Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds
Abstract
1. Introduction
2. Nitric Oxide Generation in Plant Cells
Query 1 RCVGRIQW 8
RC G IQW
Sbjct 224 RCTGKIQW 231
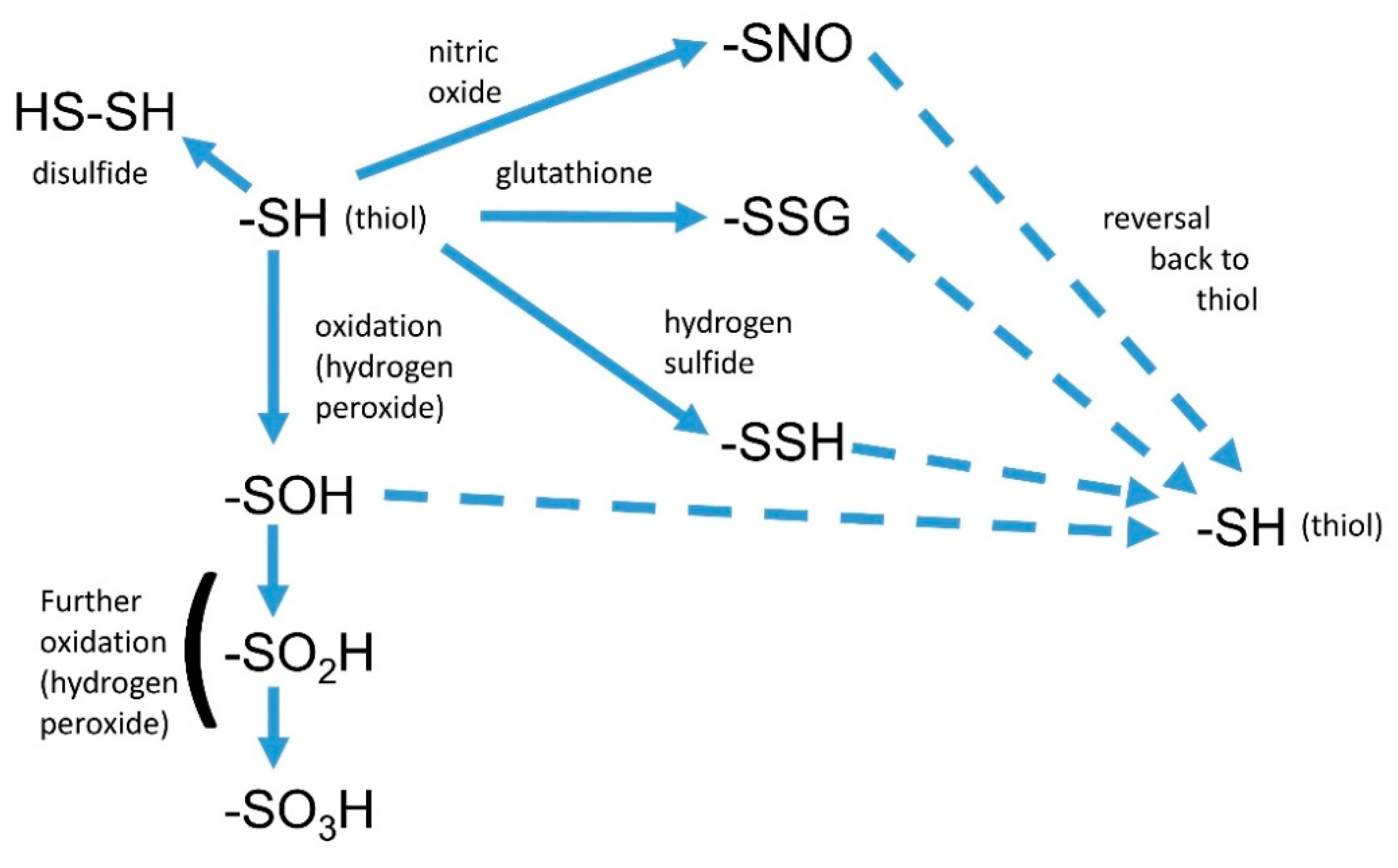
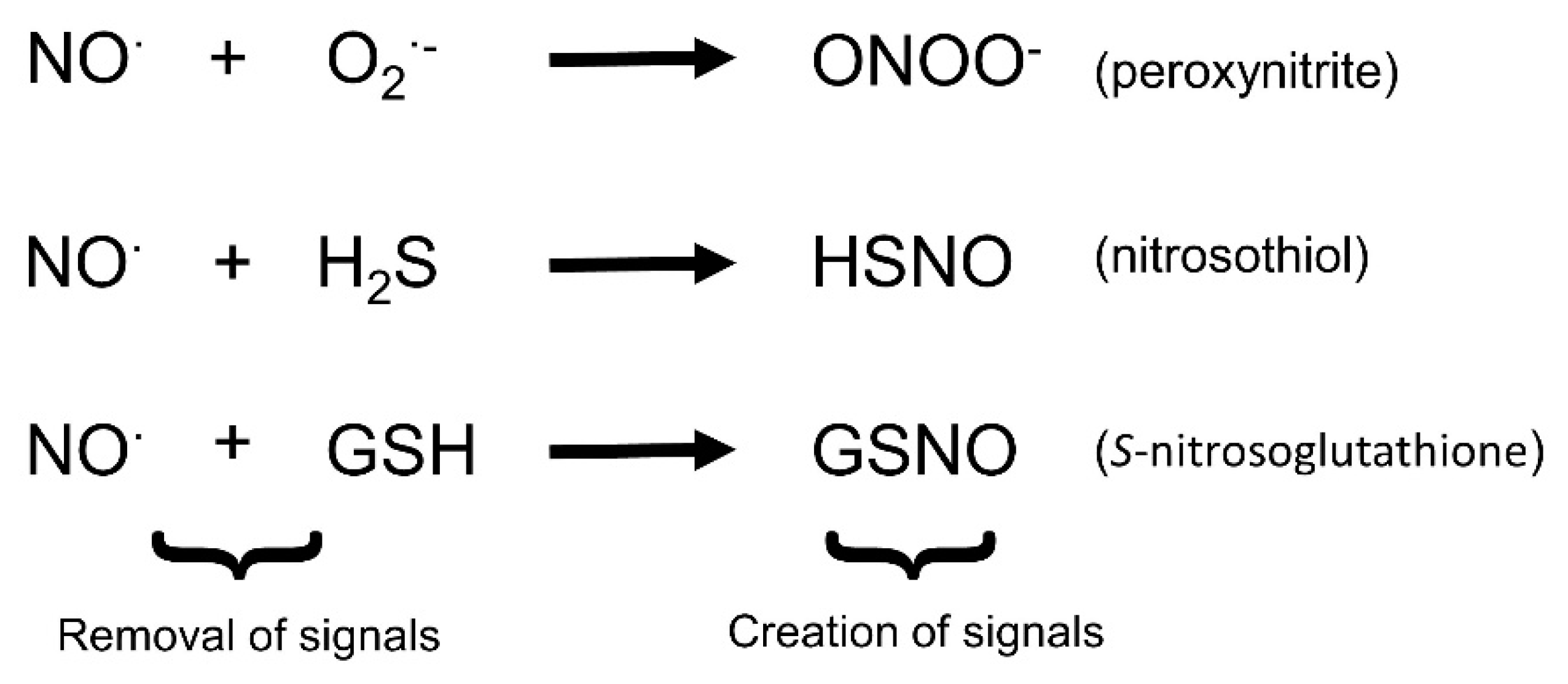
3. Interactions of Nitric Oxide with Other Reactive Signals
4. Conclusions and the Future
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palmer, R.M.J.; Ferrige, A.G.; Moncada, S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987, 327, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Laxalt, A.M.; Beligni, M.V.; Lamattina, L. Nitric oxide preserves the level of chlorophyll in potato leaves infected by Phytophthora infestans. Eur. J. Plant Pathol. 1997, 103, 643–651. [Google Scholar] [CrossRef]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant diseas resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef]
- Hancock, J.T. Harnessing evolutionary toxins for signaling: Reactive oxygen species, nitric oxide and hydrogen sulfide in plant cell regulation. Front. Plant Sci. 2017, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Arc, E.; Galland, M.; Godin, B.; Cueff, G.; Rajjou, L. Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 2013, 4, 346. [Google Scholar] [CrossRef]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef]
- Gayatri, G.; Agurla, S.; Raghavendra, A.S. Nitric oxide in guard cells as an important secondary messenger during stomatal closure. Front. Plant Sci. 2013, 4, 425. [Google Scholar] [CrossRef]
- Mur, L.A.; Carver, T.L.; Prats, E. NO way to live; the various roles of nitric oxide in plant-pathogen interactions. J. Exp. Bot. 2006, 57, 489–505. [Google Scholar] [CrossRef]
- Hiscock, S.J.; Bright, J.; McInnis, S.M.; Desikan, R.; Hancock, J.T. Signaling on the stigma: Potential new roles for ROS and NO in plant cell signaling. Plant Signal. Behav. 2007, 2, 23–24. [Google Scholar] [CrossRef]
- Kwon, E.; Feechan, A.; Yun, B.W.; Hwang, B.H.; Pallas, J.A.; Kang, J.G.; Loake, G.J. AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 2012, 236, 887–900. [Google Scholar] [CrossRef]
- Hu, J.; Yang, H.; Mu, J.; Lu, T.; Peng, J.; Deng, X.; Kong, Z.; Bao, S.; Cao, X.; Zuo, J. Nitric oxide regulates protein methylation during stress responses in plants. Mol. Cell 2017, 67, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Watanabe, N.S.; Sakihama, Y.; Cohen, M.F. An overview of methods in plant nitric oxide (NO) research: Why do we always need to use multiple methods? Meth. Mol. Biol. 2016, 1424, 1–14. [Google Scholar] [CrossRef]
- McCormick, K.; Baillie, G.S. Compartmentalisation of second messenger signalling pathways. Curr. Opin. Genet. Dev. 2014, 27, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Zaccolo, M.; Magalhães, P.; Pozzan, T. Compartmentalisation of cAMP and Ca2+ signals. Curr. Opin. Cell Biol. 2002, 14, 160–166. [Google Scholar] [CrossRef]
