Advancing Sustainable Agriculture: Molecular and Physiological Insights into Rapeseed Responsiveness to Organic Amendment Fertilization
Abstract
1. Introduction
2. Results
2.1. Plant Growth and Nitrogen Content
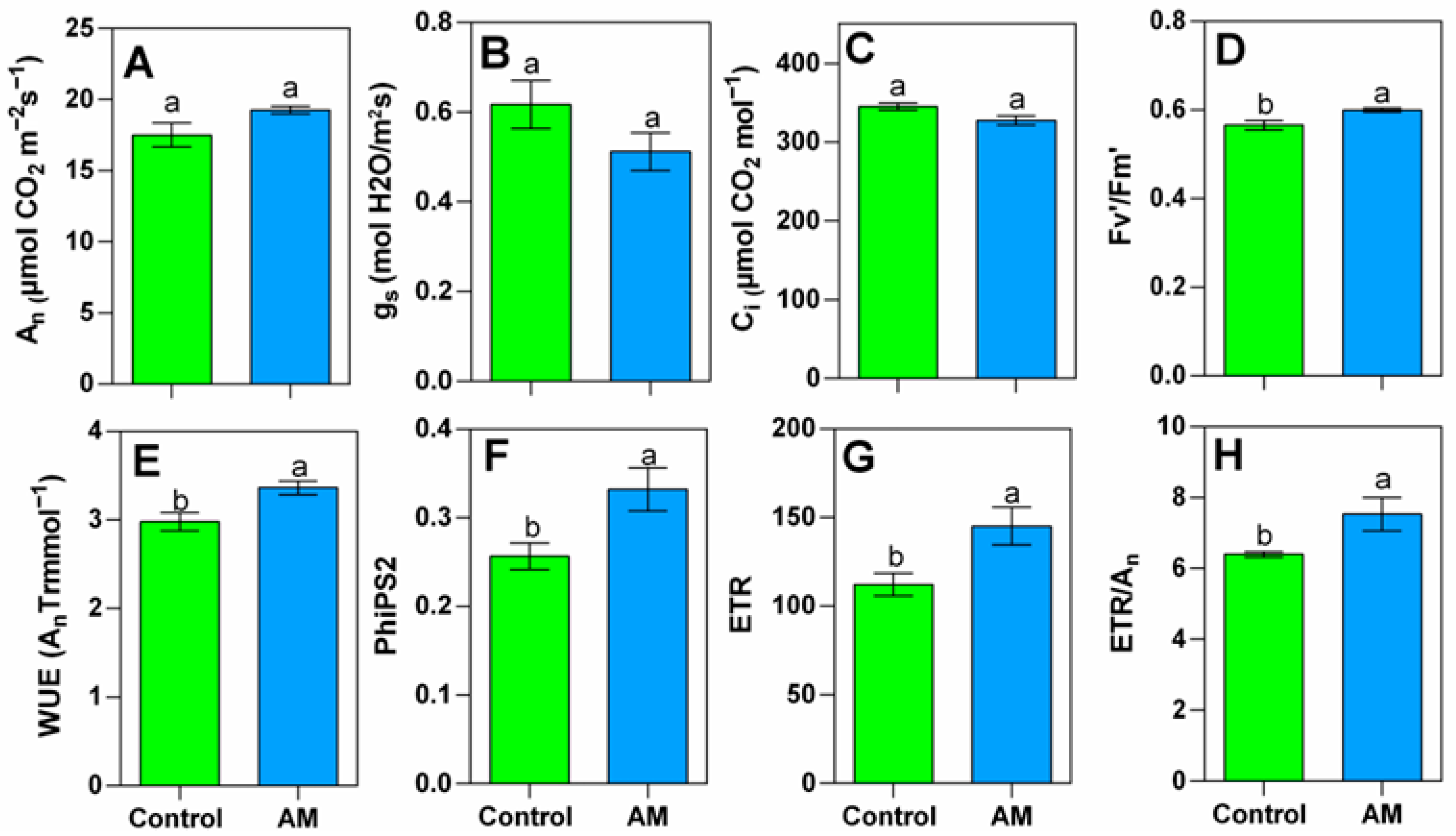
2.2. Gas Exchange and Fluorescence Determination
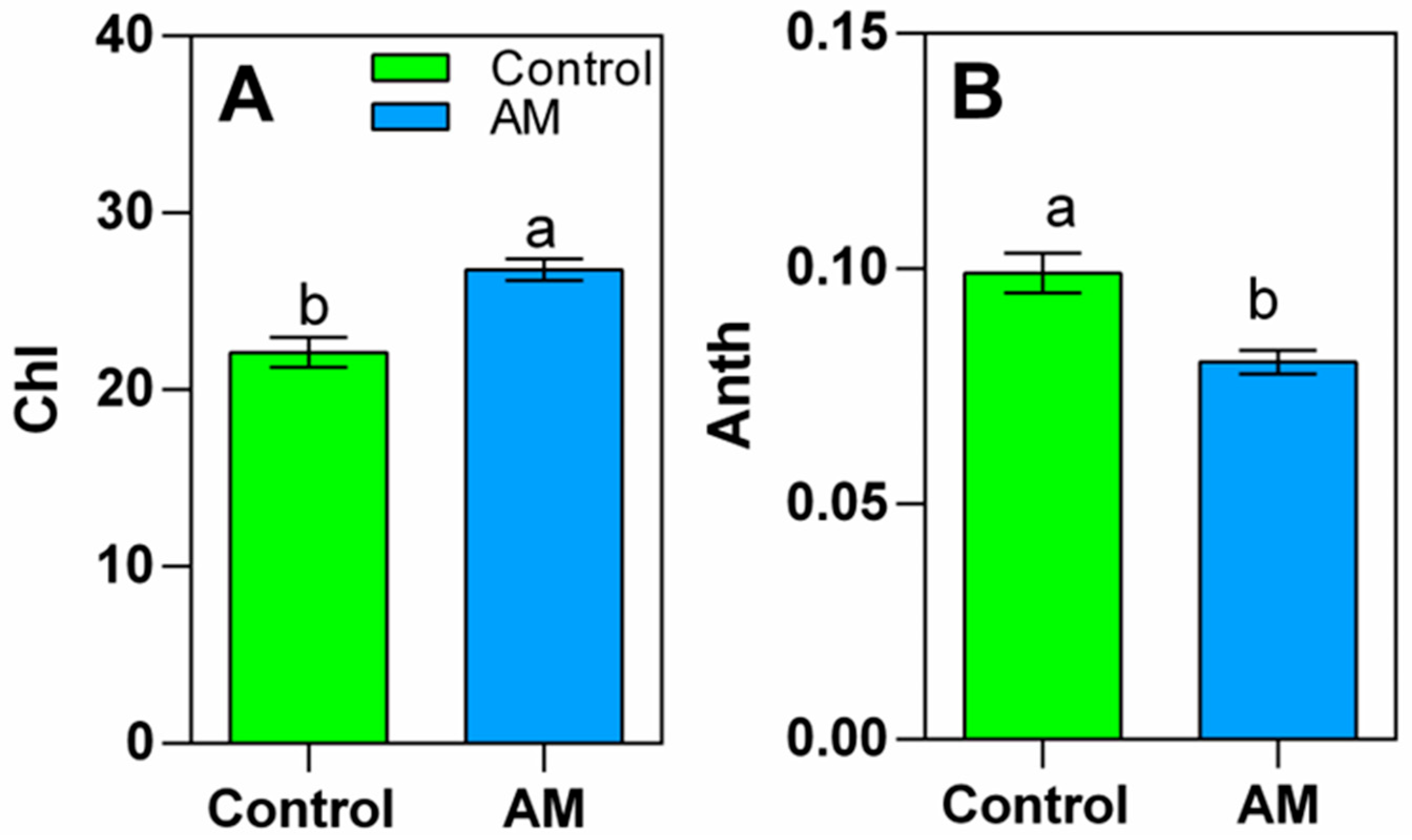
2.3. Chlorophyll and Anthocyanin Content
