Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes
Abstract
:1. Introduction
2. Nitric Oxide Generation in Plant Peroxisomes
3. Peroxisomal Proteins: Targets of NO-mediated PTMs
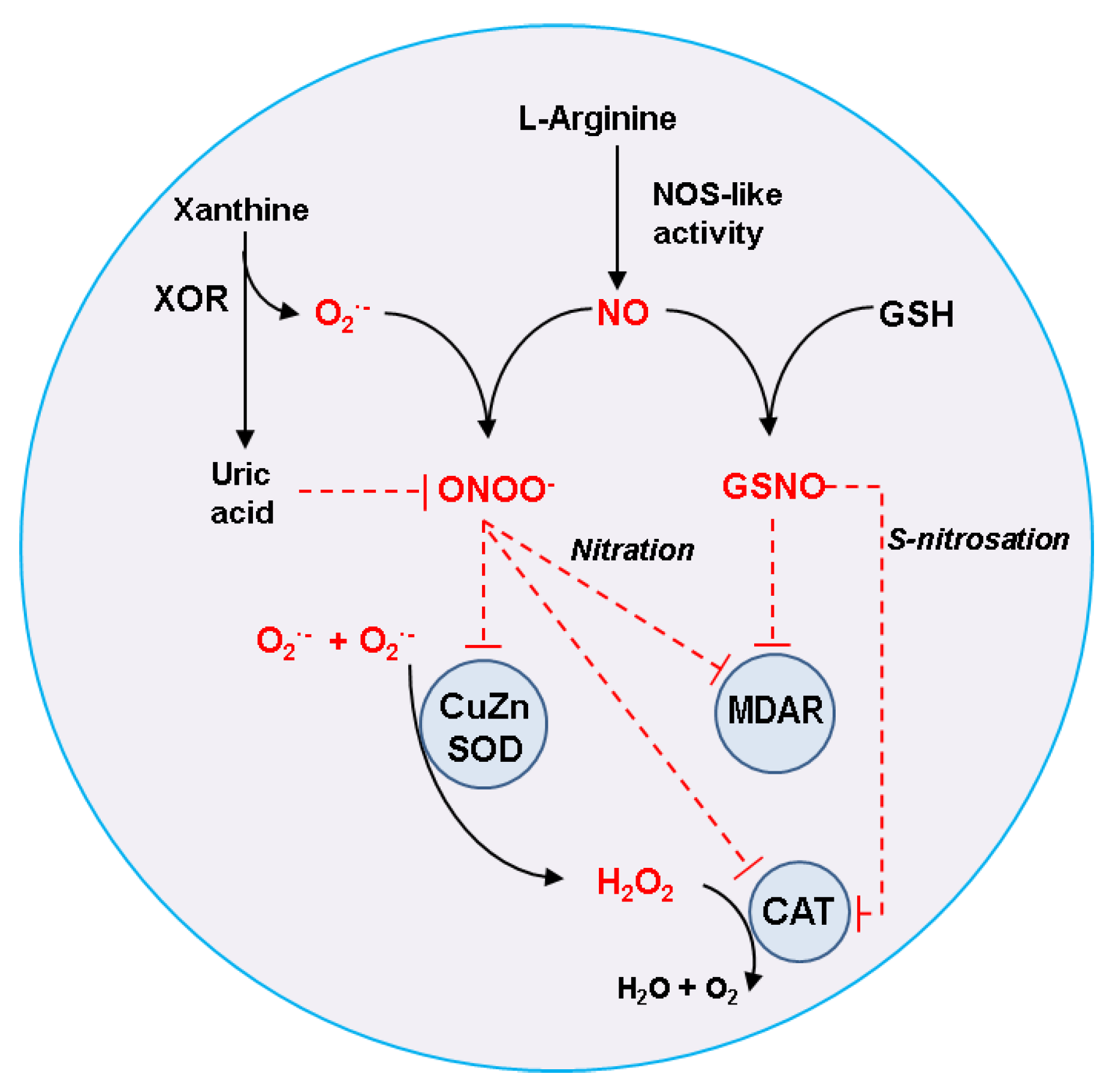
3.1. Catalase (CAT, EC 1.11.1.6)
3.2. Monodehydroascorbate Reductase (MDAR, EC 1.6.5.4)
3.3. Superoxide Dismutase (SOD; EC 1.15.1.1)
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Nishimura, M.; Hayashi, M.; Kato, A.; Yamaguchi, K.; Mano, S. Functional transformation of microbodies in higher plant cells. Cell Struct. Funct. 1996, 21, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nishimura, M. Entering a new era of research on plant peroxisomes. Curr. Opin. Plant Biol. 2003, 6, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant peroxisomes: Biogenesis and function. Plant Cell. 2012, 24, 2279–2303. [Google Scholar] [CrossRef] [PubMed]
- Sørhagen, K.; Laxa, M.; Peterhänsel, C.; Reumann, S. The emerging role of photorespiration and non-photorespiratory peroxisomal metabolism in pathogen defence. Plant Biol. (Stuttg) 2013, 15, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Goto-Yamada, S.; Mano, S.; Yamada, K.; Oikawa, K.; Hosokawa, Y.; Hara-Nishimura, I.; Nishimura, M. Dynamics of the light-dependent transition of plant peroxisomes. Plant Cell Physiol. 2015, 56, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.T.; González, K.L.; Bartel, B. Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A.; Corpas, F.J.; Sandalio, L.M.; Palma, J.M.; Gómez, M.; Barroso, J.B. Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 2002, 53, 1255–1272. [Google Scholar] [CrossRef] [PubMed]
- Weber, H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002, 7, 217–224. [Google Scholar] [CrossRef]
- del Río, L.A.; López-Huertas, E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; del Río, L.A.; Palma, J.M. Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; Palma, J.M.; Rodríguez-Ruiz, M. Plant peroxisomes: A nitro-oxidative cocktail. Redox Biol. 2017, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Peroxynitrite (ONOO−) is endogenously produced in Arabidopsis peroxisomes and is overproduced under cadmium stress. Ann. Bot. 2014, 113, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Barroso, J.B.; Valderrama, R.; Corpas, F.J. Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol Plant 2013, 35, 2635–2640. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; Carreras, A.; Quirós, M.; León, A.M.; Romero-Puertas, M.C.; Esteban, F.J.; Valderrama, R.; Palma, J.M.; Sandalio, L.M.; et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 2004, 136, 2722–2733. [Google Scholar] [CrossRef] [PubMed]
- Prado, A.M.; Porterfield, D.M.; Feijó, J.A. Nitric oxide is involved in growth regulation and reorientation of pollen tubes. Development 2004, 131, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Schlicht, M.; Ludwig-Müller, J.; Burbach, C.; Volkmann, D.; Baluska, F. Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytol. 2013, 200, 473–482. [Google Scholar] [CrossRef]