- Baillie, G.S.; Scott, J.D.; Houslay, M.D. Compartmentalisation of phosphodiesterases and protein kinase A: Opposites attract. FEBS Lett. 2005, 579, 3264–3270. [Google Scholar] [CrossRef] [PubMed]
- Bononi, A.; Missiroli, S.; Poletti, F.; Suski, J.M.; Agnoletto, C.; Bonora, M.; De Marchi, E.; Giorgi, C.; Marchi, S.; Patergnani, S.; et al. Mitochondria-associated membranes (MAMs) as hotspot Ca2+ signaling units. Adv. Exp. Med. Biol. 2012, 740, 411–437. [Google Scholar] [CrossRef]
- De Rezende, F.F.; Martins Lima, A.; Niland, S.; Wittig, I.; Heide, H.; Schröder, K.; Eble, J.A. Integrin α7β1 is a redox-regulated target of hydrogen peroxide in vascular smooth muscle cell adhesion. Free Radic. Biol. Med. 2012, 53, 521–531. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016, 171, 1581–1592. [Google Scholar] [CrossRef]
- Shapiro, A.D. Nitric oxide signaling in plants. Vitam. Horm. 2005, 72, 339–398. [Google Scholar]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Grifitths, R.; Hancock, J.; Neill, S. A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 16314–16318. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Zhao, S.; Dong, H.; Zhang, H.; Sun, L.; Miao, C. Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Cai, S.; Wang, Y.; Chen, Z.H. Loss of nitrate reductases NIA1 and NIA2 impairs stomatal closure by altering genes of core ABA signaling components in Arabidopsis. Plant Signal. Behav. 2016, 11, e1183088. [Google Scholar] [CrossRef] [PubMed]
- Millar, T.M.; Stevens, C.R.; Benjamin, N.; Eisenthal, R.; Harrison, R.; Blake, D.R. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett. 1998, 427, 225–228. [Google Scholar] [CrossRef]
- Zemojtel, T.; Fröhlich, A.; Palmieri, M.C.; Kolanczyk, M.; Mikula, I.; Wyrwicz, L.S.; Wanker, E.E.; Mundlos, S.; Vingron, M.; Martasek, P.; et al. Plant nitric oxide synthase: A never-ending story? Trends Plant. Sci. 2006, 11, 524–525. [Google Scholar] [CrossRef]
- Astier, J.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: The surprise from algae. Plant Sci. 2018, 268, 64–66. [Google Scholar] [CrossRef]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef]
- Hancock, J.T.; Neill, S.J. NO Synthase in plants? CAB Rev. 2014, 9, 1–9. [Google Scholar] [CrossRef]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric oxide synthase in plants: Where do we stand? Nitric Oxide 2017, 63, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.-S.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal. 2016, 9, re2. [Google Scholar] [CrossRef] [PubMed]