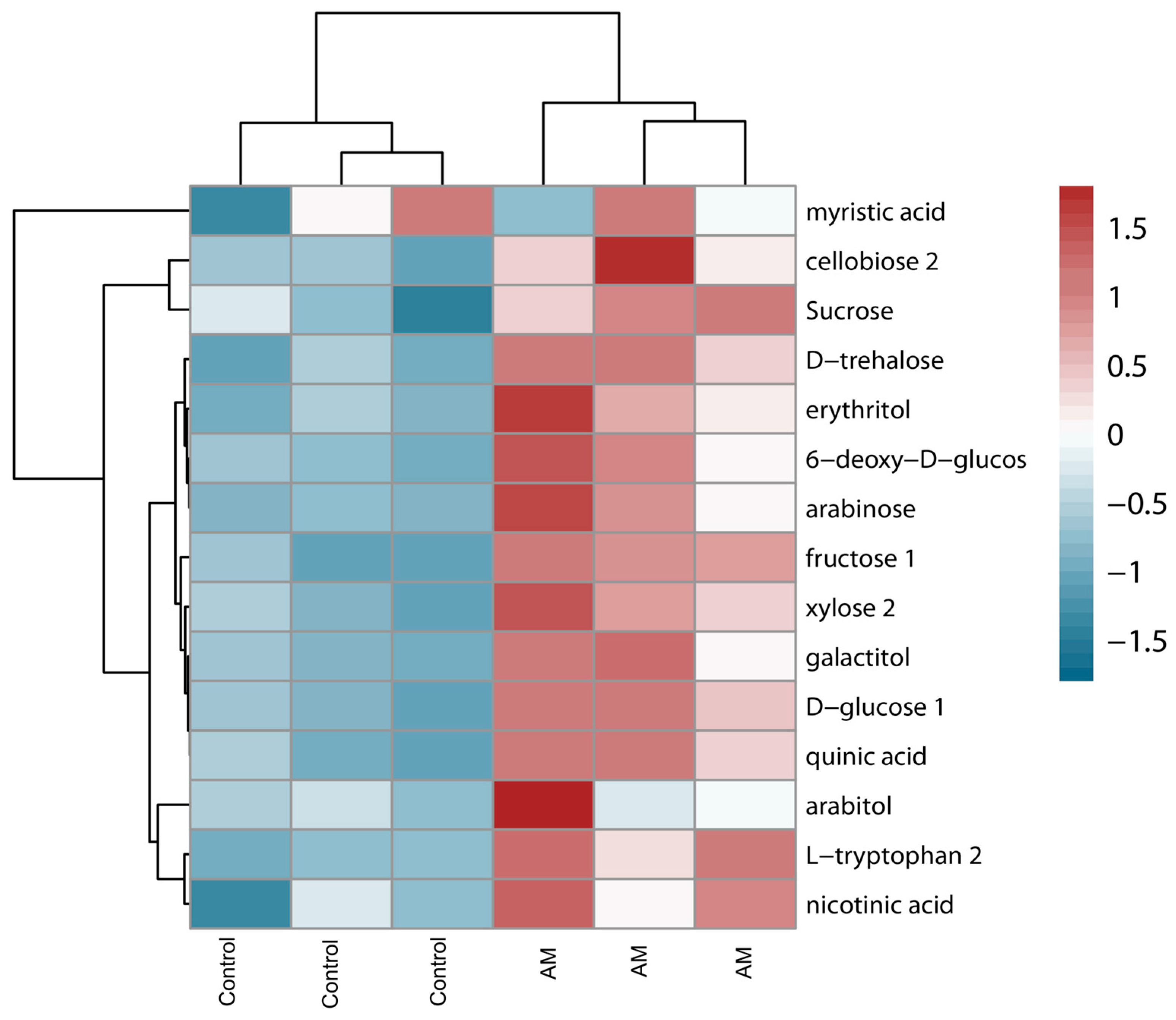
2.4. Plant Metabolism Analyses
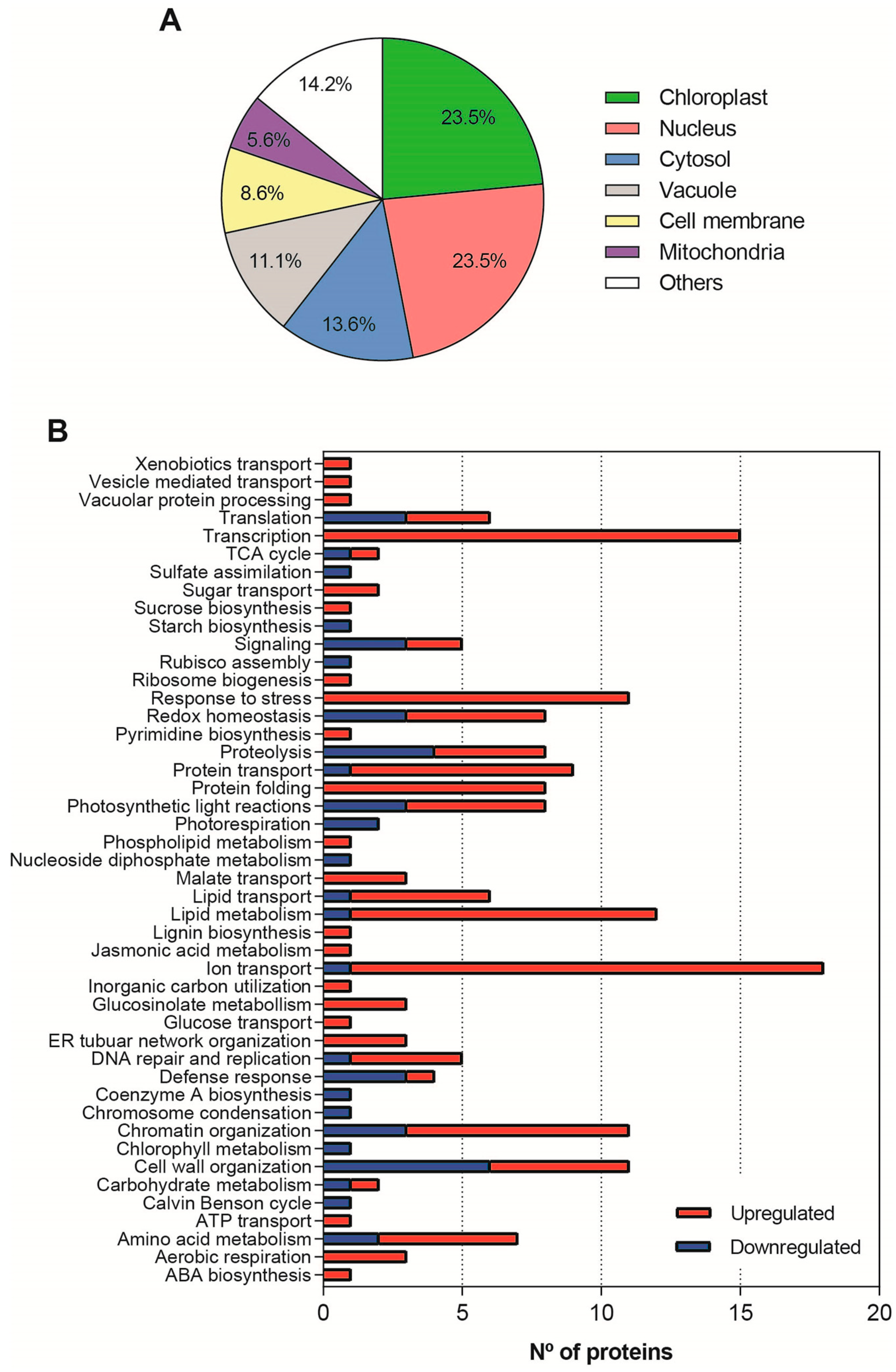
2.5. Proteomic Analyses
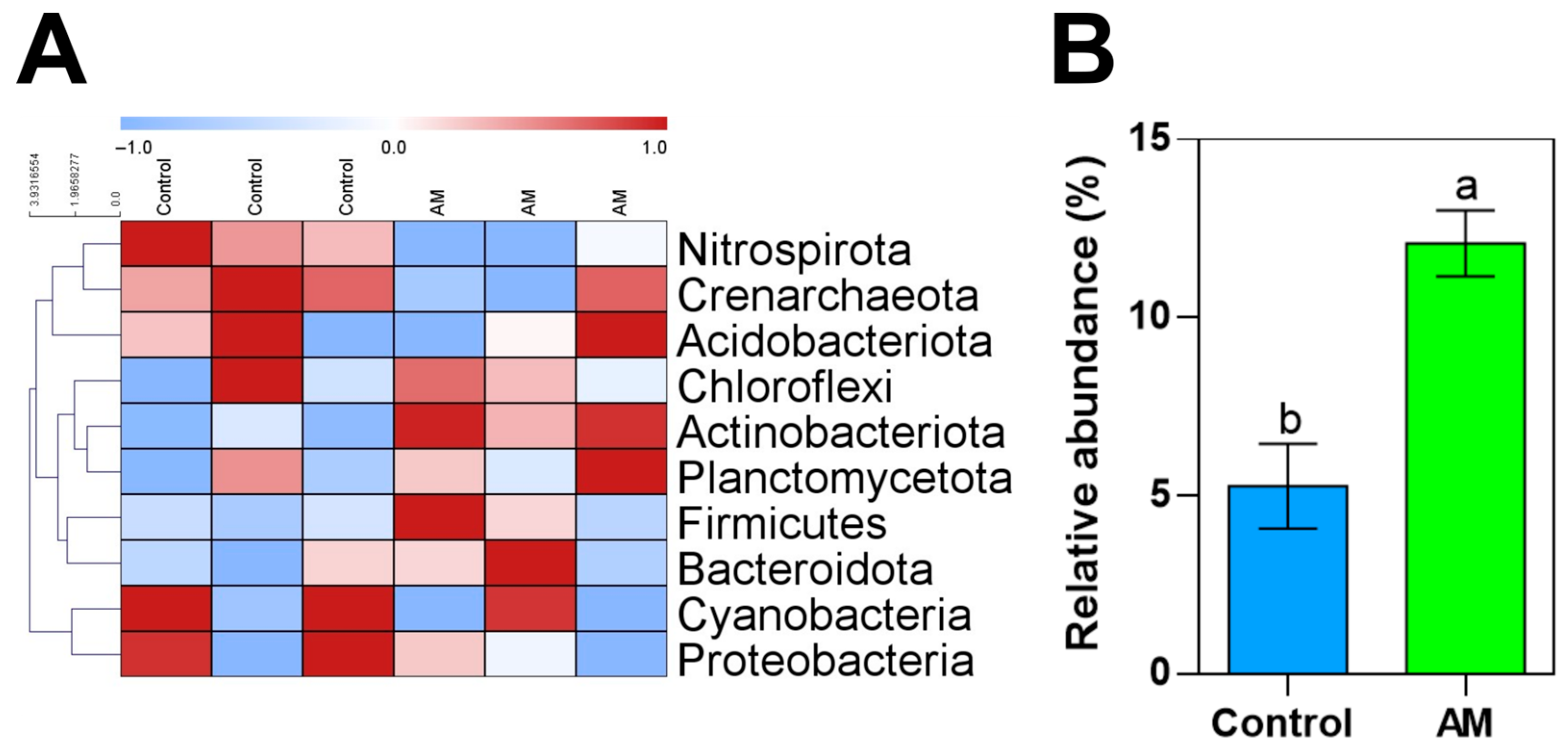
2.6. Soil Bacterial Analyses
3. Discussion
3.1. Amendment Application Contributes to Increase Leaf C and N Metabolism
3.2. Improved Oxidative Stress Regulation Mechanisms