- Corpas, F.J.; Hayashi, M.; Mano, S.; Nishimura, M.; Barroso, J.B. Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol. 2009, 151, 2083–2094. [Google Scholar] [CrossRef]
- Smiri, M.; Chaoui, A.; Rouhier, N.; Gelhaye, E.; Jacquot, J.P.; El Ferjani, E. Oxidative damage and redox change in pea seeds treated with cadmium. C R Biol. 2010, 333, 801–807. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Lead-induced stress, which triggers the production of nitric oxide (NO) and superoxide anion (O2·−) in Arabidopsis peroxisomes, affects catalase activity. Nitric Oxide. 2017, 68, 103–110. [Google Scholar] [CrossRef]
- Wilson, I.D.; Neill, S.J.; Hancock, J.T. Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 2008, 31, 622–631. [Google Scholar] [CrossRef]
- Neill, S.; Bright, J.; Desikan, R.; Hancock, J.; Harrison, J.; Wilson, I. Nitric oxide evolution and perception. J. Exp Bot. 2008, 59, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Beligni, M.V.; Lamattina, L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 2000, 210, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Gubler, F.; Jacobsen, J.V.; Jones, R.L. Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 2004, 219, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.C.; Lanteri, M.L.; Lombardo, M.C.; Lamattina, L. Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 2004, 135, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Functions of Nitric Oxide (NO) in roots during development and under adverse stress conditions. Plants (Basel) 2015, 4, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; Carreras, A.; Valderrama, R.; Palma, J.M.; León, A.M.; Sandalio, L.M.; del Río, L.A. Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta. 2006, 224, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef]
- Senthil Kumar, R.; Shen, C.H.; Wu, P.Y.; Suresh Kumar, S.; Hua, M.S.; Yeh, K.W. Nitric oxide participates in plant flowering repression by ascorbate. Sci Rep. 2016, 6, 35246. [Google Scholar] [CrossRef]
- Prado, A.M.; Colaço, R.; Moreno, N.; Silva, A.C.; Feijó, J.A. Targeting of pollen tubes to ovules is dependent on nitric oxide (NO) signaling. Mol Plant. 2008, 1, 703–714. [Google Scholar] [CrossRef]
- Zafra, A.; Rodríguez-García, M.I.; Alché Jde, D. Cellular localization of ROS and NO in olive reproductive tissues during flower development. BMC Plant Biol. 2010, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Freschi, L.; Rodríguez-Ruiz, M.; Mioto, P.T.; González-Gordo, S.; Palma, J.M. Nitro-oxidative metabolism during fruit ripening. J. Exp Bot. 2018, 69, 3449–3463. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Palma, J.M. Nitric oxide on/off in fruit ripening. Plant Biol (Stuttg). 2018, 20, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Li, M.; Kong, D.; Wang, L.; Lv, Q.; Wang, J.; Bao, F.; Gong, Q.; Xia, J.; He, Y. Nitric oxide induces cotyledon senescence involving co-operation of the NES1/MAD1 and EIN2-associated ORE1 signalling pathways in Arabidopsis. J. Exp. Bot. 2014, 65, 4051–4063. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Corpas, F.J.; Borsani, O.; Barroso, J.B.; Monza, J. Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus. Plant Sci. 2013, 201–202, 137–146. [Google Scholar] [CrossRef]
