The Suitability of Orthogonal Hosts to Study Plant Cell Wall Biosynthesis
Abstract
1. Introduction
- What determines the length of a polysaccharide?
- What determines the substitution patterns of a polysaccharide?
- What are the roles of auxiliary proteins and/or cofactors in polysaccharide synthesis?
- How is the product influenced by the supply of activated precursors such as nucleotide sugars?
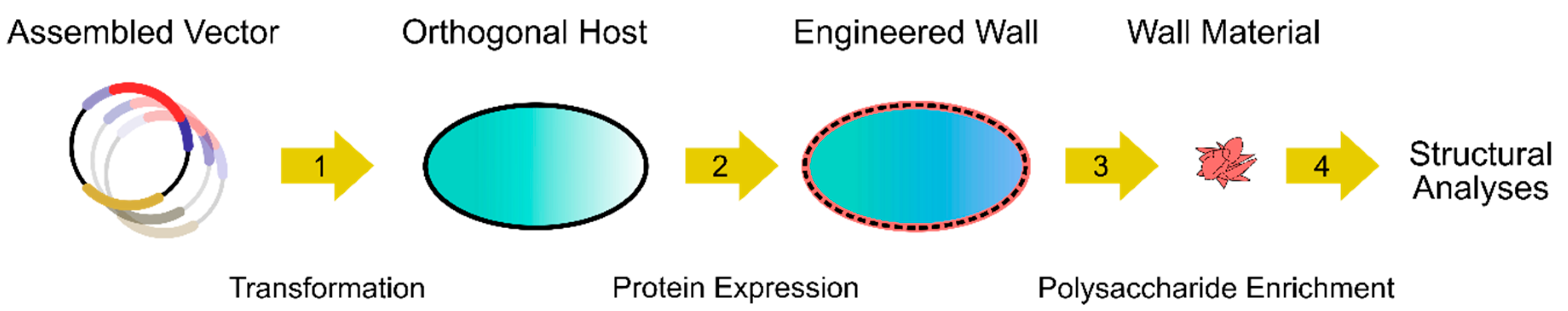
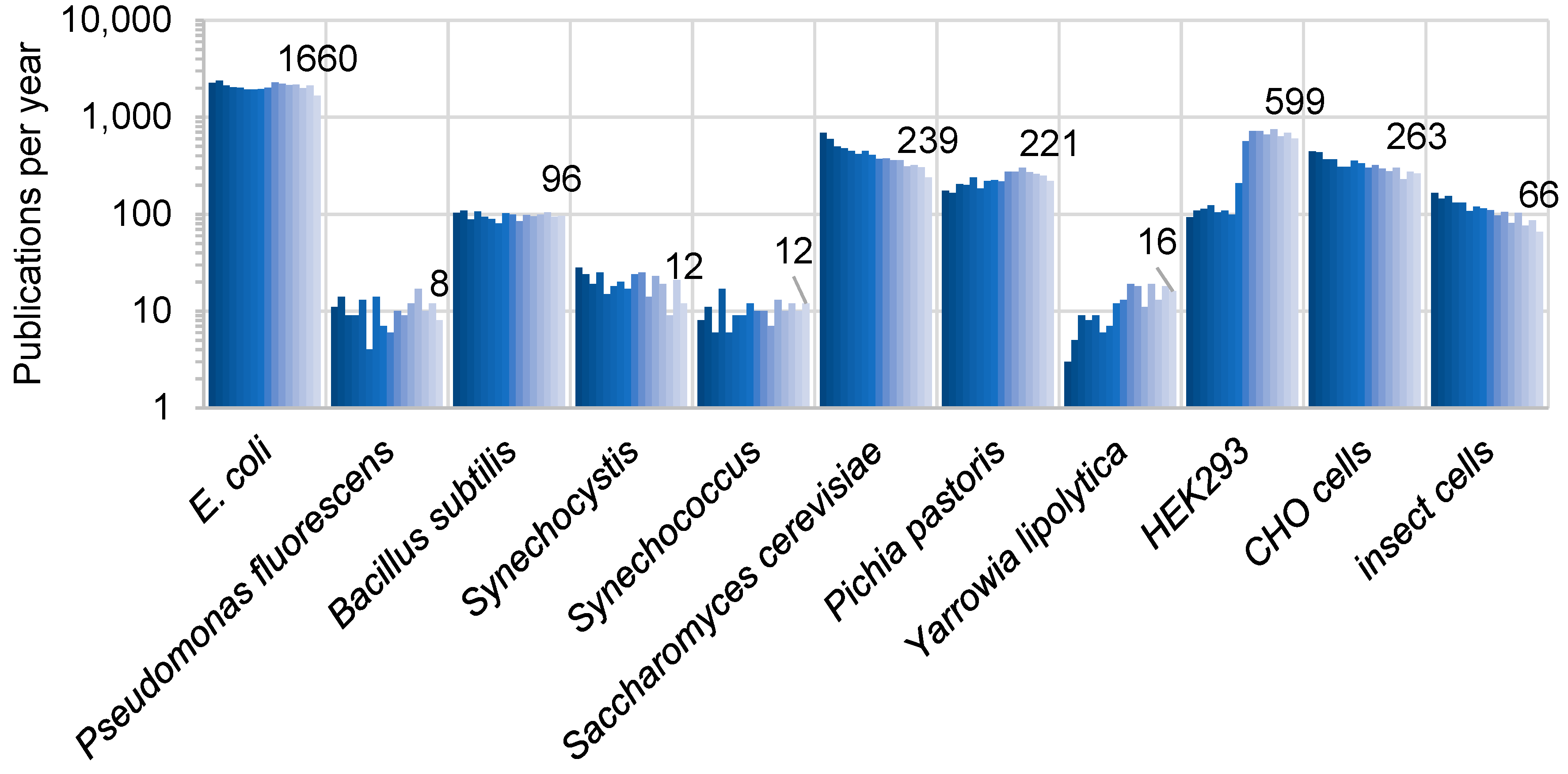
2. Criteria for the Choice of Orthologous Hosts
3. Bacteria
3.1. Gram-Negative Bacteria
3.2. Gram-Positive Bacteria
3.3. Cyanobacteria
4. Fungi
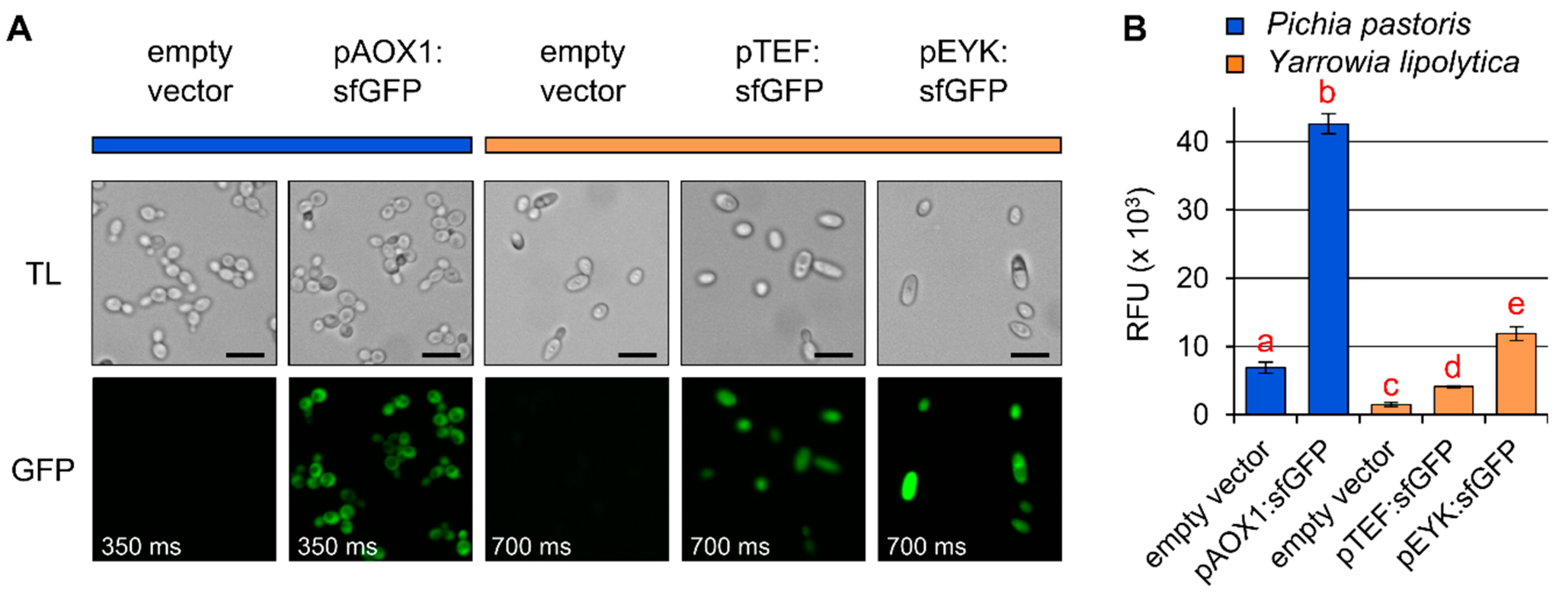
4.1. General Evaluation of Four Species
4.2. Direct Comparison of Two Hosts
5. Animal Cells
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Albersheim, P.; Darvill, A.; Roberts, K.; Sederoff, R.; Staehelin, A. Plant Cell Walls. From Chemistry to Biology; Garland Science: New York, NY, USA, 2011; ISBN 978-0-8153-1996-2. [Google Scholar]
- Amos, R.A.; Mohnen, D. Critical review of plant cell wall matrix polysaccharide glycosyltransferase activities verified by heterologous protein expression. Front. Plant Sci. 2019, 10, 915. [Google Scholar] [CrossRef] [PubMed]
- Schultink, A.; Liu, L.; Zhu, L.; Pauly, M. Structural diversity and function of xyloglucan sidechain substituents. Plants 2014, 3, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G. Hemicellulose biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Döring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef]
- Pauly, M.; Keegstra, K. Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 2016, 67, 235–259. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Yang, B.; Schmidt, M.H.-W.; Günl, M.; Usadel, B. Starting to gel: How Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int. J. Mol. Sci. 2015, 16, 3452–3473. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Engle, K.A.; Günl, M.; Dieluweit, S.; Schmidt, M.H.-W.; Yang, J.; Moremen, K.W.; Mohnen, D.; Usadel, B. Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol. 2018, 178, 1045–1064. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Günl, M.; Schmidt, M.H.-W.; Usadel, B. Highly branched xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol. 2015, 169, 2481–2495. [Google Scholar]
- Voiniciuc, C.; Schmidt, M.H.-W.; Berger, A.; Yang, B.; Ebert, B.; Scheller, H.V.; North, H.M.; Usadel, B.; Günl, M. MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol. 2015, 169, 403–420. [Google Scholar] [CrossRef]
- Beyer, H.M.; Engesser, R.; Hörner, M.; Koschmieder, J.; Beyer, P.; Timmer, J.; Zurbriggen, M.D.; Weber, W. Synthetic biology makes polymer materials count. Adv. Mater. 2018, 30, e1800472. [Google Scholar] [CrossRef] [PubMed]
- Doblin, M.S.; Johnson, K.L.; Humphries, J.; Newbigin, E.J.; Bacic, A. Are designer plant cell walls a realistic aspiration or will the plasticity of the plant’s metabolism win out? Curr. Opin. Biotechnol. 2013, 26, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Gidley, M.J.; Bacic, A.; Doblin, M.S. Cell wall biomechanics: A tractable challenge in manipulating plant cell walls ‘fit for purpose’! Curr. Opin. Biotechnol. 2018, 49, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Voiniciuc, C.; Pauly, M.; Usadel, B. Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 2018, 176, 2590–2600. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Monty, J.; Linhardt, R.J. Polysaccharide-based nanocomposites and their applications. Carbohydr. Res. 2015, 405, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Voiniciuc, C.; Dama, M.; Gawenda, N.; Stritt, F.; Pauly, M. Mechanistic insights from plant heteromannan synthesis in yeast. Proc. Natl. Acad. Sci. USA 2019, 116, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Popper, Z.A.; Michel, G.; Hervé, C.; Domozych, D.S.; Willats, W.G.T.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef]
- Nester, E.W. Agrobacterium: Nature’s genetic engineer. Front. Plant Sci. 2015, 5, 1–16. [Google Scholar] [CrossRef]
- Liepman, A.H.; Wilkerson, C.G.; Keegstra, K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 2005, 102, 2221–2226. [Google Scholar] [CrossRef]
- Giaever, G.; Nislow, C. The yeast deletion collection: A decade of functional genomics. Genetics 2014, 197, 451–465. [Google Scholar] [CrossRef]
- Pauly, M.; Andersen, L.N.; Kauppinen, S.; Kofod, L.V.; York, W.S.; Albersheim, P.; Darvill, A. A xyloglucan-specific endo- -1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 1999, 9, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Feldbrügge, M.; Kellner, R.; Schipper, K. The biotechnological use and potential of plant pathogenic smut fungi. Appl. Microbiol. Biotechnol. 2013, 97, 3253–3265. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Beyer, H.M.; Gonschorek, P.; Samodelov, S.L.; Meier, M.; Weber, W.; Zurbriggen, M.D. AQUA cloning: A versatile and simple enzyme-free cloning approach. PLoS ONE 2015, 10, e0137652. [Google Scholar] [CrossRef] [PubMed]
- Prielhofer, R.; Barrero, J.J.; Steuer, S.; Gassler, T.; Zahrl, R.; Baumann, K.; Sauer, M.; Mattanovich, D.; Gasser, B.; Marx, H. GoldenPiCS: A golden gate-derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris. BMC Syst. Biol. 2017, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Geier, M.; Fauland, P.; Vogl, T.; Glieder, A. Compact multi-enzyme pathways in P. pastoris. Chem. Commun. 2015, 51, 1643–1646. [Google Scholar] [CrossRef]
- Larroude, M.; Rossignol, T.; Nicaud, J.M.; Ledesma-Amaro, R. Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 2018, 36, 2150–2164. [Google Scholar] [CrossRef]
- Jacobs, P.P.; Geysens, S.; Vervecken, W.; Contreras, R.; Callewaert, N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat. Protoc. 2009, 4, 58–70. [Google Scholar] [CrossRef]
- Rebnegger, C.; Vos, T.; Graf, A.B.; Valli, M.; Pronk, J.T.; Daran-Lapujade, P.; Mattanovich, D. Pichia pastoris exhibits high viability and a low maintenance energy requirement at near-zero specific growth rates. Appl. Environ. Microbiol. 2016, 82, 4570–4583. [Google Scholar] [CrossRef]
- Garcia-Muse, T. Characterization of B-type cyclins in the smut fungus Ustilago maydis: Roles in morphogenesis and pathogenicity. J. Cell Sci. 2003, 117, 487–506. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, F.C.; Papkovsky, D.B. Rapid high-throughput assessment of aerobic bacteria in complex samples by fluorescence-based oxygen respirometry. Appl. Environ. Microbiol. 2006, 72, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Burdett, I.D.; Kirkwood, T.B.; Whalley, J.B. Growth kinetics of individual Bacillus subtilis cells and correlation with nucleoid extension. J. Bacteriol. 1986, 167, 219–230. [Google Scholar] [CrossRef] [PubMed]
