Cell Wall Metabolism in Response to Abiotic Stress
Abstract
:1. Introduction
| Reference | Stress | Cell Wall | Data Studied |
|---|---|---|---|
| [21] | C, Fl, L, Wd | Primary cell wall | Transcriptomic and proteomic |
| [22] | Wd | Primary and secondary cell wall | Physiological, proteomic and cell wall composition |
| [23] | Ap, HM, L, Wd | Secondary cell wall | Transcriptomic, proteomic and metabolomic |
| [24] | Wd, S, C, Ap, HM, L | Secondary cell wall | Transcriptomic, proteomic and cell wall composition |
| [25] | S | Primary cell wall | Transcriptomic and proteomic |
| [26] | S | Primary and secondary cell wall | Proteomic |
| [27] | HM | Primary cell wall | Transcriptomic and proteomic |
| [28] | HM | Primary and secondary cell wall | Transcriptomic and proteomic |
2. Water Deficit
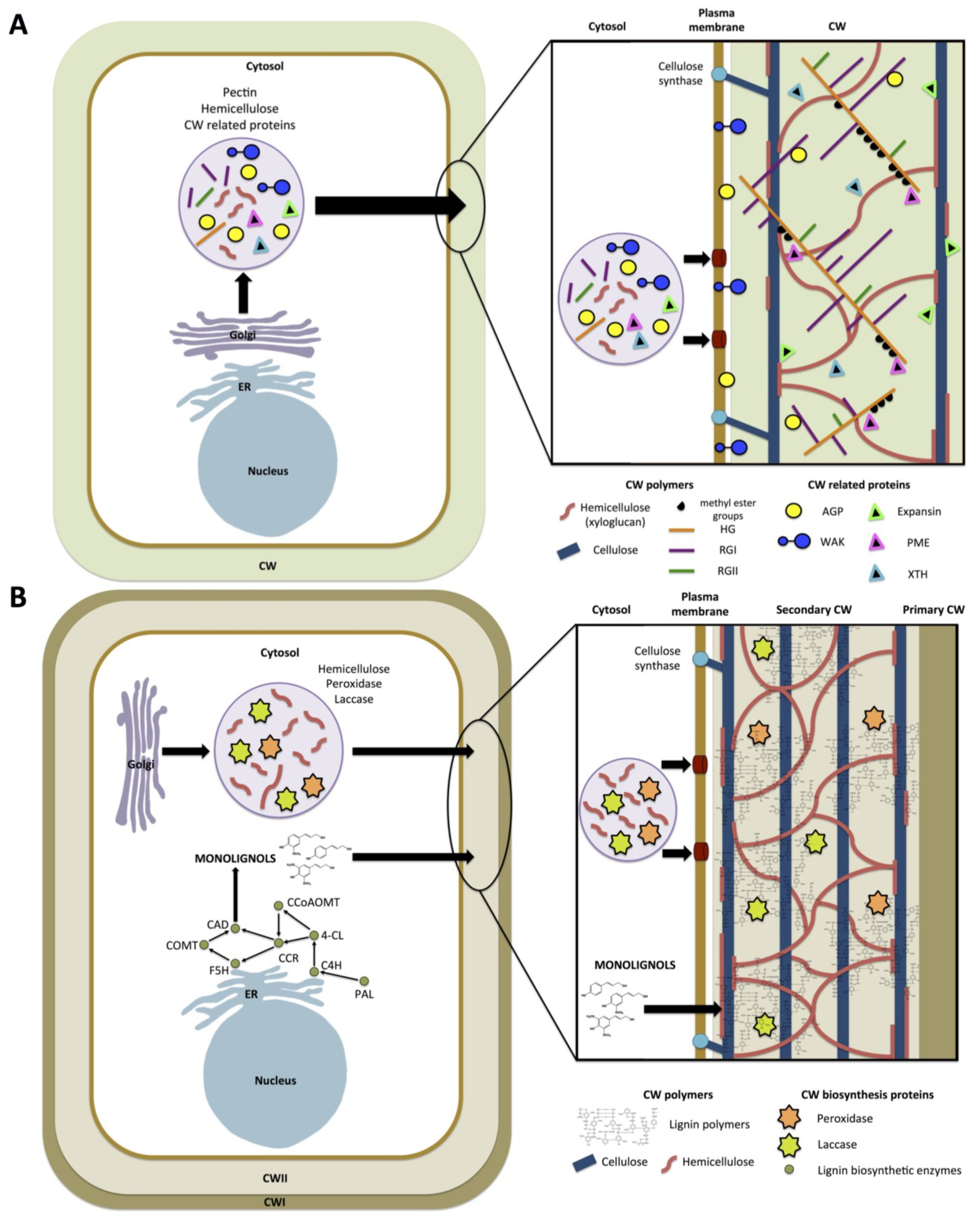
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); polygalacturonase (PG); pectin methylesterase (PME); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); polygalacturonase (PG); pectin methylesterase (PME); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); polygalacturonase (PG); pectin methylesterase (PME); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); polygalacturonase (PG); pectin methylesterase (PME); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX).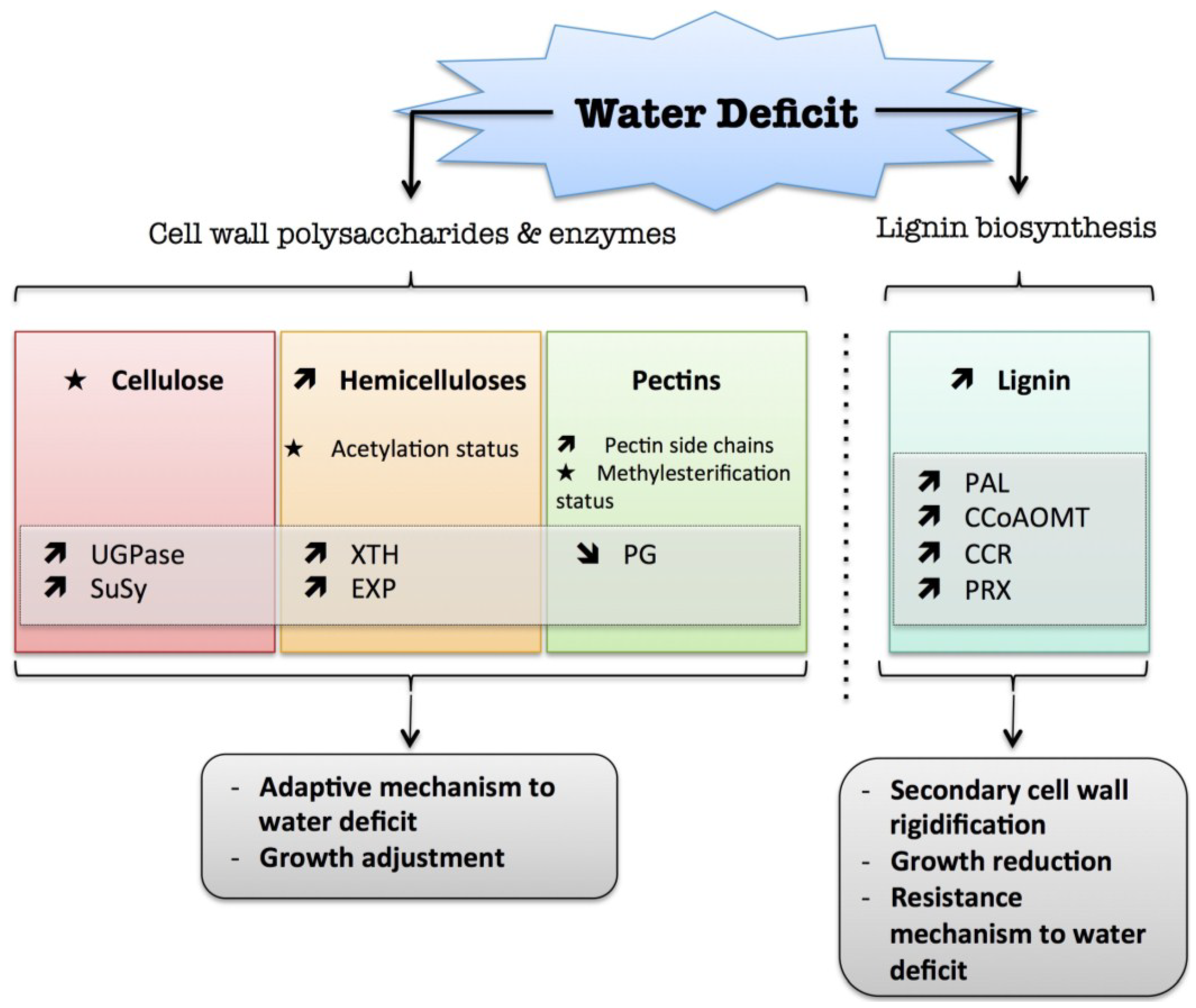
3. Flooding
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); expansin (EXP); endoglucanase (EGase); polygalacturonase (PG); polygalacturonase inhibitor protein (PGIP); pectin/pectate lyase-like (PLL); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); trans-cinnamate 4-hydroxylase (C4H); 4 coumarate CoA-ligase (4CL); ferulate 5-hydroxylase (F5H); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); expansin (EXP); endoglucanase (EGase); polygalacturonase (PG); polygalacturonase inhibitor protein (PGIP); pectin/pectate lyase-like (PLL); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); trans-cinnamate 4-hydroxylase (C4H); 4 coumarate CoA-ligase (4CL); ferulate 5-hydroxylase (F5H); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); expansin (EXP); endoglucanase (EGase); polygalacturonase (PG); polygalacturonase inhibitor protein (PGIP); pectin/pectate lyase-like (PLL); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); trans-cinnamate 4-hydroxylase (C4H); 4 coumarate CoA-ligase (4CL); ferulate 5-hydroxylase (F5H); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); expansin (EXP); endoglucanase (EGase); polygalacturonase (PG); polygalacturonase inhibitor protein (PGIP); pectin/pectate lyase-like (PLL); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); trans-cinnamate 4-hydroxylase (C4H); 4 coumarate CoA-ligase (4CL); ferulate 5-hydroxylase (F5H); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT).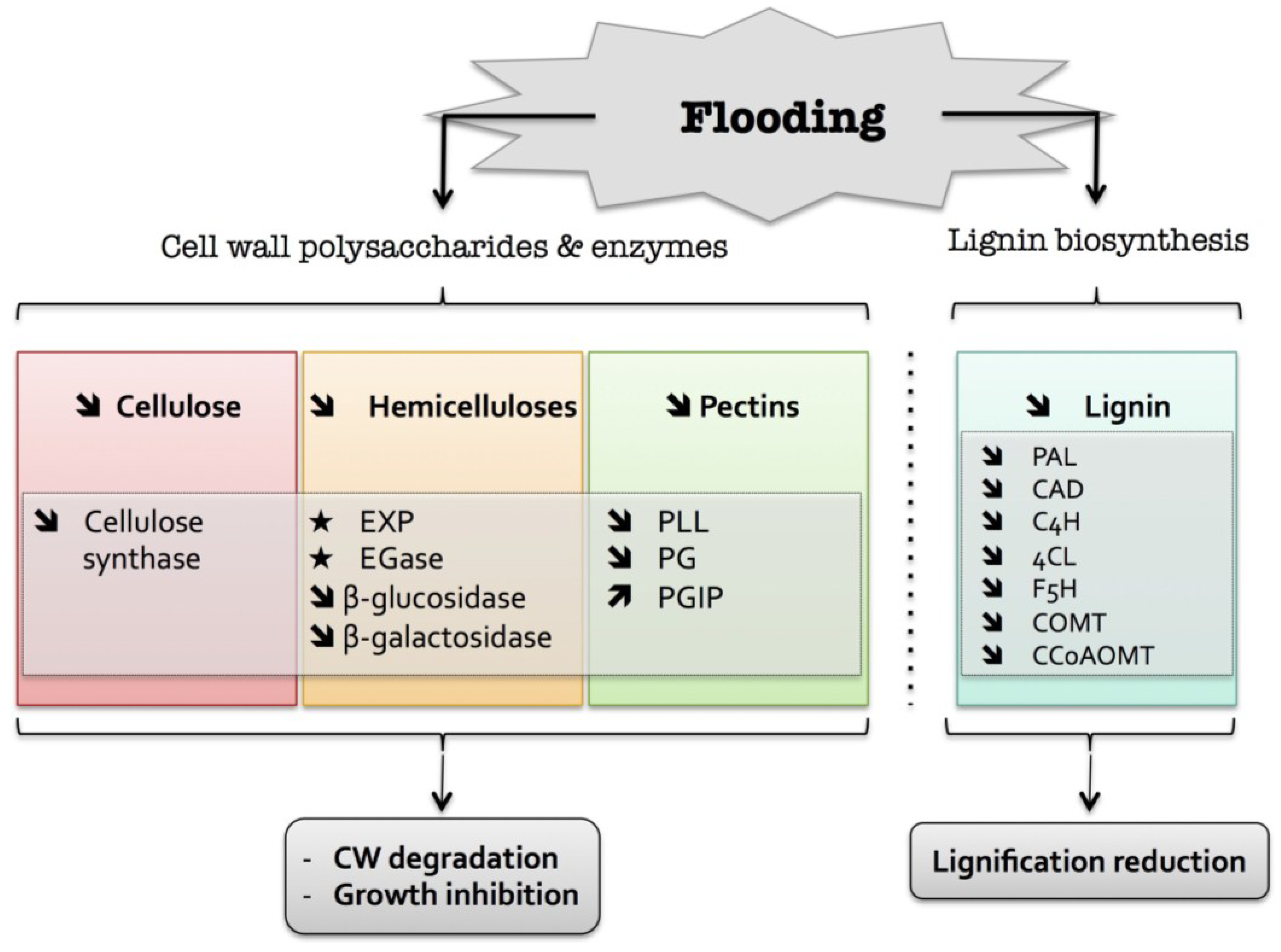
4. Heat
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); pectin methylesterase (PME); cell wall peroxidases (PRX); arabinogalactan protein (AGP).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); pectin methylesterase (PME); cell wall peroxidases (PRX); arabinogalactan protein (AGP).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); pectin methylesterase (PME); cell wall peroxidases (PRX); arabinogalactan protein (AGP).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); pectin methylesterase (PME); cell wall peroxidases (PRX); arabinogalactan protein (AGP).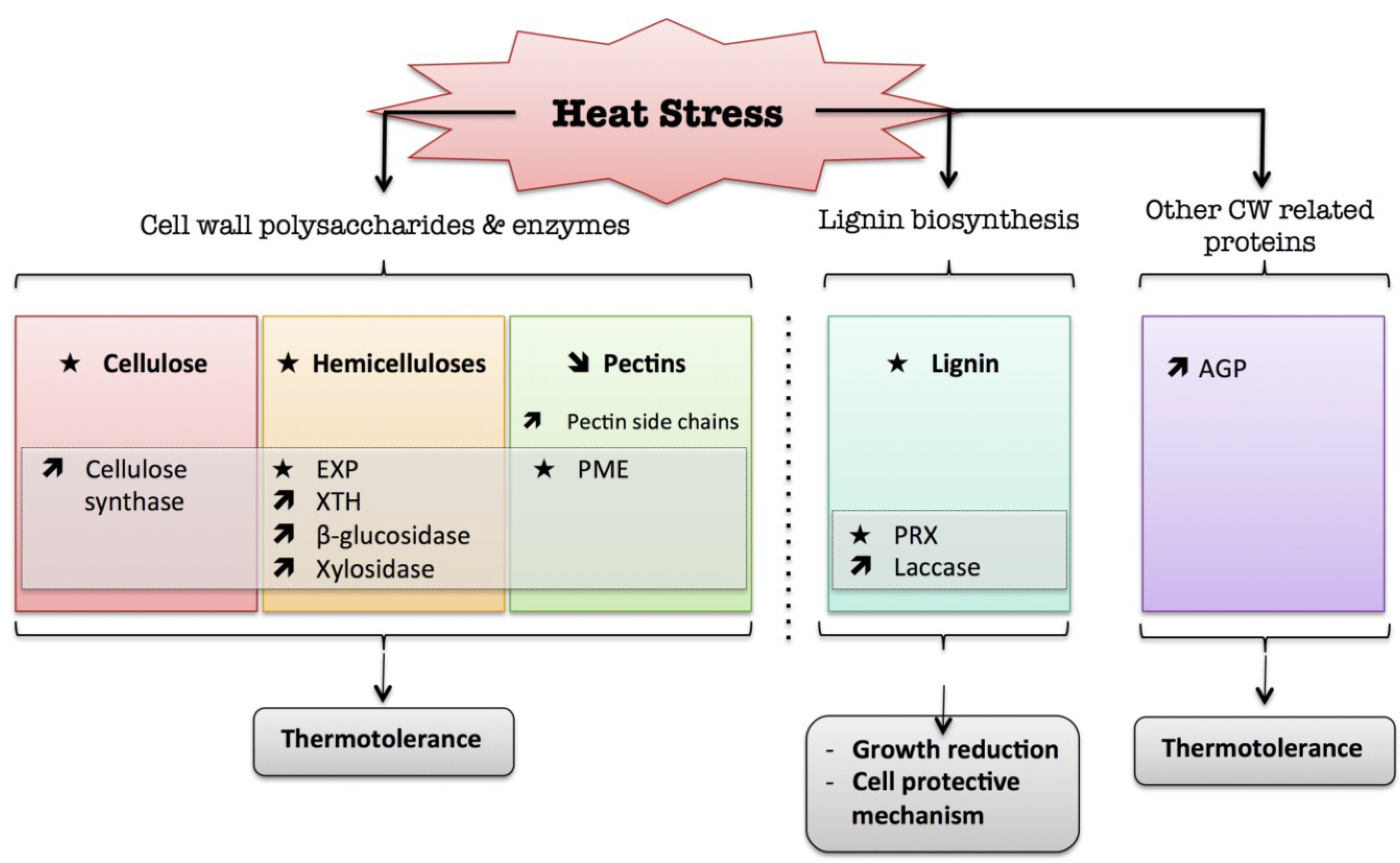
5. Cold
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); pectin methylesterase (PME); UDP-D-xylose 4-epimerase (MUR4); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX); arabinogalactan protein (AGP); wall-associated kinase (WAK).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); pectin methylesterase (PME); UDP-D-xylose 4-epimerase (MUR4); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX); arabinogalactan protein (AGP); wall-associated kinase (WAK).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); pectin methylesterase (PME); UDP-D-xylose 4-epimerase (MUR4); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX); arabinogalactan protein (AGP); wall-associated kinase (WAK).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); pectin methylesterase (PME); UDP-D-xylose 4-epimerase (MUR4); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cinnamoyl-CoA reductase (CCR); cell wall peroxidases (PRX); arabinogalactan protein (AGP); wall-associated kinase (WAK).