- Tamás, L.; Demecsová, L.; Zelinová, V. L-NAME decreases the amount of nitric oxide and enhances the toxicity of cadmium via superoxide generation in barley root tip. J. Plant Physiol. 2018, 224–225, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Butt, Y.K.-C.; Lum, J.H.-K.; Lo, S.C.-L. Proteomic identification of plant proteins probed by mammalian nitric oxide synthase antibodies. Planta 2003, 216, 762–771. [Google Scholar] [PubMed]
- Stuehr, D.; Vasquez-Vivar, J. Nitric oxide synthases- from genes to function. Nitric Oxide 2017, 63, 29. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Underbakke, E.S.; Kulp, D.W.; Schief, W.R.; Marletta, M.A. Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc. Natl. Acad. Sci. USA 2013, 110, E3577–E3586. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Misra, I.; Iyanagi, T.; Kim, J.-J. Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J. Biol. Chem. 2009, 284, 30708–30717. [Google Scholar] [CrossRef]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; López, R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef]
- Iyanagi, T. Structure and function of NADPH-cytochrome P450 reductase and nitric oxide synthase reductase domain. Biochem. Biophys. Res. Commun. 2005, 338, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Gusarov, I.; Starodubtseva, M.; Wang, Z.-Q.; McQuade, L.; Lippard, S.J.; Stuehr, D.J.; Nudler, E. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 2008, 283, 13140–13147. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Sequencing the rice genome. Plant Cell 2000, 12, 2011–2018. [Google Scholar] [CrossRef]
- Icking, A.; Matt, S.; Opitz, N.; Wiesenthal, A.; Müller-Esterl, W.; Schilling, K. NOSTRIN functions as a homotrimeric adaptor protein facilitating internalization of eNOS. J. Cell Sci. 2005, 118, 5059–5069. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Snowman, A.M.; Eliasson, M.J.; Cohen, N.A.; Snyder, S.H. CAPON: A protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron 1998, 20, 115–124. [Google Scholar] [CrossRef]
- Dedio, J.; Konig, P.; Wohlfart, P.; Schroeder, C.; Kummer, W.; Muller-Esterl, W. NOSIP, a novel modulator of endothelial nitric oxide synthase activity. FASEB J. 2001, 15, 79–89. [Google Scholar] [CrossRef]
- Lancaster, J., Jr. Nitric oxide: A brief overview of chemical and physical properties relevant to therapeutic applications. Future Sci. OA 2015, 1, FSO59. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Pazmiño, D.M.; Testillano, P.S.; Risueño, M.C.; Del Río, L.A.; Sandalio, L.M. Cellular response of pea plants to cadmium toxicity: Cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiol. 2009, 150, 229–243. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Rahman, A.; Ansary, M.M.; Watanabe, A.; Fujita, M.; Tran, L.S. Hydrogen sulfide modulates cadmium-induced physiological and biochemical responses to alleviate cadmium toxicity in rice. Sci. Rep. 2015, 5, 14078. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic identification of S-nitrosylated proteins. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Craig, T.; Whiteman, M. Competition of reactive signals and thiol modifications of proteins. J. Cell Signal. 2017, 2, 170. [Google Scholar] [CrossRef]