- van Alphen, P.; Abedini Najafabadi, H.; Branco dos Santos, F.; Hellingwerf, K.J. Increasing the photoautotrophic growth rate of Synechocystis sp. PCC 6803 by identifying the limitations of its cultivation. Biotechnol. J. 2018, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, J.; Lin, P.-C.; Chen, H.-Y.; Pakrasi, H.B. Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. mBio 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Pettolino, F.; Walsh, C.; Fincher, G.B.; Bacic, A. Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 2012, 7, 1590–1607. [Google Scholar] [CrossRef]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef]
- Vuttipongchaikij, S.; Brocklehurst, D.; Steele-King, C.; Ashford, D.A.; Gomez, L.D.; McQueen-Mason, S.J. Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol. 2012, 195, 585–595. [Google Scholar] [CrossRef]
- Welner, D.H.; Shin, D.; Tomaleri, G.P.; DeGiovanni, A.M.; Yi-Lin Tsai, A.; Tran, H.M.; Hansen, S.F.; Green, D.T.; Scheller, H.V.; Adams, P.D. Plant cell wall glycosyltransferases: High-throughput recombinant expression screening and general requirements for these challenging enzymes. PLoS ONE 2017, 12, e0177591. [Google Scholar] [CrossRef]
- Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef]
- Chakraborty, A.K.; Friebolin, H.; Stirm, S. Primary structure of the Escherichia coli serotype K30 capsular polysaccharide. J. Bacteriol. 1980, 141, 971–972. [Google Scholar] [PubMed]
- Rodriguez, M.-L.; Jann, B.; Jann, K. Comparative structural elucidation of the K18, K22, and K100 antigens of Escherichia coli as related ribosyl-ribitol phosphates. Carbohydr. Res. 1988, 173, 243–253. [Google Scholar] [CrossRef]
- Bales, P.M.; Renke, E.M.; May, S.L.; Shen, Y.; Nelson, D.C. Purification and characterization of biofilm-associated EPS exopolysaccharides from ESKAPE organisms and other pathogens. PLoS ONE 2013, 8, e67950. [Google Scholar] [CrossRef] [PubMed]
- Buldum, G.; Bismarck, A.; Mantalaris, A. Recombinant biosynthesis of bacterial cellulose in genetically modified Escherichia coli. Bioprocess Biosyst. Eng. 2018, 41, 265–279. [Google Scholar] [CrossRef]
- Retallack, D.M.; Jin, H.; Chew, L. Reliable protein production in a Pseudomonas fluorescens expression system. Protein Expr. Purif. 2012, 81, 157–165. [Google Scholar] [CrossRef]
- Chen, R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol. Adv. 2012, 30, 1102–1107. [Google Scholar] [CrossRef]
- Hazlewood, G.P.; Laurie, J.I.; Ferreira, L.M.A.; Gilbert, H.J. Pseudomonas fluorescens subsp. cellulosa: An alternative model for bacterial cellulase. J. Appl. Bacteriol. 1992, 72, 244–251. [Google Scholar]
- Braithwaite, K.L.; Black, G.W.; Hazlewood, G.P.; Ali, B.R.S.; Gilbert, H.J. A non-modular endo- β -1,4-mannanase from Pseudomonas fluorescens subspecies cellulosa. Biochem. J. 1995, 305, 1005–1010. [Google Scholar] [CrossRef]
- Tan, S.Z.; Reisch, C.R.; Prather, K.L.J. A robust CRISPR interference gene repression system in Pseudomonas. J. Bacteriol. 2018, 200, e00575-17. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef]
- Westers, L.; Westers, H.; Quax, W.J. Bacillus subtilis as cell factory for pharmaceutical proteins: A biotechnological approach to optimize the host organism. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1694, 299–310. [Google Scholar] [CrossRef] [PubMed]