6. Salt
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); glycosyl hydrolase family (GH); reversibly glycosylated polypeptide (RGP); phenylalanine ammonia-lyase (PAL); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); arabinogalactan protein (AGP); wall-associated kinase (WAK); proline-rich protein (PRP); glycine-rich protein (GRP).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); glycosyl hydrolase family (GH); reversibly glycosylated polypeptide (RGP); phenylalanine ammonia-lyase (PAL); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); arabinogalactan protein (AGP); wall-associated kinase (WAK); proline-rich protein (PRP); glycine-rich protein (GRP).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); glycosyl hydrolase family (GH); reversibly glycosylated polypeptide (RGP); phenylalanine ammonia-lyase (PAL); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); arabinogalactan protein (AGP); wall-associated kinase (WAK); proline-rich protein (PRP); glycine-rich protein (GRP).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); glycosyl hydrolase family (GH); reversibly glycosylated polypeptide (RGP); phenylalanine ammonia-lyase (PAL); caffeate O-methyltransferase (COMT); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); arabinogalactan protein (AGP); wall-associated kinase (WAK); proline-rich protein (PRP); glycine-rich protein (GRP).
7. Heavy Metals
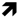 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); endoglucanase (EGase); pectin methylesterase (PME); pectin acetylesterase (PAE); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeate O-methyltransferase (COMT); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); endoglucanase (EGase); pectin methylesterase (PME); pectin acetylesterase (PAE); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeate O-methyltransferase (COMT); cell wall peroxidases (PRX).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); endoglucanase (EGase); pectin methylesterase (PME); pectin acetylesterase (PAE); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeate O-methyltransferase (COMT); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); endoglucanase (EGase); pectin methylesterase (PME); pectin acetylesterase (PAE); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeate O-methyltransferase (COMT); cell wall peroxidases (PRX).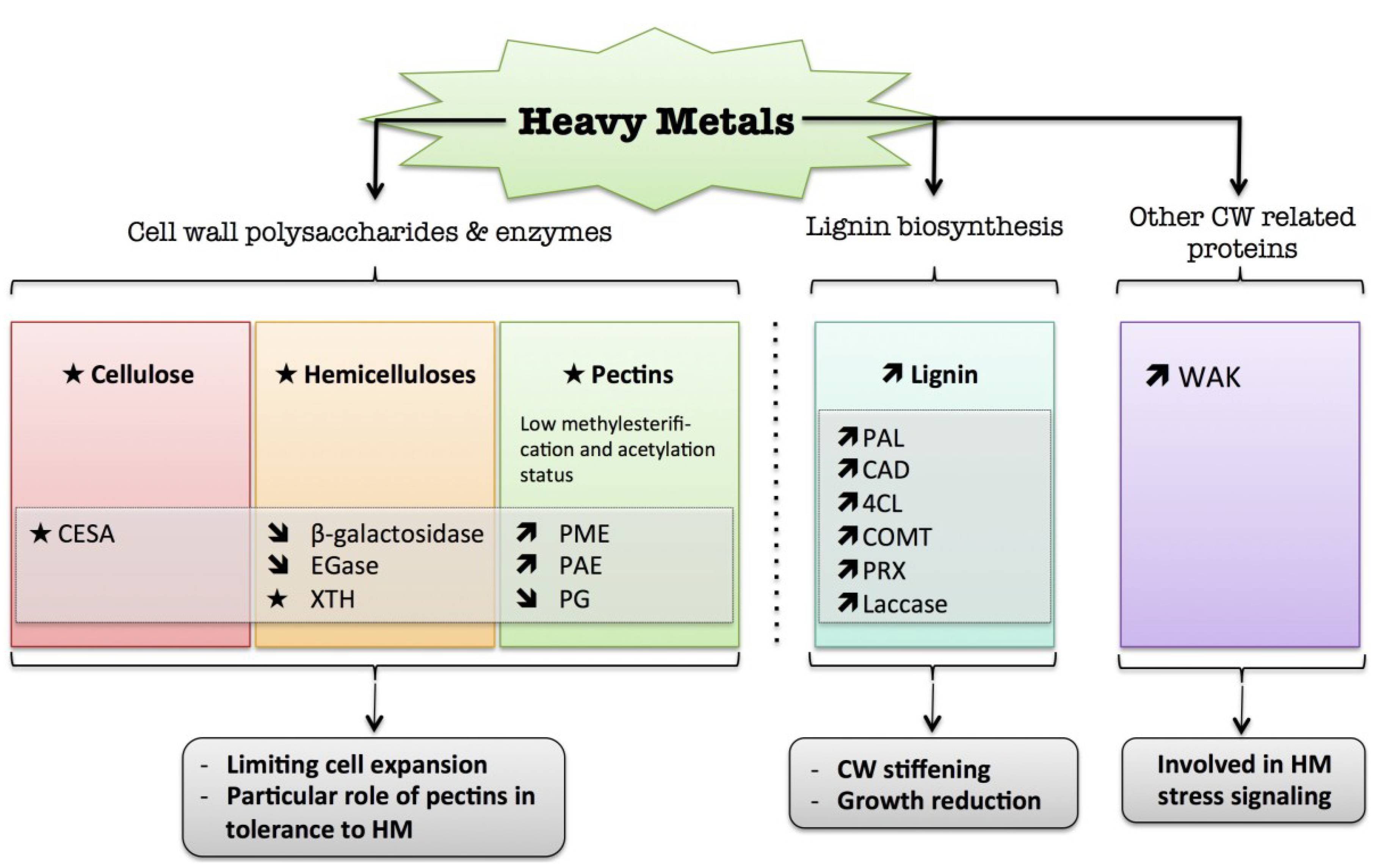
8. Light
8.1. Shade Avoidance
8.2. Light Irradiance
8.3. UV Light
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); trans-cinnamate 4-hydroxylase (C4H); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); trans-cinnamate 4-hydroxylase (C4H); cell wall peroxidases (PRX).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); trans-cinnamate 4-hydroxylase (C4H); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); 4 coumarate CoA-ligase (4CL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); trans-cinnamate 4-hydroxylase (C4H); cell wall peroxidases (PRX).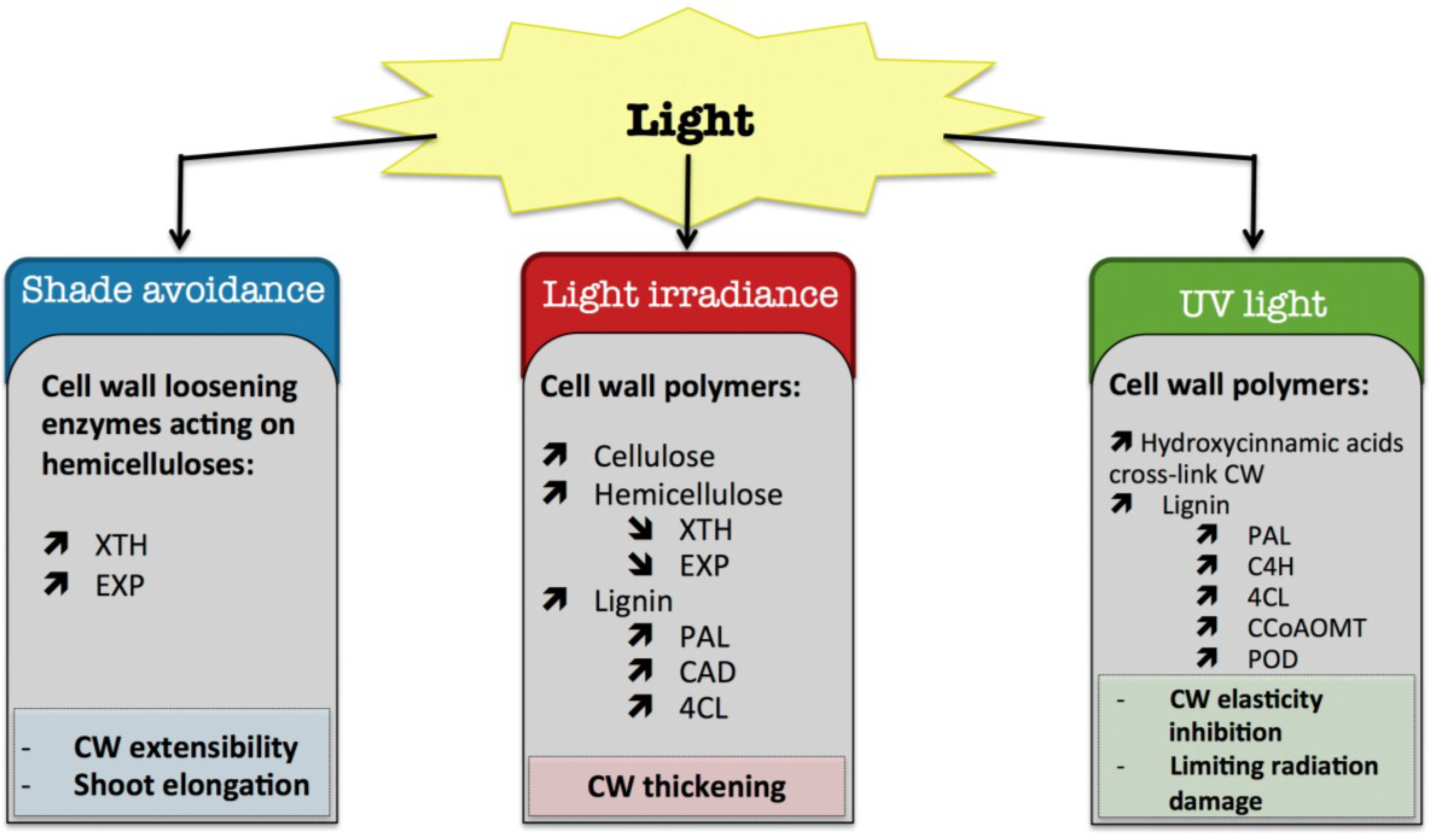
9. Air Pollutants
9.1. Ozone (O3)
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); endoglucanase (EGase); pectin methylesterase (PME); pectin/pectate lyase-like (PLL); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); glycine-rich protein (GRP); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); endoglucanase (EGase); pectin methylesterase (PME); pectin/pectate lyase-like (PLL); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); glycine-rich protein (GRP); cell wall peroxidases (PRX).