- Manai, J.; Gouia, H.; Corpas, F.J. Redox and nitric oxide homeostasis are affected in tomato (Solanum lycopersicum) roots under salinity-induced oxidative stress. J. Plant Physiol. 2014, 171, 1028–1035. [Google Scholar] [CrossRef]
- Feigl, G.; Lehotai, N.; Molnár, Á.; Ördög, A.; Rodríguez-Ruiz, M.; Palma, J.M.; Corpas, F.J.; Erdei, L.; Kolbert, Z. Zinc induces distinct changes in the metabolism of reactive oxygen and nitrogen species (ROS and RNS) in the roots of two Brassica species with different sensitivity to zinc stress. Ann Bot. 2015, 116, 613–625. [Google Scholar] [CrossRef]
- Houmani, H.; Rodríguez-Ruiz, M.; Palma, J.M.; Corpas, F.J. Mechanical wounding promotes local and long distance response in the halophyte Cakile maritima through the involvement of the ROS and RNS metabolism. Nitric Oxide. 2018, 74, 93–101. [Google Scholar] [CrossRef]
- Kharbech, O.; Houmani, H.; Chaoui, A.; Corpas, F.J. Alleviation of Cr(VI)-induced oxidative stress in maize (Zea mays L.) seedlings by NO and H2S donors through differential organ-dependent regulation of ROS and NADPH-recycling metabolisms. J. Plant Physiol. 2017, 219, 71–80. [Google Scholar] [CrossRef]
- Trapet, P.; Kulik, A.; Lamotte, O.; Jeandroz, S.; Bourque, S.; Nicolas-Francès, V.; Rosnoblet, C.; Besson-Bard, A.; Wendehenne, D. NO signaling in plant immunity: A tale of messengers. Phytochemistry 2015, 112, 72–79. [Google Scholar] [CrossRef]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: where do we stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Nitric oxide synthase-like activity in higher plants. Nitric Oxide 2017, 68, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Sandalio, L.M.; Valderrama, R.; Palma, J.M.; Lupiáñez, J.A.; del Río, L.A. Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 1999, 274, 36729–36733. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Calmodulin antagonist affects peroxisomal functionality by disrupting both peroxisomal Ca2+ and protein import. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Peroxisomal plant nitric oxide synthase (NOS) protein is imported by peroxisomal targeting signal type 2 (PTS2) in a process that depends on the cytosolic receptor PEX7 and calmodulin. FEBS Lett. 2014, 588, 2049–2054. [Google Scholar] [CrossRef]
- Gupta, K.J.; Igamberdiev, A.U. The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 2011, 11, 537–543. [Google Scholar] [CrossRef]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis endomembrane system. Mol. Plant. 2013, 6, 1419–1437. [Google Scholar] [CrossRef]
- Stolz, D.B.; Zamora, R.; Vodovotz, Y.; Loughran, P.A.; Billiar, T.R.; Kim, Y.M.; Simmons, R.L.; Watkins, S.C. Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology 2002, 36, 81–93. [Google Scholar] [CrossRef]
- Loughran, P.A.; Stolz, D.B.; Vodovotz, Y.; Watkins, S.C.; Simmons, R.L.; Billiar, T.R. Monomeric inducible nitric oxide synthase localizes to peroxisomes in hepatocytes. Proc. Natl. Acad. Sci. USA 2005, 102, 13837–13842. [Google Scholar] [CrossRef]