- van Dijl, J.M.; Hecker, M. Bacillus subtilis: From soil bacterium to super-secreting cell factory. Microb. Cell Fact. 2013, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Ducros, V.M.A.; Flint, J.E.; Roberts, S.M.; Morland, C.; Zechel, D.L.; Smith, N.; Bjørnvad, M.E.; Borchert, T.V.; Wilson, K.S.; et al. Understanding how diverse β-mannanases recognize heterogeneous substrates. Biochemistry 2009, 48, 7009–7018. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, A.; Itoh, T.; Kawamata, A.; Hashimoto, W.; Murata, K. Plant cell wall degradation by saprophytic Bacillus subtilis strains: Gene clusters responsible for rhamnogalacturonan depolymerization. Appl. Environ. Microbiol. 2007, 73, 3803–3813. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, R.; Jenkins, J.; Harris, G.; Nasser, W.; Robert-Baudouy, J. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat. Struct. Biol. 1994, 1, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, L.; Albersheim, P. Structure of plant cell walls: IX. purification and partial characterization of a wall-degrading endo-arabanase and an arabinosidase from Bacillus subtilis. Plant Physiol. 1979, 63, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Labavitch, J.M.; Freeman, L.E.; Albersheim, P. Structure of plant cell walls. Purification and characterization of a beta-1,4-galactanase which degrades a structural component of the primary cell walls of dicots. J. Biol. Chem. 1976, 251, 5904–5910. [Google Scholar]
- Choudhury, B.; Leoff, C.; Saile, E.; Wilkins, P.; Quinn, C.P.; Kannenberg, E.L.; Carlson, R.W. The structure of the major cell wall polysaccharide of Bacillus anthracis is species-specific. J. Biol. Chem. 2006, 281, 27932–27941. [Google Scholar] [CrossRef]
- Garvey, M.; Klose, H.; Fischer, R.; Lambertz, C.; Commandeur, U. Cellulases for biomass degradation: Comparing recombinant cellulase expression platforms. Trends Biotechnol. 2013, 31, 581–593. [Google Scholar] [CrossRef]
- Savakis, P.; Hellingwerf, K.J. Engineering cyanobacteria for direct biofuel production from CO2. Curr. Opin. Biotechnol. 2015, 33, 8–14. [Google Scholar] [CrossRef]
- Takenaka, H.; Yamaguchi, Y. Commercial-scale culturing of cyanobacteria: An industrial experience. In Cyanobacteria: An Economic Perspective; Wiley Blackwell: Hoboken, NJ, USA, 2013; pp. 293–301. ISBN 9781118402238. [Google Scholar]
- Pereira, H.; Páramo, J.; Silva, J.; Marques, A.; Barros, A.; Maurício, D.; Santos, T.; Schulze, P.; Barros, R.; Gouveia, L.; et al. Scale-up and large-scale production of Tetraselmis sp. CTP4 (Chlorophyta) for CO2 mitigation: From an agar plate to 100-m3 industrial photobioreactors. Sci. Rep. 2018, 8, 5112. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, F.; Carbonell, V.; Cossu, M.; Correddu, D.; Jones, P.R. Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803. PLoS ONE 2012, 7, e50470. [Google Scholar] [CrossRef] [PubMed]
- Behle, A.; Saake, P.; Axmann, I.M. Comparative analysis of inducible promoters in cyanobacteria. bioRxiv 2019, 757948. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.; Meng, H.; Zhu, Y.; Bao, G.; Zhang, Y.; Li, Y.; Ma, Y. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci. Rep. 2015, 4, 4500. [Google Scholar] [CrossRef]