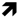 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); endoglucanase (EGase); pectin methylesterase (PME); pectin/pectate lyase-like (PLL); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); glycine-rich protein (GRP); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); sucrose synthase (SuSy); UDP-glucose pyrophosphorylase (UGPase); endoglucanase (EGase); pectin methylesterase (PME); pectin/pectate lyase-like (PLL); polygalacturonase (PG); phenylalanine ammonia-lyase (PAL); hydroxycinnamyl alcohol dehydrogenase (CAD); glycine-rich protein (GRP); cell wall peroxidases (PRX).
9.2. CO2
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); UDP-glucose pyrophosphorylase (UGPase); pectin methylesterase (PME); pectin acetylesterase (PAE); pectin/pectate lyase-like (PLL); glycosyltransferase family 43 (GT43); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); UDP-glucose pyrophosphorylase (UGPase); pectin methylesterase (PME); pectin acetylesterase (PAE); pectin/pectate lyase-like (PLL); glycosyltransferase family 43 (GT43); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cell wall peroxidases (PRX).
 ) means increased abundance and arrow (
) means increased abundance and arrow (  ) means decreased abundance of the molecules. Star (
) means decreased abundance of the molecules. Star (  ) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); UDP-glucose pyrophosphorylase (UGPase); pectin methylesterase (PME); pectin acetylesterase (PAE); pectin/pectate lyase-like (PLL); glycosyltransferase family 43 (GT43); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cell wall peroxidases (PRX).
) means contrasting data on the gene/protein or the molecule studied according to the literature cited in the review. Cell wall (CW); xyloglucan endo-β-transglucosylases/hydrolases (XET/XTH); expansin (EXP); UDP-glucose pyrophosphorylase (UGPase); pectin methylesterase (PME); pectin acetylesterase (PAE); pectin/pectate lyase-like (PLL); glycosyltransferase family 43 (GT43); phenylalanine ammonia-lyase (PAL); caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT); cell wall peroxidases (PRX).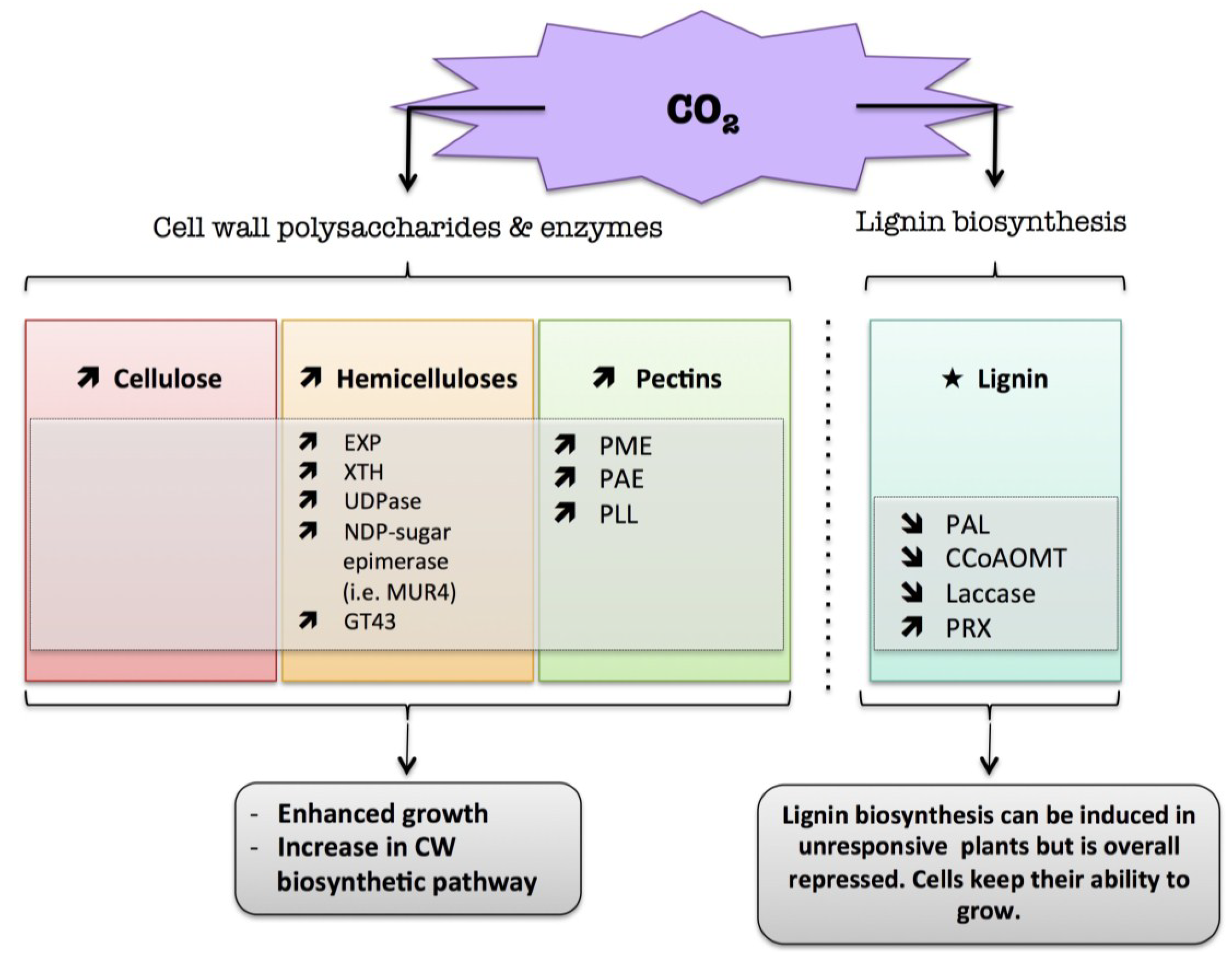
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shaik, R.; Ramakrishna, W. Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol. 2014, 164, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Shu, P.-C.; Cai, T.; Fan, X.-W.; Li, Y.-Z. Existence of abiotic stress-responsive genes within the regions of QTLs controlling maize grain yield: One of the root causes for unstability of the QTLs? S. Afr. J. Bot. 2014, 93, 231–241. [Google Scholar] [CrossRef]
- Wang, Y.; Frei, M. Stressed food—The impact of abiotic environmental stresses on crop quality. Agric. Ecosyst. Environ. 2011, 141, 271–286. [Google Scholar] [CrossRef]
- Dolferus, R. To grow or not to grow: A stressful decision for plants. Plant Sci. 2014, 229, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Potters, G.; Jansen, M.A.K.; Guisez, Y.; Pasternak, T. Stress drives plant cells to take the road towards embryogenesis. In Floriculture, Ornamental and Plant Biotechnology, Advances and Topical Issues; Teixeira da Silva, J.A., Ed.; Global Science Books Ltd.: London, UK, 2006; Volume 2, pp. 289–294. [Google Scholar]
- Wrzaczek, M.; Brosché, M.; Kangasjärvi, J. ROS signaling loops—Production, perception, regulation. Curr. Opin. Plant Biol. 2013, 16, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.; Roberts, K. Architecture of the primary cell wall. In The Cytoskeletal Basis of Plant Growth and Form; Lloyd, C.W., Ed.; Academic Press: London, UK, 1991; pp. 109–129. [Google Scholar]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; McCann, M.C. Maize and sorghum: Genetic resources for bioenergy grasses. Trends Plant Sci. 2008, 13, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 445–476. [Google Scholar] [CrossRef] [PubMed]
- Grabber, J.H.; Ralph, J.; Lapierre, C.; Barrière, Y. Genetic and molecular basis of grass cell-wall degradability. I. Lignin–cell wall matrix interactions. C. R. Biol. 2004, 327, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Sénéchal, F.; Wattier, C.; Rustérucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef] [PubMed]
- Liepman, A.H.; Wightman, R.; Geshi, N.; Turner, S.R.; Scheller, H.V. Arabidopsis a powerful model system for plant cell wall research. Plant J. 2010, 61, 1107–1121. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin Biosynthesis and Structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Tronchet, M.; Balagué, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.J.; Blaukopf, C. Irritable walls: The plant extracellular matrix and signaling. Plant Physiol. 2010, 153, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Voesenek, L.A.; Pierik, R. Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. Crit. Rev. Plant Sci. 2011, 30, 548–562. [Google Scholar] [CrossRef]
- Moore, J.P.; Vicré-Gibouin, M.; Farrant, J.M.; Driouich, A. Adaptations of higher plant cell walls to water loss: Drought vs. desiccation. Physiol. Plant 2008, 134, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.C.M.S.; Bonine, C.A.V.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Cabane, M.; Afif, D.; Hawkins, S. Lignins and abiotic stresses. Adv. Bot. Res. 2013, 61, 220. [Google Scholar]
- Zagorchev, L.; Kamenova, P.; Odjakova, M. The role of plant cell wall proteins in response to salt stress. Sci. World J. 2014. Article ID 764089. [Google Scholar]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.; Dai, S. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteomics 2013, 82, 230–253. [Google Scholar] [CrossRef] [PubMed]
- Krzeslowska, M. The wall cell in plant cell response to trace metals: Polysaccharide remodelling and its role in defense strategy. Acta Physiol. Plant. 2011, 33, 35–51. [Google Scholar] [CrossRef]
- Ovečka, M.; Takáč, T. Managing heavy metal toxicity stress in plants: Biological and biotechnological tools. Biotechnol. Adv. 2014, 32, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Kim, J.Y. Callose synthesis in higher plants. Plant Signal. Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Stass, A.; Horst, W.J. Callose in abiotic stress. In Chemistry, Biochemistry and Biology of (1→3)-β-Glucans and Related Polysaccharides; Bacic, A., Fincher, G.B., Stone, B.A., Eds.; Academic: New York, NY, USA, 2009; pp. 499–524. [Google Scholar]
- Guzmán, P.; Fernández, V.; Graça, J.; Cabral, V.; Kayali, N.; Khayet, M.; Gil, L. Chemical and structural analysis of Eucalyptus globulus and E. camaldulensis leaf cuticles: A lipidized cell wall region. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J.; Li, Z.C. Role of expansin in cell enlargement of oat coleoptiles (analysis of developmental gradients and photocontrol). Plant Physiol. 1993, 103, 1321–1328. [Google Scholar] [PubMed]
- Tardieu, F.O.; Granier, C.; Muller, B. Water deficit and growth. Co-ordinating processes without an orchestrator? Curr. Opin. Plant Biol. 2011, 14, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Zlatev, Z.; Cebola Lidon, F. An overview on drought induced changes in plant growth, water relations and photosynthesis. Emir. J. Food Agric. 2012, 24, 57–72. [Google Scholar]
- Shao, H.-B.; Chu, L.-Y.; Jaleel, C.A.; Zhao, C.-X. Water-deficit stress-induced anatomical changes in higher plants. C. R. Biol. 2008, 331, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Lisar, S.Y.S.; Motafakkerazad, R.; Hossain, M.M.; Rahman, I.M.M. Water stress in plants: Causes, effects and responses. In Water Stress; Rahman, M., Hasegawa, H., Eds.; InTech: Rijeka, Croatia, 2012; pp. 1–14. [Google Scholar]
- Tardieu, F. Any trait or trait-related allele can confer drought tolerance: Just design the right drought scenario. J. Exp. Bot. 2011, 63, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Greiner, S. Growth control by cell wall pectins. Protoplasma 2012, 249, 169–175. [Google Scholar] [CrossRef]
- Tardieu, F.; Parent, B.; Caldeira, C.F.; Welcker, C. Genetic and physiological controls of growth under water deficit. Plant Physiol. 2014, 164, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Balducci, L.; Deslauriers, A.; Giovannelli, A.; Beaulieu, M.; Delzon, S.; Rossi, S.; Rathgeber, C.B.K. How do drought and warming influence survival and wood traits of Picea mariana saplings? J. Exp. Bot. 2014, 4, 1–13. [Google Scholar]
- Hura, T.; Hura, K.; Dziurka, K.; Ostrowska, A.; Bączek-Kwinta, R.; Grzesiak, M. An increase in the content of cell wall-bound phenolics correlates with the productivity of triticale under soil drought. J. Plant Physiol. 2012, 169, 1728–1736. [Google Scholar] [CrossRef] [PubMed]
- Izanloo, A.; Condon, A.G.; Langridge, P.; Tester, M.; Schnurbusch, T. Different mechanisms of adaptation to cyclic water stress in two South Australian bread wheat cultivars. J. Exp. Bot. 2008, 59, 3327–3346. [Google Scholar] [CrossRef] [PubMed]
- Beikircher, B.; de Cesare, C.; Mayr1, S. Hydraulics of high-yield orchard trees: A case study of three Malus domestica cultivars. Tree Physiol. 2013, 33, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Chartzoulakis, K.; Patakas, A.; Kofidis, G.; Bosabalidis, A.; Nastou, A. Water stress affects leaf anatomy, gas exchange, water relations and growth of two avocado cultivars. Sci. Hortic. 2002, 95, 39–50. [Google Scholar] [CrossRef]
- Saito, T.; Terashima, I. Reversible decreases in the bulk elastic modulus of mature leaves of deciduous Quercus species subjected to two drought treatments. Plant Cell Environ. 2004, 27, 863–875. [Google Scholar] [CrossRef]
- De Diego, N.; Sampedro, M.C.; Barrio, R.J.; Saiz-Fernandez, I.; Moncalean, P.; Lacuesta, M. Solute accumulation and elastic modulus changes in six radiata pine breeds exposed to drought. Tree Physiol. 2013, 33, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.P.; Silva, H.; Ledent, J.F.; Pinto, M. Effect of drought stress on the osmotic adjustment, cell wall elasticity and cell volume of six cultivars of common beans (Phaseolus vulgaris L.). Eur. J. Agron. 2007, 26, 30–38. [Google Scholar] [CrossRef]
- Clifford, S.C.; Arndt, S.K.; Corlett, J.E.; Joshi, S.; Sankhla, N.; Popp, M.; Jones, H.G. The role of solute accumulation, osmotic adjustment and changes in cell wall elasticity in drought tolerance in Ziziphus mauritiana (Lamk.). J. Exp. Bot. 1998, 49, 967–977. [Google Scholar] [CrossRef]
- Hessini, K.; Martínez, J.P.; Gandour, M.; Albouchi, A.; Soltani, A.; Abdelly, C. Effect of water stress on growth, osmotic adjustment, cell wall elasticity and water-use efficiency in Spartina alterniflora. Environ. Exp. Bot. 2009, 67, 312–319. [Google Scholar] [CrossRef]
- Iraki, N.M.; Bressan, R.A.; Hasegawa, P.M.; Carpita, N.C. Alteration of the physical and chemical structure of the primary cell wall of growth-limited plant cells adapted to osmotic stress. Plant Physiol. 1989, 91, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Sweet, W.J.; Morrison, J.C.; Labavitch, J.M.; Matthews, M.A. Altered synthesis and composition of cell wall of grape (Vitis vinifera L.) leaves during expansion and growth inhibiting water deficit. Plant Cell Physiol. 1990, 31, 407–414. [Google Scholar]
- Piro, G.; Leucci, M.R.; Waldron, K.; Dalessandro, G. Exposure to water stress causes changes in the biosynthesis of cell wall polysaccharides in roots of wheat cultivars varying in drought tolerance. Plant Sci. 2003, 165, 559–569. [Google Scholar] [CrossRef]
- Bray, E.A. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 2004, 55, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Meng, Y.; Yang, C.; Zhou, Z.; Wang, Y.; Chen, B. Protein expression changes during cotton fiber elongation in response to drought stress and recovery. Proteomics 2014, 14, 1776–1795. [Google Scholar] [CrossRef] [PubMed]
