy | 5 | - |
| Cyperus rotundus (purple nutsedge) | Manara | 05/04/2016 | End session-post harvest | 10 | - |
| Amir | 02/08/2016 | End session-healthy | 10 | - | |
| Amir | 21/06/2017 | End session-healthy | 10 | - |
| Pairs | Primer | Sequence | Uses | Amplification | References |
|---|---|---|---|---|---|
| Pair 1 | A200a-for A200a-rev | 5′-CCGACGCCTAAAATACAGGA-3′ 5′-GGGCTTTTTAGGGCCTTTTT-3′ | Target gene | M maydis AFLP-derived species-specific fragment | [8] |
| Pair 2 | COX-F COX-R | 5′-GTATGCCACGTCGCATTCCAGA-3′ 5′-CAACTACGGATATATAAGRRCCRRAACTG-3′ | Control | Cytochrome c oxidase (COX) gene product | [24,26] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dor, S.; Degani, O. Uncovering the Host Range for Maize Pathogen Magnaporthiopsis maydis. Plants 2019, 8, 259. https://doi.org/10.3390/plants8080259
Dor S, Degani O. Uncovering the Host Range for Maize Pathogen Magnaporthiopsis maydis. Plants. 2019; 8(8):259. https://doi.org/10.3390/plants8080259
Chicago/Turabian StyleDor, Shlomit, and Ofir Degani. 2019. "Uncovering the Host Range for Maize Pathogen Magnaporthiopsis maydis" Plants 8, no. 8: 259. https://doi.org/10.3390/plants8080259