- Vasudevan, R.; Gale, G.A.R.; Schiavon, A.A.; Puzorjov, A.; Malin, J.; Gillespie, M.D.; Vavitsas, K.; Zulkower, V.; Wang, B.; Howe, C.J.; et al. CyanoGate: A modular cloning suite for engineering cyanobacteria based on the plant MoClo syntax. Plant Physiol. 2019, 180, 39–55. [Google Scholar]
- Hölzl, G.; Zähringer, U.; Warnecke, D.; Heinz, E. Glycoengineering of cyanobacterial thylakoid membranes for future studies on the role of glycolipids in photosynthesis. Plant Cell Physiol. 2005, 46, 1766–1778. [Google Scholar] [CrossRef][Green Version]
- Bentley, F.K.; Zurbriggen, A.; Melis, A. Heterologous expression of the mevalonic acid pathway in cyanobacteria enhances endogenous carbon partitioning to isoprene. Mol. Plant 2014, 7, 71–86. [Google Scholar] [CrossRef]
- Möllers, K.; Cannella, D.; Jørgensen, H.; Frigaard, N.-U. Cyanobacterial biomass as carbohydrate and nutrient feedstock for bioethanol production by yeast fermentation. Biotechnol. Biofuels 2014, 7, 64. [Google Scholar] [CrossRef]
- Chow, T.J.; Su, H.Y.; Tsai, T.Y.; Chou, H.H.; Lee, T.M.; Chang, J.S. Using recombinant cyanobacterium (Synechococcus elongatus) with increased carbohydrate productivity as feedstock for bioethanol production via separate hydrolysis and fermentation process. Bioresour. Technol. 2015, 184, 33–41. [Google Scholar] [CrossRef]
- Zhao, C.; Li, Z.; Li, T.; Zhang, Y.; Bryant, D.A.; Zhao, J. High-yield production of extracellular type-I cellulose by the cyanobacterium Synechococcus sp. PCC 7002. Cell Discov. 2015, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.; Drews, G.; Weckesser, J.; Mayer, H. Lipopolysaccharides in four strains of the unicellular cyanobacterium Synechocystis. Arch. Microbiol. 1980, 127, 217–222. [Google Scholar] [CrossRef]
- Orellana, A.; Moraga, C.; Araya, M.; Moreno, A. Overview of nucleotide sugar transporter gene family functions across multiple species. J. Mol. Biol. 2016, 428, 3150–3165. [Google Scholar] [CrossRef] [PubMed]
- Dean, N.; Zhang, Y.B.; Poster, J.B. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 31908–31914. [Google Scholar] [CrossRef]
- Baldwin, T.C.; Handford, M.G.; Yuseff, M.I.; Orellana, A.; Dupree, P. Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 2001, 13, 2283–2295. [Google Scholar] [CrossRef]
- Davis, J.; Brandizzi, F.; Liepman, A.H.; Keegstra, K. Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 2010, 64, 1028–1037. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Petersen, B.L.; Egelund, J.; Damager, I.; Faber, K.; Krüger Jensen, J.; Yang, Z.; Bennett, E.P.; Scheller, H.V.; Ulvskov, P. Assay and heterologous expression in Pichia pastoris of plant cell wall type-II membrane anchored glycosyltransferases. Glycoconj. J. 2009, 26, 1235–1246. [Google Scholar] [CrossRef]
- Gille, S.; Cheng, K.; Skinner, M.E.; Liepman, A.H.; Wilkerson, C.G.; Pauly, M. Deep sequencing of voodoo lily (Amorphophallus konjac): An approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 2011, 234, 515–526. [Google Scholar] [CrossRef]
- Egelund, J.; Petersen, B.L.; Motawia, M.S.; Damager, I.; Faik, A.; Olsen, C.E.; Ishii, T.; Clausen, H.; Ulvskov, P.; Geshi, N. Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-alpha-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 2006, 18, 2593–2607. [Google Scholar] [CrossRef]