- Loughran, P.A.; Stolz, D.B.; Barrick, S.R.; Wheeler, D.S.; Friedman, P.A.; Rachubinski, R.A.; Watkins, S.C.; Billiar, T.R. PEX7 and EBP50 target iNOS to the peroxisome in hepatocytes. Nitric Oxide 2013, 31, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Fares, A.; Rossignol, M.; Peltier, J.B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 416, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.; van Donselaar, E.; Slot, J.W.; Fries, D.M.; Blachard-Fillion, B.; Hodara, R.; Lightfoot, R.; Polydoro, M.; Spielberg, D.; Thomson, L.; et al. Subcellular localization of tyrosine-nitrated proteins is dictated by reactive oxygen species generating enzymes and by proximity to nitric oxide synthase. Free Radic. Biol. Med. 2006, 40, 1903–1913. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Leterrier, M.; Begara-Morales, J.C.; Valderrama, R.; Chaki, M.; López-Jaramillo, J.; Luque, F.; Palma, J.M.; Padilla, M.N.; Sánchez-Calvo, B.; et al. Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim. Biophys. Acta. 2013, 1830, 4981–4989. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Galisteo, A.P.; Rodríguez-Serrano, M.; Pazmiño, D.M.; Gupta, D.K.; Sandalio, L.M.; Romero-Puertas, M.C. S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J. Exp. Bot. 2012, 63, 2089–2103. [Google Scholar] [CrossRef]
- Puyaubert, J.; Fares, A.; Rézé, N.; Peltier, J.B.; Baudouin, E. Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci. 2014, 215–216, 150–156. [Google Scholar] [CrossRef]
- Abat, J.K.; Mattoo, A.K.; Deswal, R. S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 2008, 275, 2862–2872. [Google Scholar] [CrossRef]
- Tanou, G.; Job, C.; Rajjou, L.; Arc, E.; Belghazi, M.; Diamantidis, G.; Molassiotis, A.; Job, D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 2009, 60, 795–804. [Google Scholar] [CrossRef]
- Lozano-Juste, J.; Colom-Moreno, R.; León, J. In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3501–3517. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; López-Jaramillo, F.J.; Sánchez-Calvo, B.; Carreras, A.; Ortega-Muñoz, M.; Santoyo-González, F.; Corpas, F.J.; Barroso, J.B. Vinyl sulfone silica: Application of an open preactivated support to the study of transnitrosylation of plant proteins by S-nitrosoglutathione. BMC Plant Biol. 2013, 13, 61. [Google Scholar] [CrossRef]
- Chaki, M.; Álvarez de Morales, P.; Ruiz, C.; Begara-Morales, J.C.; Barroso, J.B.; Corpas, F.J.; Palma, J.M. Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Ann Bot. 2015, 116, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Holzmeister, C.; Gaupels, F.; Geerlof, A.; Sarioglu, H.; Sattler, M.; Durner, J.; Lindermayr, C. Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J. Exp. Bot. 2015, 66, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Mata-Pérez, C.; Valderrama, R.; Padilla, M.N.; López-Jaramillo, J.; Luque, F.; Corpas, F.J.; Barroso, J.B. Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation. J. Exp. Bot. 2015, 66, 5983–5996. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P. Classical catalase: ancient and modern. Arch. Biochem. Biophys. 