- Ricardi, M.M.; Gonzalez, R.M.; Zhong, S.; Dominguez, P.G.; Duffy, T.; Turjanski, P.G.; Salter, J.D.S.; Alleva, K.; Carrari, F.; Giovannoni, J.J. Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol. 2014, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, C.C.; Guo, W.D.; Li, X.B.; Lu, M.; Yu, C.L. Differential expression of cell wall related genes in the elongation zone of rice roots under water deficit. Russ. J. Plant Physiol. 2006, 53, 390–395. [Google Scholar] [CrossRef]
- Cho, S.K.; Kim, J.E.; Park, J.A.; Eom, T.J.; Kim, W.T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 2006, 580, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Seo, Y.S.; Kim, S.J.; Kim, W.T.; Shin, J.S. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Rep. 2011, 30, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Alvarez, S.; Marsh, E.L.; LeNoble, M.E.; Cho, I.J.; Sivaguru, M.; Chen, S.; Nguyen, H.T.; Wu, Y.; Schachtman, D.P.; et al. Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol. 2007, 145, 1533–1548. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; McQueen-Mason, S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett. 2004, 559, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sharp, R.E.; Durachko, D.M.; Cosgrove, D.J. Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol. 1996, 111, 765–772. [Google Scholar] [PubMed]
- Wu, Y.; Thorne, E.T.; Sharp, R.E.; Cosgrove, D.J. Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol. 2001, 126, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cosgrove, D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000, 51, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Han, Y.; Feng, Y.; Xing, S.; Zhao, M.; Chen, Y.; Wang, W. Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J. Biotechnol. 2013, 163, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Zhang, C.; Jiang, X.; Kang, M.; Yin, X.; Lu, P.; Zhang, X.; Zheng, Y.; Gao, J. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 2012, 160, 2064–2082. [Google Scholar] [CrossRef] [PubMed]
- Lü, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Hoson, T.; Kamisaka, S. Changes in amounts and molecular mass distribution of cell-wall polysaccharides of wheat (Triticum aestivum L.) coleoptiles under water. J. Plant Physiol. 1997, 151, 33–40. [Google Scholar] [CrossRef]
- Peaucelle, A.; Braybrook, S.; Höfte, H. Cell wall mechanics and growth control in plants: The role of pectins revisited. Front Plant Sci. 2012, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Leucci, M.R.; Lenucci, M.S.; Piro, G.; Dalessandro, G. Water stress and cell wall polysaccharides in the apical root zone of wheat cultivars varying in drought tolerance. J. Plant Physiol. 2008, 165, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Gribaa, A.; Dardelle, F.; Lehner, A.; Rihouey, C.; Burel, C.; Ferchichi, A.; Driouich, A.; Mollet, J.C. Effect of water deficit on the cell wall of the date palm (Phoenix dactylifera “Deglet nour”, Arecales) fruit during development. Plant Cell Environ. 2013, 36, 1056–1070. [Google Scholar] [CrossRef]
- Moore, J.P.; Nguema-Ona, E.; Fagerström, A.; Fangel, J.U.; Willats, W.G.T.; Hugo, A.; Vivier, M.A. Profiling the main cell wall polysaccharides of grapevine leaves using high throughput methods. Carbohydr. Polym. 2014, 99, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Rajamani, U.; Verma, J.; Subba, P.; Chakraborty, N.; Datta, A.; Chakraborty, S.; Chakraborty, N. Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: A proteomic approach. J. Proteome Res. 2010, 9, 3443–3464. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Schröder, R.; Hallett, I.C.; Cohen, D.; MacRae, E. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Tucker, G.A.; Robertson, N.G.; Grierson, D. The conversion of tomato fruit polygalacturonase isoenzyme 2 into isoenzyme 1 in vitro. Eur. J. Biochem. 1981, 115, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ma, Y.; Chen, N.A.; Guo, S.; Liu, H.; Guo, X.; Chong, K.; Xu, Y. Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 2013, 37, 1144–1158. [Google Scholar] [CrossRef] [PubMed]
- An, S.H.; Sohn, K.H.; Choi, H.W.; Hwang, I.S.; Lee, S.C.; Hwang, B.K. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 2008, 228, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chantreau, M.; Sibout, R.; Hawkins, S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013, 4, 220. [Google Scholar] [PubMed]
- Terzi, R.; Güler, N.S.; Çaliskan, N.; Kadioglu, A. Lignification response for rolled leaves of Ctenanthe setosa under long-term drought stress. Turk. J. Biol. 2013, 37, 614–619. [Google Scholar] [CrossRef]
- Yoshimura, K.; Masuda, A.; Kuwano, M.; Yokota, A.; Akashi, K. Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol. 2007, 49, 226–241. [Google Scholar] [CrossRef]
- Fan, L. Progressive inhibition by water deficit of cell wall extensibility and growth along the elongation zone of maize roots is related to increased lignin metabolism and progressive stelar accumulation of wall phenolics. Plant Physiol. 2006, 140, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Moura-Sobczak, J.; Souza, U.; Mazzafera, P. Drought stress and changes in the lignin content and composition in Eucalyptus. BMC Proc. 2011, 5, 103. [Google Scholar] [CrossRef]
- Lin, C.C.; Kao, C.H. Osmotic stress-induced changes in cell wall peroxidase activity and hydrogen peroxide level in roots of rice seedlings. Plant Growth Regul. 2002, 37, 177–184. [Google Scholar] [CrossRef]
- Lee, B.R.; Kim, K.Y.; Jung, W.J.; Avice, J.C.; Ourry, A.; Kim, T.H. Peroxidases and lignification in relation to the intensity of water-deficit stress in white clover (Trifolium repens L.). J. Exp. Bot. 2007, 58, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Bacon, M.A.; Thompson, D.S.; Davies, W.J. Can cell wall peroxidase activity explain the leaf growth response of Lolium temulentum L. during drought? J. Exp. Bot. 1997, 48, 2075–2085. [Google Scholar] [CrossRef]
- Hura, T.; Hura, K.; Ostrowska, A.; Grzesiak, M.; Dziurka, K. The cell wall-bound phenolics as a biochemical indicator of soil drought resistance in winter triticale. Plant Soil Environ. 2013, 59, 189–195. [Google Scholar] [CrossRef]
- Christianson, J.A.; Llewellyn, D.J.; Dennis, E.S.; Wilson, I.W. Globalgene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol. 2010, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.J.; Oyanagi, A.; Komatsu, S. Cell wall proteome of wheat roots under flooding stress using gel-based and LC MS/MS-based proteomics approaches. Biochim. Biophys. 2010, 1804, 124–136. [Google Scholar] [CrossRef]
- Salavati, A.; Khatoon, A.; Nanjo, Y.; Komatsu, S. Analysis of proteomic changes in roots of soybean seedlings during recovery after flooding. J. Proteomics 2012, 75, 878–893. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, Y.; Nakamura, T.; Komatsu, S. Identification of indicator proteins associated with flooding injury in soybean seedlings using label-free quantitative proteomics. J. Proteome Res. 2013, 12, 4758–4798. [Google Scholar] [CrossRef]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell. 2013, 25, 4941–4955. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, P.G.; Alves, J.D.; Magalhăes, P.C.; Magalhăes, M.M.; Lima, L.C.O.; Oliveira, L.E.M. Flooding tolerance and cell wall alterations in maize mesocotyl during hypoxia. Pesq. Agropec. Bras. 2001, 36, 1027–1035. [Google Scholar] [CrossRef]
- Ooume, K.; Inoue, Y.; Soga, K.; Wakabayashi, K.; Fujii, S.; Yamamoto, R.; Hoson, T. Cellular basis of growth suppression by submergence in azuki bean epicotyls. Ann. Bot. 2009, 103, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Ferreira, C.S.; Piedade, M.T.F.; Tine, M.A.S.; Rossatto, D.R.; Parolin, P.; Buckeridge, M.S. The role of carbohydrates in seed germination and seedling establishment of Himatanthus sucuuba, an Amazonian tree with populations adapted to flooded and non-flooded conditions. Ann. Bot. 2009, 104, 1111–1119. [Google Scholar]
- Nanjo, Y.; Maruyama, K.; Yasue, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Komatsu, S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol. Biol. 2011, 77, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Kreuzwieser, J.; Hauberg, J.; Howell, K.A.; Carroll, A.; Rennenberg, H.; Millar, A.H.; Whelan, J. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009, 149, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Kobayashi, Y.; Nishizawa, K.; Nanjo, Y.; Furukawa, K. Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 2010, 39, 1435–1449. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.E. Breeding for heat tolerance. Plant Breed. Rev. 1992, 10, 129–168. [Google Scholar]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Dolferus, R.; Ji, X.; Richards, R.A. Abiotic stress and control of grain number in cereals. Plant Sci. 2011, 181, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Crafts-Brandner, S.J. Sensitivity of Photosynthesis in a C4 Plant, Maize, to Heat Stress. Plant Physiol. 2002, 129, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Knight, M.R. Protection against Heat Stress-Induced Oxidative Damage in Arabidopsis Involves Calcium, Abscisic Acid, Ethylene, and Salicylic Acid. Plant Physiol. 2002, 128, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.-D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Sarker, M.; Saifuzzaman, M.; Teixeira da Silva, J.A.; Lozovskaya, M.V.; Akhter, M.M. Evaluation of growth, yield, relative performance and heat susceptibility of eight wheat (Triticum aestivum L.) genotypes grown under heat stress. Int. J. Plant Prod. 2013, 7, 615–636. [Google Scholar]
- Liu, X.; Huang, B. Root physiological factors involved in creeping bentgrass response to high soil temperatures. Environ. Exp. Bot. 2005, 53, 233–245. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Suwa, R.; Hakata, H.; Hara, H.; El-Shemy, H.A.; Adu-Gyamfi, J.J.; Nguyen, N.T.; Kanai, S.; Lightfoot, D.A.; Mohapatra, P.K.; Fujita, K. High temperature effects on photosynthate partitioning and sugar metabolism during ear expansion in maize (Zea mays L.) genotypes. Plant Physiol. Biochem. 2010, 48, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.B.; Santos, dos, T.B.; Vieira, L.G.E.; de Lourdes Lúcio Ferrarese, M.; Ferrarese-Filho, O.; Donatti, L.; Boeger, M.R.T.; de Oliveira Petkowicz, C.L. Heat stress causes alterations in the cell-wall polymers and anatomy of coffee leaves (Coffea arabica L.). Carbohydr. Polym. 2013, 93, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Guimarães, L.; Vieira, A.; Chaves, I.; Pinheiro, C.; Queiroz, V.; Renaut, J.; Ricardo, C.P. Effect of greenhouse conditions on the leaf apoplastic proteome of Coffea arabica plants. J. Proteomics 2014, 104, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Rakszegi, M.; Lovegrove, A.; Balla, K.; Láng, L.; Bedő, Z.; Veisz, O.; Shewry, P.R. Effect of heat and drought stress on the structure and composition of arabinoxylan and β-glucan in wheat grain. Carbohydr. Polym. 2014, 102, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.A.; Lim, C.J.; Hong, J.K.; Park, C.Y.; Cheong, Y.H.; Chung, W.S.; Lee, K.O.; Lee, S.Y.; Cho, M.J.; Lim, C.O. Identification of cell wall genes modified by a permissive high temperature in Chinese cabbage. Plant Sci. 2006, 171, 175–182. [Google Scholar] [CrossRef]
- Rienth, M.; Torregrosa, L.; Luchaire, N.; Chatbanyong, R.; Lecourieux, D.; Kelly, M.T.; Romieu, C. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol. 2013, 14, 108–108. [Google Scholar] [CrossRef]