- Purushotham, P.; Cho, S.H.; Díaz-Moreno, S.M.; Kumar, M.; Nixon, B.T.; Bulone, V.; Zimmer, J. A single heterologously expressed plant cellulose synthase isoform is sufficient for cellulose microfibril formation in vitro. Proc. Natl. Acad. Sci. USA 2016, 113, 11360–11365. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Purushotham, P.; Fang, C.; Maranas, C.; Díaz-Moreno, S.M.; Bulone, V.; Zimmer, J.; Kumar, M.; Nixon, B.T. Synthesis and self-assembly of cellulose microfibrils from reconstituted cellulose synthase. Plant Physiol. 2017, 175, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Obst, U.; Lu, T.K.; Sieber, V. A modular toolkit for generating Pichia pastoris secretion libraries. ACS Synth. Biol. 2017, 6, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; Engel, J.; Jin, E.; Holdridge, B.; Xu, P. YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metab. Eng. Commun. 2017, 5, 68–77. [Google Scholar] [CrossRef]
- Egermeier, M.; Sauer, M.; Marx, H. Golden Gate-based metabolic engineering strategy for wild-type strains of Yarrowia lipolytica. FEMS Microbiol. Lett. 2019, 366, 1–9. [Google Scholar] [CrossRef]
- Holkenbrink, C.; Dam, M.I.; Kildegaard, K.R.; Beder, J.; Dahlin, J.; Doménech Belda, D.; Borodina, I. EasyCloneYALI: CRISPR/Cas9-based synthetic toolbox for engineering of the yeast Yarrowia lipolytica. Biotechnol. J. 2018, 13, 1–8. [Google Scholar] [CrossRef]
- Vieira Gomes, A.; Souza Carmo, T.; Silva Carvalho, L.; Mendonça Bahia, F.; Parachin, N. Comparison of yeasts as hosts for recombinant protein production. Microorganisms 2018, 6, 38. [Google Scholar] [CrossRef]
- Tiels, P.; Baranova, E.; Piens, K.; De Visscher, C.; Pynaert, G.; Nerinckx, W.; Stout, J.; Fudalej, F.; Hulpiau, P.; Tännler, S.; et al. A bacterial glycosidase enables mannose-6-phosphate modification and improved cellular uptake of yeast-produced recombinant human lysosomal enzymes. Nat. Biotechnol. 2012, 30, 1225–1231. [Google Scholar] [CrossRef]
- Kollár, R.; Petráková, E.; Ashwell, G.; Robbins, P.W.; Cabib, E. Architecture of the yeast cell wall. J. Biol. Chem. 1995, 270, 1170–1178. [Google Scholar] [CrossRef]
- Vega, R.; Domínguez, A. Cell wall composition of the yeast and mycelial forms of Yarrowia lipolytica. Arch. Microbiol. 1986, 144, 124–130. [Google Scholar] [CrossRef]
- Lazar, Z.; Gamboa-Meléndez, H.; Le Coq, A.-M.C.; Neuvéglise, C.; Nicaud, J.-M. Awakening the endogenous Leloir pathway for efficient galactose utilization by Yarrowia lipolytica. Biotechnol. Biofuels 2015, 8, 185. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.; Perez-Martin, J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008, 18, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J.; Leon, C.G.; Carabez-Trejo, A.; Reyes-Salinas, E. Structure and chemical composition of the cell walls from the haploid yeast and mycelial forms of Ustilago maydis. Fungal Genet. Biol. 1996, 20, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Sandal, T.; Kamp-Hansen, P.; Dalbøge, H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 1998, 14, 1267–1283. [Google Scholar] [CrossRef]