- Sirover, M.A. Subcellular dynamics of multifunctional protein regulation: Mechanisms of GAPDH intracellular translocation. J. Cell Biochem. 2012, 113, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Holtgrefe, S.; Gohlke, J.; Starmann, J.; Druce, S.; Klocke, S.; Altmann, B.; Wojtera, J.; Lindermayr, C.; Scheibe, R. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant 2008, 133, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Henson, D.; Nyirenda, M.; Desikan, R.; Harrison, J.; Lewis, M.; Hughes, J.; Neill, S.J. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Aroca, A.; Schneider, M.; Scheibe, R.; Gotor, C.; Romero, L.C. Hydrogen Sulfide Regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 2017, 58, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.; Pead, S.; Whiteman, M.; Wood, M.E.; Wilson, I.D.; Ladomery, M.R.; Teklic, T.; Lisjak, M.; Hancock, J.T. Detection of thiol modifications by hydrogen sulfide. Methods Enzymol. 2015, 555, 233–251. [Google Scholar] [CrossRef]
- Williams, E.; Whiteman, M.; Wood, M.E.; Wilson, I.D.; Ladomery, M.R.; Allainguillaume, J.; Teklic, T.; Lisjak, M.; Hancock, J.T. Investigating ROS, RNS and H2S sensitive signalling proteins. In Redox Signal Transduction: Methods and Protocols; Hancock, J.T., Conway, M., Eds.; Springer: Berlin, Germany, 2019; in press. [Google Scholar]
- Yu, M.; Yun, B.W.; Spoel, S.H.; Loake, G.J. A sleigh ride through the SNO: Regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 2012, 15, 424–430. [Google Scholar] [CrossRef]
- Kolbert, Z.; Feigl, G.; Bordé, Á.; Molnár, Á.; Erdei, L. Protein tyrosine nitration in plants: Present knowledge, computational prediction and future perspectives. Plant Physiol. Bioch. 2017, 113, 56–63. [Google Scholar] [CrossRef]
- Holzmeister, C.; Gaupels, F.; Geerlof, A.; Sarioglu, H.; Sattler, M.; Durner, J.; Lindermayr, C. Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J. Exp. Bot. 2015, 66, 989–999. [Google Scholar] [CrossRef]
- Speckmann, B.; Steinbrenner, H.; Grune, T.; Klotz, L.O. Peroxynitrite: From interception to signaling. Arch Biochem. Biophys. 2016, 595, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Bauer, G. Increasing the endogenous NO level causes catalase inactivation and reactivation of intercellular apoptosis signaling specifically in tumor cells. Redox Biol. 2015, 6, 353–371. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Whiteman, M. Cellular redox environment and its influence on redox signalling molecules. React. Oxyg. Species 2018, 5, 78–85. [Google Scholar]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef]