- Xu, J.; Belanger, F.; Huang, B. Differential gene expression in shoots and roots under heat stress for a geothermal and non-thermal Agrostis grass species contrasting in heat tolerance. Environ. Exp. Bot. 2008, 63, 240–247. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, X.; Shi, Y.; Xu, J.; Huang, B. Transgenic Tobacco Plants Overexpressing a Grass PpEXP1 Gene Exhibit Enhanced Tolerance to Heat Stress. PLoS One 2014, 9, e100792. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.; Fan, C.; Yang, Q.; Li, X.; Wan, B.; Dong, Y.; Wang, X.; Zhou, Y. Identification of Heat Responsive Genes in Brassica napus Siliques at the Seed-Filling Stage through Transcriptional Profiling. PLoS One 2014, 9, e101914. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ci, D.; Tian, M.; Zhang, D. Comparison of the physiological effects and transcriptome responses of Populus simonii under different abiotic stresses. Plant Mol. Biol. 2014, 86, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Chen, H.; Li, X.J.; Yang, M.F.; Liu, G.S.; Shen, S.H. A comparative proteomic analysis of rice seedlings under various high-temperature stresses. Biochim. Biophys. Acta 2009, 1794, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Gulen, H.; Eris, A. Effect of heat stress on peroxidase activity and total protein content in strawberry plants. Plant Sci. 2004, 166, 739–744. [Google Scholar] [CrossRef]
- Stitt, M.; Hurry, V. A plant for all seasons: Alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr. Opin. Plant Biol. 2002, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Goulas, E.; Schubert, M.; Kieselbach, T.; Kleczkowski, L.A.; Gardeström, P.; Schröder, W.; Hurry, V. The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J. 2006, 47, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.-C.; Chien, W.-F.; Chao, C.-H.; Lu, M.-K. Effects of cold stress on alterations of physiochemical and chemical properties of rice polysaccharides. Carbohydr. Polym. 2010, 80, 373–376. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genomics 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.A.; Park, S.H.; Huh, G.H.; Paek, K.-H.; Shin, J.S.; Bae, J.M. Growth retardation and differential regulation of expansin genes in chilling-stressed sweet potato. Plant Biotechnol. Rep. 2009, 3, 75–85. [Google Scholar] [CrossRef]
- Stefanowska, M.; Kuras, M.; Kubacka-Zębalska, M.; Kacperska, A. Low Temperature Affects Pattern of Leaf Growth and Structure of Cell Walls in Winter Oilseed Rape (Brassica napus L., var oleifera L.). Ann. Bot. 1999, 84, 313–319. [Google Scholar] [CrossRef]
- Pearce, R.S. Molecular analysis of acclimation to cold. Plant Growth Regul. 1999, 29, 47–76. [Google Scholar] [CrossRef]
- Warren, G.J. Cold stress: Manipulating freezing tolerance in plants. Curr. Biol. 1998, 8, R514–R516. [Google Scholar] [CrossRef] [PubMed]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation - what is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Hajela, R.K.; Horvath, D.P.; Gilmour, S.J.; Thomashow, M.F. Molecular cloning and expression of cor (cold regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990, 93, 1246–1252. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Dong, C.H.; Zhu, J.K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.; Ren, F.; Guo, Q.-Q.; Hu, X.-P.; Li, X.-B. A novel cold-regulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant Cell. Rep. 2011, 30, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Janska, A.; Marsik, P.; Zelenkova, S.; Ovesna, J. Cold stress and acclimation-what is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Levitt, J. Chilling, freezing, and high temperature stresses. In Responses of Plants to Environmental Stresses, 2nd ed.; Academic Press: Orlando, FL, USA, 1980. [Google Scholar]
- Kume, S.; Kobayashi, F.; Ishibashi, M.; Ohno, R.; Nakamura, C.; Takumi, S. Differential and coordinated expression of Cbf and Cor/Lea genes during long-term cold acclimation in two wheat cultivars showing distinct levels of freezing tolerance. Genes Genet. Syst. 2005, 80, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Monroy, A.F.; Dryanova, A.; Malette, B.; Oren, D.H.; Farajalla, M.R.; Liu, W.; Danyluk, J.; Ubayasena, L.W.C.; Kane, K.; Scoles, G.J.; et al. Regulatory gene candidates and gene expression analysis of cold acclimation in winter and spring wheat. Plant Mol. Biol. 2007, 64, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, S.; Kuroda, K. Cryo-scanning electron microscopic study on freezing behavior of xylem ray parenchyma cells in hardwood species. Micron 2000, 31, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Pearce, R.S.; Fuller, M.P. Freezing of barley studied by infrared video thermography. Plant Physiol. 2001, 125, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Kuroda, K.; Jitsuyama, Y.; Takezawa, D.; Arakawa, K.; Fujikawa, S. Roles of the plasma membrane and the cell wall in the responses of plant cells to freezing. Planta 2002, 215, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Solecka, D.; Zebrowski, J.; Kacperska, A. Are involved in cold acclimation and de-acclimation of winter oil-seed rape plants? Ann. Bot. 2008, 101, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Rajashekar, C.B.; Lafta, A. Cell-wall changes and cell tension in response to cold acclimation and exogenous abscisic acid in leaves and cell cultures. Plant Physiol. 1996, 111, 605–612. [Google Scholar] [PubMed]
- Kubacka-Zebalska, M.; Kacperska, A. Low temperature-induced modifications of cell wall content and polysaccharide composition in leaves of winter oilseed rape (Brassica napus L. var oleifera L.). Plant Sci. 1999, 148, 59–67. [Google Scholar] [CrossRef]
- Weiser, R.L.; Wallner, S.J.; Waddell, J.W. Cell wall and extensin mRNA changes during cold acclimation of pea seedlings. Plant Physiol. 1990, 93, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.P.; Hayashi, A.H.; Braga, M.R.; Nievola, C.C. Biochemical and anatomical responses related to the in vitro survival of the tropical bromeliad Nidularium minutum to low temperatures. Plant Physiol. Biochem. 2013, 71, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, L.; Domon, J.M.; Klimek, J.F.; Fournet, F.; Sellier, H.; Gillet, F.; Pelloux, J.; Lejeune-Hénaut, I.; Carpita, N.C.; Rayon, C. Structural alteration of cell wall pectins accompanies pea development in response to cold. Phytochemistry 2014, 104, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Thonar, C.; Liners, F.; van Cutsem, P. Polymorphism and modulation of cell wall esterase enzyme activities in the chicory root during the growing season. J. Exp. Bot. 2006, 57, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y. Use of serial analysis of gene expression technology to reveal changes in gene expression in arabidopsis pollen undergoing cold stress. Plant Physiol. 2003, 132, 517–529. [Google Scholar] [CrossRef] [PubMed]
- He, Z.H.; Fujiki, M.; Kohorn, B.D. A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 1996, 271, 19789–19793. [Google Scholar] [CrossRef] [PubMed]
- He, Z.H.; Cheeseman, I.; He, D.; Kohorn, B.D. A cluster of five cell wall-associated receptor kinase genes, Wak1–5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 1999, 39, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Ruzvidzo, O.; Morse, M.; Donaldson, L.; Kwezi, L.; Gehring, C. The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One 2010, 5, e8904. [Google Scholar] [CrossRef] [PubMed]
- Kohorn, B.D.; Kohorn, S.L. The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 2012, 3, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Huang, L.; Wang, W.; Zhao, X.; Zhu, L.; Fu, B.; Li, Z. Genome-wide gene expression profiling of introgressed indica rice alleles associated with seedling cold tolerance improvement in a japonica rice background. BMC Genomics 2012, 13, 461. [Google Scholar] [CrossRef] [PubMed]
- Zabotin, A.I.; Barisheva, T.S.; Zabotina, O.A.; Larskaya, I.A.; Lozovaya, V.V.; Beldman, G.; Voragen, A.G.J. Alterations in cell walls of winter wheat roots during low temperature acclimation. J. Plant Physiol. 1998, 152, 473–479. [Google Scholar] [CrossRef]
- Ko, J.H.; Prassinos, C.; Keathley, D.; Han, K.H. Novel aspects of transcriptional regulation in the winter survival and maintenance mechanism of poplar. Tree Physiol. 2011, 31, 208–225. [Google Scholar] [CrossRef] [PubMed]
- Domon, J.-M.; Baldwin, L.; Acket, S.; Caudeville, E.; Arnoult, S.; Zub, H.; Gillet, F.; Lejeune-Hénaut, I.; Brancourt-Hulmel, M.; Pelloux, J.; et al. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry 2013, 85, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Zabotin, A.I.; Barisheva, T.S.; Trofimova, O.I.; Toroschina, T.E.; Larskaya, I.A.; Zabotina, O.A. Oligosaccharin and ABA synergistically affect the acquisition of freezing tolerance in winter wheat. Plant Physiol. Biochem. 2009, 47, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Imin, N.; Kerim, T.; Rolfe, B.G.; Weinman, J.J. Effect of early cold stress on the maturation of rice anthers. Proteomics 2004, 4, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Ogawa, M.; Kuwahara, A.; Hanada, A.; Kamiya, Y.; Yamaguchi, S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 2004, 16, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Jin, R.; Chan, Z. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014, 203, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; J<em>iang, Y.; Chen, R.; Xu, Z.; Gao, X. Isolation of a novel xyloglucan endotransglucosylase (OsXET9) gene from rice and analysis of the response of this gene to abiotic stresses. Afr. J. Biotechnol. 2011, 10, 1–11. [Google Scholar]
- Zhu, Y.-N.; Shi, D.-Q.; Ruan, M.-B.; Zhang, L.-L.; Meng, Z.-H.; Liu, J.; Yang, W.-C. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS One 2013, 8, e80218. [Google Scholar] [CrossRef] [PubMed]
- Evers, D.; Legay, S.; Lamoureux, D.; Hausman, J.F.; Hoffmann, L.; Renaut, J. Towards a synthetic view of potato cold and salt stress response by transcriptomic and proteomic analyses. Plant Mol. Biol. 2012, 78, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Meng, M.; Geisler, M.; Johansson, H.; Mellerowicz, E.J.; Karpinski, S.; Kleczkowski, L.A. Differential tissue/organ-dependent expression of two sucrose- and cold-responsive genes for UDP-glucose pyrophosphorylase in Populus. Gene 2007, 389, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.Y.; Huang, G.Q.; Sun, X.; Li, P.; Zhao, L.L.; Zhang, D.J.; Li, X.B. GhAGP31, a cotton non-classical arabinogalactan protein, is involved in response to cold stress during early seedling development. Plant Biol. 2011, 14, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Solecka, D.; Kacperska, A. Activity of L-phenylalanine ammonia-lyase in winter rape leaves treated with cold. Acta Biochim. Pol. 1993, 40, 113–118. [Google Scholar] [PubMed]
- Solecka, D.; Boudet, A.M.; Kacperska, A. Phenylpropanoid and anthocyanin changes in low-temperature treated winter oilseed rape leaves. Plant Physiol. Biochem. 1999, 37, 491–496. [Google Scholar] [CrossRef]
- Solecka, D.; Kacperska, A. Phenylpropanoid deficiency affects the course of plant acclimation to cold. Physiol. Plant 2003, 119, 253–262. [Google Scholar] [CrossRef]
- Stefanowska, M.; Kuras, M.; Kacperska, A. Low temperature-induced modifications in cell ultrastructure and localization of phenolics in winter oilseed rape (Brassica napus L. var oleifera L.) leaves. Ann. Bot. 2002, 90, 637–645. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Jeong, J.C.; Lee, H.-S.; Kwak, S.-S. Comparative characterization of sweet potato antioxidant genes from expressed sequence tags of dehydration-treated fibrous roots under different abiotic stress conditions. Mol. Biol. Rep. 2012, 40, 2887–2896. [Google Scholar] [CrossRef] [PubMed]