- Trassaert, M.; Vandermies, M.; Carly, F.; Denies, O.; Thomas, S.; Fickers, P.; Nicaud, J.M. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microb. Cell Fact. 2017, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Quijano, R.R.; Bezzate, S.; Bordon-Pallier, F.; Gaillardin, C. Tagging morphogenetic genes by insertional mutagenesis in the yeast Yarrowia lipolytica. J. Bacteriol. 2001, 183, 3098–3107. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Jiang, X.; Zheng, H.; Qi, Q.; Hou, J. Homology-independent genome integration enables rapid library construction for enzyme expression and pathway optimization in Yarrowia lipolytica. Biotechnol. Bioeng. 2019, 116, 354–363. [Google Scholar] [CrossRef]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef]
- Urbanowicz, B.R.; Peña, M.J.; Moniz, H.A.; Moremen, K.W.; York, W.S. Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 2014, 80, 197–206. [Google Scholar] [CrossRef]
- Culbertson, A.T.; Ehrlich, J.J.; Choe, J.-Y.; Honzatko, R.B.; Zabotina, O.A. Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers. Proc. Natl. Acad. Sci. USA 2018, 115, 6064–6069. [Google Scholar] [CrossRef]
- Chen, M.-T.; Lin, S.; Shandil, I.; Andrews, D.; Stadheim, T.A.; Choi, B.-K. Generation of diploid Pichia pastoris strains by mating and their application for recombinant protein production. Microb. Cell Fact. 2012, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.E. Glycosylation. Curr. Opin. Biotechnol. 1993, 4, 596–602. [Google Scholar] [CrossRef]
- York, W.S.; Darvill, A.G.; McNeil, M.; Stevenson, T.T.; Albersheim, P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1986, 118, 3–40. [Google Scholar]
- Gardner, S.L.; Burrell, M.M.; Fry, S.C. Screening of Arabidopsis thaliana stems for variation in cell wall polysaccharides. Phytochemistry 2002, 60, 241–254. [Google Scholar] [CrossRef]
- Florea, M.; Hagemann, H.; Santosa, G.; Abbott, J.; Micklem, C.N.; Spencer-Milnes, X.; de Arroyo Garcia, L.; Paschou, D.; Lazenbatt, C.; Kong, D.; et al. Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc. Natl. Acad. Sci. USA 2016, 113, E3431–E3440. [Google Scholar] [CrossRef]
| Attribute/Species | E. coli | Pseudomonas | Bacillus | Synechocystis | Synechococcus | Saccharomyces | Pichia | Yarrowia | Ustilago | HEK293 | CHO |
|---|---|---|---|---|---|---|---|---|---|---|---|
| species classification | Bacteria | Fungi | Animalia | ||||||||
| plant CWGTs expressed | yes | - | - | - | - | - | yes | - | - | yes | - |
| other plant GTs | yes | - | + | - | yes | yes | yes | yes | - | yes | yes |
| plant polysaccharide degradation | - | yes | yes | - | - | - | - | - | yes | - | - |
| eukaryotic PTMs | - | - | - | - | - | yes | yes | yes | yes | yes | yes |
| available strains | ++++ | ++ | ++++ | + | + | ++++ | +++ | ++ | + | +++ | +++ |
| knockout library | yes | - | yes | - | - | yes | - | - | - | yes* | - |
| available vectors | ++++ | +++ | ++++ | ++ | ++ | ++++ | +++ | ++ | + | +++ | +++ |
| photosynthetic | - | - | - | yes | yes | - | - | - | - | - | - |
| cultivation cost | $ | $ | $ | $$$ | $$$ | $$ | $$ | $$ | $$ | $$$$ | $$$$ |
| doubling time | ++++ | ++++ | ++++ | ++ | ++ | +++ | +++ | +++ | +++ | + | + |
| E. coli | Pseudo- monas | Bacillus | Synecho- cystis | Synecho- coccus | Saccharo- myces | Pichia | Yarrowia | Ustilago | CHO | |
|---|---|---|---|---|---|---|---|---|---|---|
| t-Glc | 4.4 ± 0.4 | 5.5 ± 0.3 | 12.9 ± 2.7 | 11.5 ± 0.2 | 1.6 ± 0.2 | 42.2 ± 2.5 | 8.0 ± 1.0 | 6.6 ± 1.7 | 7.4 ± 0.1 | 4.4 ± 0.7 |
| 3-Glc | 1.9 ± 0.0 | 51.8 ± 3.1 | 4.9 ± 0.7 | 5.3 ± 1.4 | 1.9 ± 0.6 | 8.9 ± 0.7 | ||||