2012, 525, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, C.; Calderon, P.B. Catalase, a remarkable enzyme: Targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 2017, 398, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C. Reversible binding and inhibition of catalase by nitric oxide. Eur. J. Biochem. 1995, 232, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Purwar, N.; McGarry, J.M.; Kostera, J.; Pacheco, A.A.; Schmidt, M. Interaction of nitric oxide with catalase: Structural and kinetic analysis. Biochemistry 2011, 50, 4491–4503. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.; Durner, J.; Navarre, D.A.; Klessig, D.F. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant Microbe Interact. 2000, 13, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Hernández, J.A.; del Río, L.A.; Sevilla, F. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 1997, 114, 275–284. [Google Scholar] [CrossRef]
- Kuźniak, E.; Skłodowska, M. Compartment-specific role of the ascorbate-glutathione cycle in the response of tomato leaf cells to Botrytis cinerea infection. J. Exp. Bot. 2005, 56, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.M.; Jiménez, A.; Sandalio, L.M.; Corpas, F.J.; Lundqvist, M.; Gómez, M.; Sevilla, F.; del Río, L.A. Antioxidative enzymes from chloroplasts, mitochondria, and peroxisomes during leaf senescence of nodulated pea plants. J. Exp. Bot. 2006, 57, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Río, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005, 43, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995, 64, 97–112. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A.; Corpas, F.J.; López-Huertas, E.; Palma, J.M. Plant superoxide dismutases: Function under abiotic stress conditions. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D., Palma, J., Corpas, F., Eds.; Springer: Cham, Switzerland, 2018; pp. 1–26. [Google Scholar]
- del Río, L.A.; Lyon, D.S.; Olah, I.; Glick, B.; Salin, M.L. Immunocytochemical evidence for a peroxisomal localization of manganese superoxide dismutase in leaf protoplasts from a higher plant. Planta 1983, 158, 216–224. [Google Scholar] [CrossRef]
- Keller, G.A.; Warner, T.G.; Steimer, K.S.; Hallewell, R.A. Cu,Zn superoxide dismutase is a peroxisomal enzyme in human fibroblasts and hepatoma cells. Proc. Natl. Acad. Sci. USA 1991, 88, 7381–8735. [Google Scholar] [CrossRef]
- Alvarez, B.; Demicheli, V.; Durán, R.; Trujillo, M.; Cerveñansky, C.; Freeman, B.A.; Radi, R. Inactivation of human Cu,Zn superoxide dismutase by peroxynitrite and formation of histidinyl radical. Free Radic. Biol. Med. 2004, 37, 813–822. [Google Scholar] [CrossRef]
- Demicheli, V.; Quijano, C.; Alvarez, B.; Radi, R. Inactivation and nitration of human superoxide dismutase (SOD) by fluxes of nitric oxide and superoxide. Free Radic. Biol. Med. 2007, 42, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; del Río, L.A. Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 2001, 6, 145–150. [Google Scholar] [CrossRef]