- Badowiec, A.; Swigonska, S.; Weidner, S. Changes in the protein patterns in pea (Pisum sativum L.) roots under the influence of long- and short-term chilling stress and post-stress recovery. Plant Physiol. Biochem. 2013, 71, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Badowiec, A.; Weidner, S. Proteomic changes in the roots of germinating Phaseolus vulgaris seeds in response to chilling stress and post-stress recovery. J. Plant Physiol. 2014, 171, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Szabolcs, I. Salt-Affected Soils; CRC Press: Boca Raton, FL, USA, 1998; p. 274. [Google Scholar]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.-K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. (NY) 2006, 27, 141–177. [Google Scholar]
- Uddin, M.N.; Hanstein, S.; Faust, F.; Eitenmüller, P.T.; Pitann, B.; Schubert, S. Diferulic acids in the cell wall may contribute to the suppression of shoot growth in the first phase of salt stress in maize. Phytochemistry 2014, 102, 126–136. [Google Scholar] [CrossRef] [PubMed]
- De Lima, R.B.; dos Santos, T.B.; Vieira, L.G.E.; de Lourdes Lúcio Ferrarese, M.; Ferrarese-Filho, O.; Donatti, L.; Boeger, M.R.T.; de Oliveira Petkowicz, C.L. Salt stress alters the cell wall polysaccharides and anatomy of coffee (Coffea arabica L.) leaf cells. Carbohydr. Polym. 2014, 112, 686–694. [Google Scholar] [CrossRef]
- Neves, G.Y.S.; Marchiosi, R.; Ferrarese, M.L.L.; Siqueira-Soares, R.C.; Ferrarese-Filho, O. Root growth inhibition and lignification induced by salt stress in soybean. J. Agron. Crop. Sci. 2010, 196, 467–473. [Google Scholar] [CrossRef]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 2012, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Munns, R. Salinity Stress: Physiological constraints and adaptive mechanisms. In Plant Stress Physiology; Shabala, S., Ed.; CAB International: Oxford, UK; Oxford, MS, USA, 2012; pp. 59–93. [Google Scholar]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Kacperska, A. Sensor types in signal transduction pathways in plant cells responding to abiotic stressors: Do they depend on stress intensity? Physiol. Plant. 2004, 122, 159–168. [Google Scholar] [CrossRef]
- Hou, X.; Tong, H.; Selby, J.; Dewitt, J.; Peng, X.; He, Z.-H. Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 2005, 139, 1704–1716. [Google Scholar] [CrossRef] [PubMed]
- Lippold, F.; Sanchez, D.H.; Musialak, M.; Schlereth, A.; Scheible, W.-R.; Hincha, D.K.; Udvardi, M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009, 149, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Schippers, J.H.M.; Mieulet, D.; Obata, T.; Fernie, A.R.; Guiderdoni, E.; Mueller-Roeber, B. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J. 2013, 76, 258–273. [Google Scholar] [PubMed]
- Cominelli, E.; Sala, T.; Calvi, D.; Gusmaroli, G.; Tonelli, C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 2008, 53, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, A.; Li, F.; Zhao, M.; Wang, W. Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Biochem. 2012, 54, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.R.; Lee, H.J.; Kim, K.H.; Hong, S.-W.; Lee, S.J.; Lee, H. Ectopic expression of Expansin3 or Expansinbeta1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol. Lett. 2008, 30, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, C.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to salt stress. PLoS One 2014, 9, e97878. [Google Scholar] [CrossRef] [PubMed]
- Kieliszewski, M.J.; Lamport, D.T.A.; Tan, L.; Cannon, M.C. Hydroxyproline-rich glycoproteins: Form and function. In Annual Plant Reviews; Wiley-Blackwell: Oxford, UK, 2010; pp. 321–342. [Google Scholar]
- Lamport, D.T.A.; Kieliszewski, M.J.; Showalter, A.M. Salt stress upregulates periplasmic arabinogalactan proteins: Using salt stress to analyse AGP function. New Phytol. 2006, 169, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Brinker, M.; Brosche, M.; Vinocur, B.; Abo-Ogiala, A.; Fayyaz, P.; Janz, D.; Ottow, E.A.; Cullmann, A.D.; Saborowski, J.; Kangasjarvi, J.; et al. Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 2010, 154, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhao, J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 2647–2668. [Google Scholar] [CrossRef] [PubMed]
- Tseng, I.-C.; Hong, C.-Y.; Yu, S.-M.; Ho, T.-H.D. Abscisic acid- and stress-induced highly proline-rich glycoproteins regulate root growth in rice. Plant Physiol. 2013, 163, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Long, R.; Yang, Q.; Kang, J.; Zhang, T.; Wang, H.; Li, M.; Zhang, Z. Overexpression of a novel salt stress-induced glycine-rich protein gene from alfalfa causes salt and ABA sensitivity in Arabidopsis. Plant Cell Rep. 2013, 32, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lee, B.-H.; Dellinger, M.; Cui, X.; Zhang, C.; Wu, S.; Nothnagel, E.A.; Zhu, J.-K. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [PubMed]
- Kav, N.N.V.; Srivastava, S.; Goonewardene, L.; Blade, S.F. Proteome-level changes in the roots of Pisum sativum in response to salinity. Ann. Appl. Biol. 2004, 145, 217–230. [Google Scholar] [CrossRef]
- Xu, C.; Sibicky, T.; Huang, B. Protein profile analysis of salt-responsive proteins in leaves and roots in two cultivars of creeping bentgrass differing in salinity tolerance. Plant Cell Rep. 2010, 29, 595–615. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Harris, N.S.; Deyholos, M.K. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 2007, 58, 3591–3607. [Google Scholar] [CrossRef] [PubMed]
- Mustard, J.; Renault, S. Effects of NaCl on water relations and cell wall elasticity and composition of red-osier dogwood (Cornus stolonifera) seedlings. Physiol. Plant. 2004, 121, 265–271. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Li, X.; Zheng, Y.; Matsuura, A.; Abe, J.; Eneji, A.E.; Tanimoto, E.; Inanaga, S. Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agro. Crop Sci. 2014, 200, 212–218. [Google Scholar] [CrossRef]
- Muszyńska, A.; Jarocka, K.; Kurczynska, E.U. Plasma membrane and cell wall properties of an aspen hybrid (Populus tremula × tremuloides) parenchyma cells under the influence of salt stress. Acta Physiol. Plant 2014, 36, 1155–1165. [Google Scholar] [CrossRef]
- Uddin, M.N.; Hanstein, S.; Leubner, R.; Schubert, S. Leaf cell-wall components as influenced in the first phase of salt stress in three maize (Zea mays L.) hybrids differing in salt resistance. J. Agro Crop Sci. 2013, 199, 405–415. [Google Scholar] [CrossRef]
- Sánchez-Aguayo, I.; Rodríguez-Galán, J.M.; García, R.; Torreblanca, J.; Pardo, J.M. Salt stress enhances xylem development and expression of S-adenosyl-L-methionine synthase in lignifying tissues of tomato plants. Planta 2004, 220, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, M.-H. Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato (Solanum lycopersicum L.). Mol. Biol. Rep. 2009, 36, 1579–1585. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-J.; Yang, M.-F.; Chen, H.; Qu, L.-Q.; Chen, F.; Shen, S.-H. Abscisic acid pretreatment enhances salt tolerance of rice seedlings: Proteomic evidence. Biochim. Biophys. Acta 2010, 1804, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Salekdeh, G.H.; Siopongco, J.; Wade, L.J.; Ghareyazie, B.; Bennett, J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2002, 2, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, E.J.; Yang, E.J.; Lee, J.E.; Park, A.R.; Song, W.H.; Park, O.K. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 2004, 16, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.R.; Flowers, S.A.; Rao, G.; Welfare, K.; Senanayake, N.; Flowers, T.J. Silicon reduces sodium uptake in rice (Oryza sativa L.) in saline conditions and this is accounted for by a reduction in the transpirational bypass flow. Plant Cell Environ. 1999, 22, 559–565. [Google Scholar] [CrossRef]
- Yang, J.; Yen, H.E. Early salt stress effects on the changes in chemical composition in leaves of ice plant and Arabidopsis. A Fourier Transform Infrared Spectroscopy Study. Plant Physiol. 2002, 130, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Hayatsu, M.; Suzuki, S.; Hasegawa, A.; Tsuchiya, S.; Sasamoto, H. Effect of NaCl on ionic content and distribution in suspension-cultured cells of the halophyte Sonneratia alba vs. the glycophyte Oryza sativa. J. Plant Physiol. 2014, 171, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cheng, T.; Wang, P.; Liu, W.; Xiao, J.; Yang, Y.; Hu, X.; Jiang, Z.; Zhang, S.; Shi, J. Salinity-induced changes in protein expression in the halophytic plant Nitraria sphaerocarpa. J. Proteomics 2012, 75, 5226–5243. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Nie, L.; Jiang, P.; Feng, J.; Lv, S.; Chen, X.; Bao, H.; Guo, J.; Tai, F.; Wang, J.; et al. Transcriptome Analysis of Salicornia europaea under saline conditions revealed the adaptive primary metabolic pathways as early events to facilitate salt adaptation. PLoS One 2013, 8, e80595. [Google Scholar] [CrossRef] [PubMed]
- D'Emilio, M.; Caggiano, R.; Macchiato, M.; Ragosta, M.; Sabia, S. Soil heavy metal contamination in an industrial area: Analysis of the data collected during a decade. Environ. Monit. Asses. 2012, 185, 5951–5964. [Google Scholar] [CrossRef]
- Kelepertzis, E. Accumulation of heavy metals in agricultural soils of Mediterranean: Insights from Argolida basin, Peloponnese, Greece. Geoderma 2014, 221, 82–90. [Google Scholar] [CrossRef]
- Doncheva, S.; Georgieva, K.; Vassileva, V.; Stoyanova, Z.; Popov, N.; Ignatov, G. Effects of succinate on manganese toxicity in pea plants. J. Plant Nutr. 2005, 28, 47–62. [Google Scholar] [CrossRef]
- Huang, T.L.; Nguyen, Q.T.T.; Fu, S.F.; Lin, C.Y.; Chen, Y.C.; Huang, H.J. Transcriptomic changes and signalling pathway induced by arsenic stress in rice roots. Plant Mol. Biol. 2012, 80, 587–608. [Google Scholar] [CrossRef] [PubMed]
- Feigl, G.; Kumar, D.; Lehotai, N.; Tugyi, N.; Molnor, A.; Ordog, A.; Szepesi, A.; Gemes, K.; Laskay, G.; Erdei, L.; et al. Physiological and morphological responses of the root system of Indian mustard (Brassica jumcea L. Czern.) and rapeseed (Brassica napus L.) to copper stress. Ecotox. Environ. Safe 2013, 91, 179–189. [Google Scholar] [CrossRef]
- Appenroth, K.J. What are “heavy metals” in plant sciences? Acta Physiol. Plant 2010, 32, 615–619. [Google Scholar] [CrossRef]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Briat, J.F. Metal-ion-mediated oxidative stress and its control. In Oxidative Stress in Plants; Montagu, M., Inzé, D., Eds.; Taylor and Francis: London, UK, 2002; pp. 171–189. [Google Scholar]
- Lander, H.M. An essential role for free radicals and derived species in signal transduction. FASEB J. 1997, 11, 118–124. [Google Scholar] [PubMed]
- Opdenakker, K.; Remans, T.; Keunen, E.; Vangronsveld, J.; Cuypers, A. Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environ. Exp. Bot. 2012, 83, 53–61. [Google Scholar] [CrossRef]
- .Krzeslowska, M.; Lenartowska, M.; Mellerowicz, E.J.; Samardakiewicz, S.; Wosny, A. Pectinous cell wall thickenings formation-a response of moss protonema cells to Pb. Environ. Exp. Bot. 2009, 65, 119–131. [Google Scholar] [CrossRef]
- Krzeslowska, M.; Lenartowska, M.; Samardakiewicz, S.; Bilski, H.; Wosny, A. Lead deposited in the cell wall of Funaria hygrometrica protonema is not stable-a remobilization can occur. Environ. Pollut. 2010, 158, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Vuletic, M.; Hadsi-Taskovic Sukalovic, V.; Markovic, K.; Kravic, N.; Vucinic, Z.; Maksimovic, V. Differential response of antioxidative systems of maize roots cell walls to osmotic and heavy metal stress. Plant Biol. 2014, 16, 88–96. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, C. Zinc distribution and zinc-binding forms in Phragmites australis under zinc pollution. J. Plant Physiol. 2008, 165, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.K.M.Z.; Koyama, H.; Hara, T. Growth and cell wall properties of two wheat cultivars differing in their sensitivity to aluminum stress. J. Plant Physiol. 2006, 163, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Polec-Powlak, K.; Ruzik, R.; Lipiec, E.; Ciurzynska, M.; Gawronska, H. Investigation of Pb(II) binding to pectin in Arabidopsis thaliana. J. Anal. Atom. Spectrom. 2007, 22, 968–972. [Google Scholar] [CrossRef]