| 6-Glc | 15.2 ± 0.7 | 7.0 ± 1.2 | 7.8 ± 1.6 | 10.0 ± 1.2 | 10.8 ± 1.0 | |||||
| 2,3-Glc | 0.7 ± 0.0 | 2.3 ± 0.1 | 2.1 ± 1.0 | 0.4 ± 0.1 | 0.5 ± 0.1 | |||||
| 3,6-Glc | 1.6 ± 0.0 | 4.2 ± 0.5 | 2.5 ± 0.4 | 2.3 ± 0.6 | 0.7 ± 0.4 | 4.5 ± 0.2 | 6.3 ± 0.6 | |||
| 3,4-Glc | 24.1 ± 0.7 | |||||||||
| 4-Glc | 2.7 ± 0.1 | 1.5 ± 0.4 | 2.9 ± 0.8 | 12.2 ± 0.2 | 2.9 ± 1.1 | 3.9 ± 0.4 | 14.3 ± 3.0 | 25.5 ± 3.8 | 30.4 ± 0.9 | 14.3 ± 4.1 |
| 4,6-Glc | 7.0 ± 0.4 | |||||||||
| t-Man | 1.1 ± 0.2 | 2.1 ± 0.9 | 0.7 ± 0.0 | 7.6 ± 1.1 | 9.3 ± 1.1 | 7.1 ± 0.2 | 1.9 ± 0.2 | |||
| 2-Man | 5.4 ± 0.3 | 6.5 ± 2.8 | 1.5 ± 1.1 | 11.5 ± 0.5 | 40.4 ± 0.9 | 19.2 ± 2.7 | 1.3 ± 0.1 | 3.1 ± 0.5 | ||
| 3-Man | 1.0 ± 0.1 | |||||||||
| 4-Man | 21.8 ± 1.6 | 0.5 ± 0.0 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.2 ± 0.0 | |||||
| 6-Man | 1.1 ± 0.1 | 1.1 ± 0.1 | 0.8 ± 0.0 | 8.2 ± 0.8 | 0.6 ± 0.1 | |||||
| 4,6-Man | 2.5 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | ||||||
| 2,3-Man | 6.2 ± 0.3 | 2.9 ± 0.3 | 1.1 ± 0.1 | 0.4 ± 0.1 | 0.2 ± 0.0 | |||||
| 3,6-Man | 0.6 ± 0.1 | 1.0 ± 0.2 | 0.4 ± 0.0 | 0.5 ± 0.0 | ||||||
| 2,6-Man | 4.4 ± 0.0 | 4.4 ± 0.4 | 8.1 ± 0.8 | 12.3 ± 0.4 | 0.2 ± 0.1 | |||||
| t-Gal | 1.4 ± 0.5 | 1.9 ± 0.4 | 4.5 ± 0.2 | 3.5 ± 0.2 | 5.7 ± 0.3 | 6.6 ± 0.1 | ||||
| 2-Gal | 1.6 ± 0.0 | |||||||||
| 3-Gal | 27.0 ± 5.6 | 35.8 ± 21.6 | 46.6 ± 1.7 | |||||||
| 6-Gal | 30.5 ± 12.9 | 0.2 ± 0.0 | 9.0 ± 2.7 | |||||||
| 2,4-Gal | 5.5 ± 0.8 | |||||||||
| 4,6-Gal | 2.2 ± 0.3 | |||||||||
| t-Rib | 5.5 ± 1.8 | 9.1 ± 0.9 | ||||||||
| 2-Ribf | 34.6 ± 4.7 | 5.0 ± 0.1 | 1.1 ± 0.2 | 0.2 ± 0.0 | ||||||
| 3-Ribf | 7.7 ± 0.4 | |||||||||
| t-Xyl | 0.7 ± 0.2 | |||||||||
| 4-Xyl | 25.8 ± 0.8 | |||||||||
| 3,4-Xyl | 3.6 ± 0.3 | 4.5 ± 0.5 | ||||||||
| 2,3-Hexa | 13.6 ± 0.9 | |||||||||
| 2,6-Hexa | 5.6 ± 0.6 | 1.9 ± 0.6 | 8.1 ± 0.7 | |||||||
| 3,4-Hexa | 1.3 ± 0.2 | |||||||||
| 3,6-Hexa | 2.6 ± 1.3 | |||||||||
| 4,6-Hexa | 1.3 ± 0.1 | |||||||||
| 2,3,4-Hexa | 1.7 ± 0.1 | |||||||||
| 2,3,6-Hexa | 1.7 ± 0.4 | 1.0 ± 0.1 | 0.5 ± 0.0 | |||||||
| 3,4,6-Hexa | 0.9 ± 0.2 | |||||||||
| t-Rha | 0.5 ± 0.1 | |||||||||
| 2-Rha | 15.0 ± 1.6 | |||||||||
| 3-Rha | 25.6 ± 1.3 | |||||||||
| 3,4-Rha | 13.7 ± 0.6 | |||||||||
| 2,3-Rha | 3.5 ± 0.3 | |||||||||
| 2,4-Rha | 6.6 ± 1.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauly, M.; Gawenda, N.; Wagner, C.; Fischbach, P.; Ramírez, V.; Axmann, I.M.; Voiniciuc, C. The Suitability of Orthogonal Hosts to Study Plant Cell Wall Biosynthesis. Plants 2019, 8, 516. https://doi.org/10.3390/plants8110516
Pauly M, Gawenda N, Wagner C, Fischbach P, Ramírez V, Axmann IM, Voiniciuc C. The Suitability of Orthogonal Hosts to Study Plant Cell Wall Biosynthesis. Plants. 2019; 8(11):516. https://doi.org/10.3390/plants8110516
Chicago/Turabian StylePauly, Markus, Niklas Gawenda, Christine Wagner, Patrick Fischbach, Vicente Ramírez, Ilka M. Axmann, and Cătălin Voiniciuc. 2019. "The Suitability of Orthogonal Hosts to Study Plant Cell Wall Biosynthesis" Plants 8, no. 11: 516. https://doi.org/10.3390/plants8110516
APA StylePauly, M., Gawenda, N., Wagner, C., Fischbach, P., Ramírez, V., Axmann, I. M., & Voiniciuc, C. (2019). The Suitability of Orthogonal Hosts to Study Plant Cell Wall Biosynthesis. Plants, 8(11), 516. https://doi.org/10.3390/plants8110516