- Hancock, J.T. Considerations of the importance of redox state on reactive nitrogen species action. J. Exp. Bot. 2019, in press. [Google Scholar]
- Yun, B.W.; Skelly, M.J.; Yin, M.; Yu, M.; Mun, B.G.; Lee, S.U.; Hussain, A.; Spoel, S.H.; Loake, G.J. Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 2016, 211, 516–526. [Google Scholar] [CrossRef]
- Hogg, N.; Singh, R.J.; Kalyanaraman, B. The role of glutathione in the transport and catabolism of nitric oxide. FEBS Lett. 1996, 382, 223–228. [Google Scholar] [CrossRef]
- Feechan, A.; Kwon, E.; Yun, B.W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef]
- Lee, U.; Wie, C.; Fernandez, B.O.; Feelisch, M.; Vierling, E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 2008, 20, 786–802. [Google Scholar] [CrossRef]
- Lisjak, M.; Teklic, T.; Wilson, I.D.; Whiteman, M.; Hancock, J.T. Hydrogen sulfide: Environmental factor or signaling molecule? Plant Cell Environ. 2013, 36, 1607–1616. [Google Scholar] [CrossRef]
- Olas, B. Hydrogen sulfide and signaling pathways. Clin. Chim. Acta 2015, 439, 212–218. [Google Scholar] [CrossRef]
- Kimura, H. Hydrogen sulfide and polysulfide signaling. Antioxid. Redox Signal. 2017, 27, 619–621. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, M.; Li, L.; Kostetski, I.; Chu, S.H.; Siau, J.L.; Bhatia, M.; Moore, P.K. Evidence for the formation of a novel nitrosothiol from the gaseous mediators nitric oxide and hydrogen sulphide. Biochem. Biophys. Res. Commun. 2006, 343, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Goto, Y.; Kimura, H. Hydrogen sulfide increases glutathione production and suppresses oxidative stress in mitochondria. Antioxid. Redox Signal. 2010, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.R.; Veal, D.; Whiteman, M.; Hancock, J.T. Hydrogen gas and its role in cell signaling. CAB Rev. 2017, 12, 1–3. [Google Scholar] [CrossRef]
- Ohta, S. Molecular hydrogen as a novel antioxidant: Overview of the advantages of hydrogen for medical applications. Methods Enzymol. 2015, 555, 289–317. [Google Scholar] [PubMed]
- Buntkowsky, G.; Walaszek, B.; Adamczyk, A.; Xu, Y.; Limbach, H.-H.; Chaudret, B. Mechanisms of nuclear spin initiated para-H2 to ortho–H2 conversion. Phys. Chem. Chem. Phys. 2006, 8, 1929–1935. [Google Scholar] [CrossRef]