- Amenos, M.; Corrales, I.; Poschenrieder, C.; Illes, P.; Baluska, F.; Barcelo, J. Different effects of aluminum on the actin cytoskeleton and Brefeldin A-sensitive vesicle recycling in root apex cells of two maize varieties differing in root elongation rate and aluminum tolerance. Plant Cell Physiol. 2009, 50, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Horst, W.J.; Wang, Y.; Eticha, D. The role of the root apoplast in Aluminum-induced inhibition of root elongation and in Aluminum resistance of plant. Rev. Ann. Bot. 2010, 106, 185–197. [Google Scholar] [CrossRef]
- Vaculík, M.; Lux, A.; Luxová, M.; Tanimoto, E.; Lichtscheidl, I. Silicon mitigates cadmium inhibitory effects in young maize plants. Environ. Exp. Bot. 2009, 67, 52–58. [Google Scholar] [CrossRef]
- Gu, J.; Yin, X.; Stomph, T.J.; Wang, H.; Struik, P.C. Physiological basis of genetic variation in leaf photosynthesis among rice (Oryza sativa L.) introgression lines under drought and well-watered conditions. J. Exp. Bot. 2012, 63, 5137–5153. [Google Scholar] [CrossRef] [PubMed]
- Eticha, D.; Stass, A.; Horst, W.J. Cell-wall pectin and its degree of methylation in the maize root apex: Significance for genotypic differences in aluminium resistance. Plant Cell Env. 2005, 28, 1410–1420. [Google Scholar] [CrossRef]
- Baluska, F.; Liners, F.; Hlavacka, A.; Schlicht, M.; van Cutsem, P.; McCurdy, D.W.; Menzel, D. Cell wall pectins and xyloglu-cans are internalised into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 2005, 225, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Pelloux, J.; Rusterucci, C.; Mellerowicz, E.J. New insight into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 1363–1372. [Google Scholar]
- Willats, W.G.T.; Knox, J.P.; Mikkelsen, J.D. Pectin: New insights into an old polymer are starting to gel. Trends Food Sci. Technol. 2006, 17, 97–104. [Google Scholar] [CrossRef]
- Dronnet, V.M.; Renard, C.M.G.C.; Axelos, M.A.V.; Thibault, J.F. Heavy metal binding by pectins: Selectivity, quantification and characterization. Carbohydr. Polym. 1996, 30, 253–263. [Google Scholar] [CrossRef]
- Paynel, F.; Schaumann, A.; Arkoun, M.; Douchiche, O.; Morvan, C. Temporal regulation of cell wall pectin methylesterase and peroxidase isoforms in cadmium-treated flax hypocotyls. Ann. Bot. 2009, 104, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Douchiche, O.; Rihouey, C.; Schaumann, A.; Driouich, A.; Morvan, C. Cadmium-induced alterations of the structural features of pectins in flax hypocotyls. Planta 2007, 225, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Douchiche, O.; Driouich, A.; Morvan, C. Spatial regulation of cell wall structure in response to heavy metal stress: Cadmium-induced alteration of the methyl-esterification pattern of homogalacturonans. Ann. Bot. 2010, 105, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z-B.; Eticha, D.; Rao, I.M.; Horst, W.J. Alteration of Cell-wall Porosity is Involved in Osmotic Stress-induced Enhancement of Aluminium Resistance in Common Bean (Phaseolus vulgaris L.). J. Exp. Bot. 2010, 61, 3245–3258. [Google Scholar] [CrossRef] [PubMed]
- El-Moneim, D.A.; Contreras, R.; Silva-Navas, J.S.; Gallego, F.J.; Figueiras, A.M.; Benito, C. Pectin methylesterase gene and aluminum tolerance in Secale cereale. Environ. Exp. Bot. 2014, 107, 125–133. [Google Scholar] [CrossRef]
- Colzi, I.; Doumett, S.; Del Bubba, M.; Fornaini, J.; Arnetoli, A.; Gabbrielli, R.; Gonnelli, C. On the role of the cell wall in the phenomenon of copper tolerance in Silene paradoxa L. Environ. Exp. Bot. 2011, 72, 77–83. [Google Scholar] [CrossRef]
- Chandran, D.; Sharopova, N.; Ivashuta, S.; Gantt, J.S.; VandenBosch, K.A.; Samac, D.A. Transcriptome profiling identified novel genes associated with aluminium toxicity, resistance and tolerance in Medicago truncatula. Planta 2008, 228, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B.; Eticha, D.; Rotter, B.; Rao, I.M.; Horst, W.J. Physiological and molecular analysis of polyethylene glycol-induced reduction of aluminium accumulation in the root tips of common bean (Phaseolus vulgaris). New Phytol. 2011, 192, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Sa, G.; Sun, J.; Shen, Z.; Zhao, R.; Ding, M.; Deng, S.; Lu, Y.; Zhang, Y.; Shen, X.; Chen, S. Overexpression of Populus euphratica xyloglucan endotransglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco. Environ. Exp. Bot. 2014, 100, 74–83. [Google Scholar] [CrossRef]
- Song, X.Q.; Liu, L.F.; Jiang, Y.J.; Zhang, B.C.; Gao, Y.P.; Liu, X.L.; Lin, Q.S.; Ling, H.Q.; Zhou, Y.H. Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol. Plant. 2013, 6, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Shen, C.; Wang, Y.; Huang, C.; Shi, J. New insights into regulation of proteome and polysaccharide in cell wall of Elsholtzia splendens in response to copper stress. PLoS One 2014, 9, e109573. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Sun, L.; Yang, X.; Liu, J.X. Transcriptomic analysis of cadmium stress response in the heavy metal hyperaccumulator Sedum alfredii hance. PLoS One 2013, 8, e64643. [Google Scholar] [CrossRef] [PubMed]
- Tabbì, G.; Fry, S.C.; Bonomo, R.P. ESR study of the non-enzymic scission of xyloglucan by an ascorbate-H2O2-copper system: The involvement of the hydroxyl radical and the degradation of ascorbate. J. Inorg. Biochem. 2001, 84, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Kärkönen, A.; Fry, S.C. Effect of ascorbate and its oxidation products on H2O2 production in cell-suspension cultures of Picea abies and in the absence of cells. J. Exp. Bot. 2006, 57, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Claus, H. Laccases: Structure, reaction, distribution. Micron 2004, 35, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.B.; Singh, N.; Shohael, A.M.; Hahn, E.J.; Paek, K.Y. Phenolics metabolism and lignin synthesis in root suspension cultures of Panax ginseng in response to copper stress. Plant Sci. 2006, 171, 147–154. [Google Scholar] [CrossRef]
- Keuskamp, D.H.; Keller, M.M.; Ballaré, C.L.; Pierik, R. Blue light regulated shade avoidance. Plant Signal. Behav. 2012, 7, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A. Shade avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Keuskamp, D.H.; Kooke, R.; Voesenek, L.A.; Pierik, R. Interactions between auxin, microtubules and XTHs mediate green shade-induced petiole elongation in Arabidopsis. PLoS One 2014, 9, e90587. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell. 2001, 13, 2589–2607. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Chinnappa, C.C.; Voesenek, L.A.C.J.; Pierik, R. The regulation of cell wall extensibility during shade avoidance: A study using two contrasting ecotypes of Stellaria longipes. Plant Physiol. 2008, 148, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Chinnappa, C.C.; Staal, M.; Elzenga, J.T.; Yokoyama, R.; Nishitani, K.; Voesenek, L.A.; Pierik, R. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 2010, 154, 978–990. [Google Scholar]
- Masuda, Y.; Kamisaka, S.; Yanagisawa, H.; Suzuki, H. Effect of light on growth and metabolic activities in pea seedlings I. changes in cell wall polysaccharides during growth in the dark and in the light. Biochem. Physiol. Pflanzen. 1981, 176, 23–34. [Google Scholar] [CrossRef]
- Wargent, J.J.; Gegas, V.C.; Jenkins, G.I.; Doonan, J.H.; Paul, N.D. UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 2009, 183, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Kigel, J.; Cosgrove, D.J. Photoinhibition of stem elongation by blue and red light. Plant Physiol. 1991, 95, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Raffo, M.D.; Ponce, N.M.; Sozzi, G.O.; Vicente, A.R.; Stortz, C.A. Compositional changes in “Bartlett” pear (Pyrus communis L.) cell wall polysaccharides as affected by sunlight conditions. J. Agric. Food Chem. 2011, 59, 12155–12162. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Chatterjee, M.; Burman, N.; Khurana, J.P. Cryptochrome 1 regulates growth and development in Brassica through alteration in the expression of genes involved in light, phytohormone and stress signalling. Plant Cell Environ. 2014, 37, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Syros, T.D.; Yupsanis, T.A.; Economou, A.S. Expression of peroxidases during seedling growth in Ebenus cretica L. as affected by light and temperature treatments. Plant Growth Regul. 2005, 46, 143–151. [Google Scholar] [CrossRef]
- Ali, M.B.; Khatun, S.; Hahn, E.J.; Paek, K.Y. Enhancement of phenylpropanoid enzymes and lignin in Phalaenopsis orchid and their influence on plant acclimatisation at different levels of photosynthetic photon flux. Plant Growth Regul. 2005, 49, 137–146. [Google Scholar] [CrossRef]
- Kimura, M.; Yamamoto, Y.; Seki, M.; Sakurai, T.; Sato, M.; Abe, T.; Yoshida, S.; Manabe, K.; Shinozaki, K.; Matsui, M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003, 77, 226–233. [Google Scholar] [PubMed]
- Ruhland, C.T.; Xiong, F.S.; Clark, W.D.; Day, T.A. The influence of ultraviolet-B radiation on growth, hydroxycinnamic acids and flavonoids of Deschampsia antarctica during Springtime ozone depletion in Antarctica. Photochem. Photobiol. 2005, 5, 1086–1093. [Google Scholar] [CrossRef]
- Lesniewska, E.; Adrian, M.; Klinguer, A.; Pugin, A. Cell wall modification in grapevine cells in response to UV stress investigated by atomic force microscopy. Ultramicroscopy 2004, 100, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Pristov, J.B.; Spasić, M.; Spasojević, M. Converting low dose radiation to redox signaling. Plant Signal Behav. 2013, 8, e23151. [Google Scholar] [CrossRef] [PubMed]
- Messenger, D.J.; McLeod, A.R.; Fry, S.C. The role of ultraviolet radiation, photosensitizers, reactive oxygen species and ester groups in mechanisms of methane formation from pectin. Plant Cell Environ. 2009, 32, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pristov, J.B.; Jovanović, S.V.; Mitrović, A.; Spasojević, I. UV-irradiation provokes generation of superoxide on cell wall polygalacturonic acid. Physiol. Plant. 2013, 148, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Kalantari, S.; Makhlouf, J.; Arul, J. Impact of UV-C Irradiation on the cell wall-degrading enzymes during ripening of tomato (Lycopersicon esculentum L.) fruit. J. Agric. Food Chem. 2000, 48, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Bu, J.; Yua, Y.; Aisikaera, G.; Yinga, T. Postharvest UV-C irradiation inhibits the production of ethylene andthe activity of cell wall-degrading enzymes during softening of tomato (Lycopersicon esculentum L.) fruit. Postharvest Biol. Technol. 2013, 86, 337–345. [Google Scholar] [CrossRef]
- Yamasaki, S.; Noguchi, N.; Mimaki, K. Continuous UV-B irradiation induces morphological changes and the accumulation of polyphenolic compounds on the surface of cucumber cotyledons. J. Radiat. Res. 2007, 48, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Zagoskina, N.V.; Dubravina, G.A.; Alyavina, A.K.; Goncharuk, E.A. Effect of ultraviolet (UV-B) radiation on the formation and localization of phenolic compounds in tea plant callus cultures. Russ. J. Plant Phys. 2003, 50, 270–275. [Google Scholar] [CrossRef]
- Hilal, M.; Parrado, M.F.; Rosa, M.; Gallardo, M.; Orce, L.; Massa, E.D.; González, J.A.; Prado, F.E. Epidermal lignin deposition in quinoa cotyledons in response to UV-B radiation. Photochem. Photobiol. 2004, 79, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Pontin, M.A.; Piccoli, P.N.; Francisco, R.; Bottini, R.; Martinez-Zapater, J.M.; Lijavetzky, D. Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol. 2010, 10, 224–237. [Google Scholar] [CrossRef]
- Yokawa, K.; Baluška, F. Pectins, ROS homeostasis and UV-B responses in plant roots. Phytochemitry 2014, in press. [Google Scholar]