- del Río, L.A.; Sandalio, L.M.; Corpas, F.J.; Palma, J.M.; Barroso, J.B. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scanvenging, and role in cell signaling. Plant Physiol. 2006, 141, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Palma, J.M.; Sandalio, L.M.; Valderrama, R.; Barroso, J.B.; del Río, L.A. Peroxisomal xanthine oxidoreductase: Characterization of the enzyme from pea (Pisum sativum L.) leaves. J. Plant Physiol. 2008, 165, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Sueta, G.; Campolo, N.; Trujillo, M.; Bartesaghi, S.; Carballal, S.; Romero, N.; Alvarez, B.; Radi, R. Biochemistry of peroxynitrite and protein tyrosine nitration. Chem. Rev. 2018, 118, 1338–1408. [Google Scholar] [CrossRef] [PubMed]
- Broniowska, K.A.; Diers, A.R.; Hogg, N. S-nitrosoglutathione. Biochim. Biophys. Acta. 2013, 1830, 3173–3181. [Google Scholar] [CrossRef] [PubMed]
- Alamillo, J.M.; García-Olmedo, F. Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 2001, 25, 529–540. [Google Scholar] [CrossRef]
- Signorelli, S.; Imparatta, C.; Rodríguez-Ruiz, M.; Borsani, O.; Corpas, F.J.; Monza, J. In vivo and in vitro approaches demonstrate proline is not directly involved in the protection against superoxide, nitric oxide, nitrogen dioxide and peroxynitrite. Funct. Plant Biol. 2016, 43, 870–879. [Google Scholar] [CrossRef]
- del Río, L.A.; Palma, J.M.; Sandalio, L.M.; Corpas, F.J.; Pastori, G.M.; Bueno, P.; López-Huertas, E. Peroxisomes as a source of superoxide and hydrogen peroxide in stressed plants. Biochem. Soc. Trans. 1996, 24, 434–438. [Google Scholar] [CrossRef]
| Requirements | Peroxisomal NOS-Like Protein |
|---|---|
| Substrate | L-Arginine |
| Cofactor requirement | NADPH, Ca2+, FAD, FMN, BH4 |
| Sensitivity to inhibitor | Aminiguanidine, L-NNA, L-NAME, L-NMMA |
| Peroxisomal targeting signal (PTS) | Type 2 (PTS2) |
| Dependence of peroxisomal protein import system | PEX5, PEX7, PEX12, PEX13, Ca2+, CaM |
| Localization | Matrix |
| Peroxisomal Enzyme | NO-Derived PTM | References |
|---|---|---|
| 3-ketoacyl-CoA thiolase 1 | S-nitrosation | [52] |
| Hydroxypyruvate reductase | S-nitrosation/nitration | [54,55,56] |
| Glycolate oxidase | S-nitrosation/nitration | [55,57,58] |
| Malate dehydrogenase | S-nitrosation/nitration | [55,59] |
| Catalase | S-nitrosation/nitration | [55,56,60,61] |
| CuZn superoxide dismutase (CSD3) | Nitration | [62] |
| Monodehydroascorbate reductase | S-nitrosation/nitration | [63] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corpas, F.J.; del Río, L.A.; Palma, J.M. Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes. Plants 2019, 8, 37. https://doi.org/10.3390/plants8020037
Corpas FJ, del Río LA, Palma JM. Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes. Plants. 2019; 8(2):37. https://doi.org/10.3390/plants8020037
Chicago/Turabian StyleCorpas, Francisco J., Luis A. del Río, and José M. Palma. 2019. "Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes" Plants 8, no. 2: 37. https://doi.org/10.3390/plants8020037
APA StyleCorpas, F. J., del Río, L. A., & Palma, J. M. (2019). Impact of Nitric Oxide (NO) on the ROS Metabolism of Peroxisomes. Plants, 8(2), 37. https://doi.org/10.3390/plants8020037