- Hancock, J.T.; Hancock, T.H. Hydrogen gas, ROS metabolism and cell signaling: Are hydrogen spin states important? React. Oxyg. Species 2018, 6, 389–395. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, W.; Qi, F.; Cui, W.; Xie, Y.; Shen, W. Hydrogen-rich water regulates cucumber adventitious root development in a heme oxygenase-1/carbon monoxide-dependent manner. J. Plant Physiol. 2014, 171, 1–8. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Cai, J.; Shen, Z.; Cui, J. Hydrogen-rich water enhances cadmium tolerance in Chinese cabbage by reducing cadmium uptake and increasing antioxidant capacities. J. Plant Physiol 2015, 175, 174–182. [Google Scholar] [CrossRef]
- Zeng, J.; Ye, Z.; Sun, X. Progress in the study of biological effects of hydrogen on higher plants and its promising application in agriculture. Med. Gas Res. 2014, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liao, W.; Wang, M.; Niu, L.; Xu, Q.; Jin, X. Nitric oxide is required for hydrogen gas-induced adventitious root formation in cucumber. J. Plant Physiol. 2016, 195, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liao, W.; Niu, L.; Wang, M.; Ma, Z. Nitric oxide is involved in hydrogen gas-induced cell cycle activation during adventitious root formation in cucumber. BMC Plant Biol. 2016, 16, 146. [Google Scholar] [CrossRef] [PubMed]
- Šírová, J.; Sedlářová, M.; Piterková, J.; Luhová, L.; Petřivalský, M. The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci. 2011, 181, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Correa-Aragunde, N.; Graziano, M.; Lamattina, L. Nitric oxide plays a central role in determining lateral root development in tomato. Planta 2004, 218, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.; Desikan, R.; Hurst, R.D.; Hancock, J.T.; Neill, S.J. NO way back: Nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 2000, 24, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Whiteman, M. Hydrogen sulfide and cell signaling: Team player or referee? Plant Physiol. Biochem. 2014, 78, 37–42. [Google Scholar] [CrossRef] [PubMed]

| Source of Sequence | Sequence Name | Sequence | Reference/ Source for Sequence | Significant Find/Comment |
|---|---|---|---|---|
| NCBI | NOS signature | [GR]-C-[IV]-G-R-[ILS]-x-W | prosite.expasy.org/ PS60001 | No significant sequence identified using ScanProsite/ previously found in a range of species including Staphylococcus, insects and mammals. |
| Rat nNOS | NOS signature | -RCVGRIQW- | [37] |
|
| Rat nNOS | FMN subdomain 538–547 | -PELVLEVPIR- | [37] | No significant sequence identified |
| Rat nNOS | FMN subdomain 582–605 | -CPFSGWYMGTEIGVRDYCDNSRYN- | [37] | No significant sequence identified |
| Rat nNOS | Haem domain 80–102 | -ALEVLRGIASETHVVLILRGPEG- | [37] | No significant sequence identified |
| Rat nNOS | Haem domain 187–203 | -TKANLQDIGEHDELLKE- | [37] | No significant sequence identified |
| Rat nNOS | Haem domain 366–386 | -YSSIKRFGSKAHMDRLEEVNK- | [37] | No significant sequence identified |