- Lindroth, R.L. Impacts of elevated atmospheric CO2 and O3 on forests: Phytochemistry, trophic interactions, and ecosystem dynamics. J. Chem. Ecol. 2010, 36, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Munné-Bosch, S.; Queval, G.; Foyer, C.H. The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol. 2013, 16, 5–19. [Google Scholar] [CrossRef]
- Betzelberger, A.M.; Yendrek, C.R.; Sun, J.; Leisner, C.P.; Nelson, R.L.; Ort, D.R.; Ainsworth, E.A. Ozone exposure response for U.S. soybean cultivars: Linear reductions in photosynthetic potential, biomass and yield. Plant Physiol. 2012, 160, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Emberson, L.D.; Bueker, P.; Ashmore, M.R.; Mills, G.; Jackson, L.S.; Agrawal, M.; Atikuzzaman, M.D.; Cinderby, S.; Engardt, M.; Jamir, C.; et al. A comparison of North American and Asian exposure-response data for ozone effects on crop yields. Atmos. Environ. 2009, 43, 1945–1953. [Google Scholar] [CrossRef]
- Wilkinson, S.; Davies, W.J. Drought, ozone, ABA and ethylene: New insights from cell to plant to community. Plant Cell Environ. 2010, 33, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Racherla, P.N.; Adams, P.J. The response of surface ozone to climate change over the eastern United States. Atmos. Chem. Phys. 2008, 8, 871–885. [Google Scholar] [CrossRef]
- Günthardt-Goerg, M.S.; Mc Quattie, C.J.; Scheidegger, C.; Rhiner, C.; Matyssek, R. Ozone-induced cytochemical and ultrastructural changes in leaf mesophyll cell walls. Can. J. For. Res. 1997, 27, 453–463. [Google Scholar] [CrossRef]
- Günthardt-Goerg, M.S. Different responses to ozone of tobacco, poplar, birch, and alder. J. Plant Physiol. 1996, 148, 207–214. [Google Scholar] [CrossRef]
- Paoletti, E.; de Marco, A.; Beddows, D.C.S.; Harrison, R.M.; Manning, W.J. Ozone levels in European and USA cities are increasing more than at rural sites, while peak values are decreasing. Environ. Pollut. 2014, 192, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, E.; Contran, N.; Bernasconi, P.; Günthardt-Goerg, M.S.; Wollenveider, P. Structural and physiological responses to ozone in Manna ash (Fraxinus ornus L.) leaves of seedlings and mature trees under controlled and ambient condition. Sci. Total Environ. 2010, 408, 2014–2024. [Google Scholar] [CrossRef] [PubMed]
- Richet, N.; Afif, D.; Huber, F.; Pollet, B.; Banvoy, J.; el Zein, R.; Lapierre, C.; Dizengremel, P.; Perré, P.; Cabané, M. Cellulose and lignin biosynthesis is altered by ozone in wood of hybrid poplar (Populus tremula × alba). J. Exp. Bot. 2011, 62, 3575–3586. [Google Scholar] [CrossRef] [PubMed]
- Bussotti, F.; Agati, G.; Desotgiu, R.; Matteini, P.; Tani, C. Ozone foliar symptoms in woody plant species assessed with ultrastructural and fluorescence analysis. New Phytol. 2005, 166, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Bussotti, F.; Pancrazi, M.; Matteucci, G.; Gerosa, G. Leaf morphology and chemistry in Fagus sylvatica L. (beech) trees as affected by site factors and ozone: Results from Level II permanent monitoring plots in Italy. Tree Physiol. 2005, 25, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Keutgen, A.J.; Pawelzik, E. Contribution of amino acids to strawberry fruit quality and their relevance as stress indicators under NaCl salinity. Food Chem. 2008, 111, 642–647. [Google Scholar] [CrossRef]
- Kanter, U; Heller, W.; Durner, J.; Winkler, J.B.; Engel, M.; Behrendt, H.; Holzinger, A.; Braun, P.; Hauser, H.; Ferreira, F.; et al. Molecular and immunological characterization of ragweed (Ambrosia artemisiifolia L.) pollen after exposure of the plants to elevated ozone over a whole growing season. PLoS One 2013, 8, e61518. [Google Scholar] [CrossRef] [PubMed]
- Minas, I.S.; Vicente, O.R.; Dhanapal, O.P.; Manganaris, G.A.; Goulas, V.; Vasilakakis, M.; Crisosto, H.; Molassiotis, A. Ozone-induced kiwifruit ripening delay is mediated by ethylene biosynthesis inhibition and cell wall dismantling regulation. Plant Sci. 2014, 29, 76–85. [Google Scholar] [CrossRef]
- Oksanen, E.; Häikiö, E.; Sober, J.; Karnosky, D.F. Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol. 2003, 161, 791–799. [Google Scholar] [CrossRef]
- Cho, K.; Shibato, J.; Kubo, A.; Kohno, Y.; Satoh, K.; Kikuchi, S.; Sarkar, A.; Agrawal, G.K.; Rakwal, R. Comparative analysis of seed transcriptomes of ambient ozone-fumigated two different rice cultivars. Plant Signal Behav. 2013, 8, e26300. [Google Scholar] [CrossRef] [PubMed]
- Cabané, M.; Pireaux, J.C.; Leger, E.; Weber, E.; Dizengremel, P.; Pollet, B.; Lapierre, C. Condensed lignins are synthesized in poplar leaves exposed to ozone. Plant Physiol. 2004, 134, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.K.; Davis, K.R. Ozone-induced expression of stress-related genes in Arabidopsis thaliana. Plant Physiol. 1994, 105, 1089–1096. [Google Scholar] [PubMed]
- Eckey-Kaltenbach, H.; Ernst, D.; Heller, W.; Sandermann, H. Biochemical plant responses to ozone: IV. Cross-induction of defensive pathways in parsley (Petroselinum crispum L.) plants. Plant Physiol. 1994, 104, 67–74. [Google Scholar] [PubMed]
- Wiese, C.B.; Pell, E.J. Oxidative modification of the cell wall in tomato plants exposed to ozone. Plant Physiol. Biochem. 2003, 41, 375–382. [Google Scholar] [CrossRef]
- Ranieri, A.; Castagna, A.; Pacini, J.; Baldan, B.; Mensuali Sodi, A.; Soldatini, G.F. Early production and scavenging of hydrogen peroxide in the apoplast of sunflowers plants exposed to ozone. J. Exp. Bot. 2003, 54, 2529–2540. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, A.; Castagna, A.; Soldatini, G.F. Differential stimulation of ascorbate peroxidase isoforms by ozone exposure in sunflower plants. J. Plant Physiol. 2000, 156, 266–271. [Google Scholar] [CrossRef]
- Keutgen, A.J.; Pawelzik, E. Apoplastic antioxidative system responses to ozone stress in strawberry leaves. J. Plant Physiol. 2008, 165, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Meehl, G.A.; Stocker, W.D.; Collins, P.; Friedlingstein, A.T.; Gaye, J.M.; Gregory, A.; Kitoh, R.; Knutti, J.M.; Murphy, A.; Noda, S.C.B.; et al. Global Climate Projections. In Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., Averyt, K.B., Tignor, M., Miller, H.L., Eds.; Cambridge University Press: Cambridge, UK, New York, NY, USA, 2007. [Google Scholar]
- Leakey, A.D.B.; Bernacchi, C.J.; Dohleman, F.G.; Ort, D.R.; Long, S.P. Will photosynthesis of maize (Zea mays) in the US Corn Belt increase in future CO2 rich atmospheres? An analysis of diurnal courses of CO2 uptake under free-air concentration enrichment (FACE). Glob. Chang. Biol. 2004, 10, 951–962. [Google Scholar] [CrossRef]
- De Souza, A.P.; Arundale, R.A.; Dohleman, F.G.; Long, S.P.; Buckeridge, M.S. Will the exceptional productivity of Miscanthus x giganteus increase further under rising atmospheric CO2? Agricult. For. Meteorol. 2013, 171, 82–92. [Google Scholar] [CrossRef]
- King, J.S.; Pregitzer, K.S.; Zak, D.R.; Karnosky, D.F.; Isebrands, J.G.; Dickson, R.E.; Hendrey, G.R.; Sober, J. Fine root biomass and fluxes of soil carbon in young stands of paper birch and trembling aspen as affected by elevated atmospheric CO2 and tropospheric O3. Oecologia 2001, 128, 237–250. [Google Scholar] [CrossRef]
- Oksanen, E.; Sôber, J.; Karnosky, D.F. Interactions of elevated CO2 and ozone in leaf morphology of aspen (Populus tremuloides) and birch (Betula papyrifera) in aspen FACE experiment. Environ. Pollut. 2001, 115, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Kubiske, M.E.; Quinn, V.S.; Marquart, P.E.; Karnosky, D.F. Effects of elevated atmospheric CO2 and/or O3 on intra- and inter-specific competitive ability of aspen. Plant Biol. 2007, 9, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Cseke, L.J.; Tsai, C.J.; Rogers, A.; Nelsen, M.P.; White, H.L.; Karnosky, D.F.; P Odila, G.K. Transcriptomic comparison in the leaves of two aspen genotypes having similar carbon assimilation rates but different partitioning patterns under elevated (CO2). New Phytol. 2009, 182, 891–911. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Gou, J.; Yordanov, Y.; Zhang, H.; Thakur, R.; Jones, W.; Burton, A. Global transcriptome profiling of aspen trees under elevated (CO2) for identifying potential molecular mechanisms responsible for enhanced growth. J. Plant Res. 2013, 126, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Kostiainen, K.; Saranpää, P.; Lundqvist, S.O.; Kubiske, M.E.; Vapaavuori, E. Wood properties of Populus and Betula in long-term exposure to elevated CO2 and O3. Plant Cell Environ. 2014, 37, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Labbé, N.; Warren, J.M.; Elder, T.; Rials, T.G. Chemical and anatomical changes in Liquidambar styraciflua L. xylem after long term exposure to elevated CO2. Environ. Pollut. 2015, 17, 179–185. [Google Scholar] [CrossRef]
- Gibeaut, D.M.; Cramer, G.R.; Seemann, J.R. Growth, cell walls, and UDP-Glc dehydrogenase activity of Arabidopsis thaliana grown in elevated carbon dioxide. J. Plant Physiol. 2001, 158, 569–576. [Google Scholar] [CrossRef]
- Duan, Z.; Homma, A.; Kobayashi, M.; Nagata, M.; Kaneko, Y.; Fujiki, Y.; Nishida, I. Photoassimilation assimilate translocation and plasmodesmal biogenesis in the source leaves of Arabidopsis thaliana grown under an increased atmospheric CO2 concentration. Plant Cell. Physiol. 2014, 55, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Perez-Lopez, U.; Robredo, A.; Lacuesta, M.; Munoz-Rueda, A.; Mena-Petite, A. Atmospheric CO2 concentration influences the contributions of osmolyte accumulation and cell wall elasticity to salt tolerance in barley cultivars. J. Plant Physiol. 2010, 167, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Huyskens-Keil, S.; Herppich, W. High CO2 effects on postharvest biochemical and textural properties of white asparagus (Asparagus officinalis L.) spears. Postharvest Biol. Technol. 2013, 75, 45–53. [Google Scholar] [CrossRef]
- Chen, S.; Olbrich, A.; Langenfeld-Heyser, R.; Fritz, E.; Polle, A. Quantitative X-ray microanalysis of hydrogen peroxide within plant cells. Microsc. Res. Techniq. 2009, 72, 49–60. [Google Scholar] [CrossRef]
- Ookawara, R.; Satoh, S.; Yoshioka, T.; Ishizawa, K. Expression of α-expansin and xyloglucan endotransglucosylase/hydrolase genes associated with shoot elongation enhanced by anoxia, ethylene and carbon dioxide in arrowhead (Sagittaria pygmaea Miq.) tubers. Ann. Bot. 2005, 96, 693–702. [Google Scholar] [CrossRef] [PubMed]
- May, P.; Liao, W.; Wu, Y.; Shuai, B.; Mc Combie, W.R.; Zhang, M.Q.; Liu, Q.A. The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, M.; Velasquez, S.M.; Jamet, E.; Estevez, J.M.; Albenne, C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: Toward a model highlighting their contribution to plant cell wall architecture. Front. Plant Sci. 2014, 5, 395. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.J.; Xue, H.; Acet, T. The Arabidopsis thaliana fasciclin like arabinogalactan protein 4 gene acts synergistically with abscisic acid signaling to control root growth. Ann. Bot. 2014, 114, 1125–1133. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112-166. https://doi.org/10.3390/plants4010112
Le Gall H, Philippe F, Domon J-M, Gillet F, Pelloux J, Rayon C. Cell Wall Metabolism in Response to Abiotic Stress. Plants. 2015; 4(1):112-166. https://doi.org/10.3390/plants4010112
Chicago/Turabian StyleLe Gall, Hyacinthe, Florian Philippe, Jean-Marc Domon, Françoise Gillet, Jérôme Pelloux, and Catherine Rayon. 2015. "Cell Wall Metabolism in Response to Abiotic Stress" Plants 4, no. 1: 112-166. https://doi.org/10.3390/plants4010112
APA StyleLe Gall, H., Philippe, F., Domon, J.-M., Gillet, F., Pelloux, J., & Rayon, C. (2015). Cell Wall Metabolism in Response to Abiotic Stress. Plants, 4(1), 112-166. https://doi.org/10.3390/plants4010112