| Rat nNOS | Haem domain 396–465 | -LKDTELIYGAKHAWRNASRCVGRIQW SKLQVFDARDCTTAHGMFNYICNHVKY ATNKGNLRSAITIFPQR- | [37] | No significant sequence identified /NOS consensus sequence underlined, but not found in plants. |
| Rat nNOS | Haem domain 471–485 | -DFRVWNSQLIRYAGY- | [37] | No significant sequence identified |
| Rat nNOS | CaM domain 20–36 | -LFKRKVGGLGFLVKERV- | [37] | No significant sequence identified |
| Rat nNOS | CaM domain 105–124 | -THLETTFTGDGTPKTIRVTQ- | [37] | No significant sequence identified |
| Human iNOS | 509–537 | -KRREIPLKVLVKAVLFACMLMRKTMASRV- | [38] | Poor homology in some Oryza sequences/R536 important in human (underlined)/ -SRV- present |
| Rat nNOS | 725–753 | -KRRAIGFKKLAEAVKFSAKLMGQAMAKRV- | [38] | Poor homology in some Oryza sequences /R752 important (underlined) in rat |
| Mouse iNOS/FMN domain | 532–694 | -VRATV…PKRFT- | Derived from [37] |
|
| eNOS (human) | 566–585 | -LVLVVTSTFGNGPPENGES- | Derived from human Clustal Omega e/i/n NOS | No significant sequence identified |
| eNOS (human) | 952–980 | -EIHKTVAVLAYRTGDGLGPLHYGVCSTWL- | Derived from human Clustal Omega e/i/n NOS | Evidence of being part of a oxidoeductase or P450 reductase in plants: for example XP_015696451.1 & XP_006653834.1(both have 45% identical over 96% coverage) from Oryza: CAA46814.1 & NP_194183.1 (both 67% identical over 41% coverage) from Arabidopsisl. |
| Nitric Oxide Synthase Related Proteins/Peptides | ||||
| Nostrin isoform 2 [Homo sapiens]: NP_001034813 | Full sequence | MRDPLT…NTATKA | https://www.ncbi.nlm.nih.gov/protein/NP_001034813.2/ & [45] |
|
| Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein isoform 1 [Homo sapiens]. NP_055512 | Full sequence | MPSKT…DDEIAV | https://www.ncbi.nlm.nih.gov/protein/NP_055512 & [46] | No significant sequences identified/PH-like superfamily predicted. |
| Nitric oxide synthase-interacting protein isoform 1 [Homo sapiens]. NP_057037 | Full sequence | MTRHG…SRPVMGA | https://www.ncbi.nlm.nih.gov/protein/NP_057037 & [47] |
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hancock, J.T.; Neill, S.J. Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds. Plants 2019, 8, 41. https://doi.org/10.3390/plants8020041
Hancock JT, Neill SJ. Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds. Plants. 2019; 8(2):41. https://doi.org/10.3390/plants8020041
Chicago/Turabian StyleHancock, John T., and Steven J. Neill. 2019. "Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds" Plants 8, no. 2: 41. https://doi.org/10.3390/plants8020041
APA StyleHancock, J. T., & Neill, S. J. (2019). Nitric Oxide: Its Generation and Interactions with Other Reactive Signaling Compounds. Plants, 8(2), 41. https://doi.org/10.3390/plants8020041