3.3. Impact of Organic Amendment in Soil Microbiota
4. Materials and Methods
4.1. Plant Growth and Experimental Design
4.2. N Content
4.3. Gas Exchange and Chlorophyll Fluorescence
4.4. Chlorophyll and Anthocyanin Content Analysis
4.5. Proteomic Profile
4.6. Gas Chromatography–Mass Spectrometry (GC-MS) Analyses
4.7. Genomic Analyses of Soil Bacteria
4.8. ABA Quantification
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| An | Net photosynthetic CO2 assimilation |
| gs | Stomatal conductance |
| Ci | Sub-stomatal CO2 concentration |
| E | Transpiration rate |
| PPFD | Photosynthetic photon flux density |
| ETR | Electron transport rate |
| Fv’/Fm’ | Intrinsic quantum yield of PSII photochemistry |
| PhiPS2 | Photosystem II efficiency |
| WUE | Water use efficiency |
| Chl | Chlorophyll |
| Anth | Anthocyanin |
| ROS | Reactive oxygen species |
| TCA | Tricarboxylic acid |
| ppm | Part per million |
| CO2 | Carbon dioxide |
| ICP/OES | Inductively coupled plasma/optical emission spectrometry |
| N | Nitrogen |
| C | Carbon |
| ATP | Adenosine triphosphate |
| ADP | Adenosine diphosphate |
| DNA | Deoxyribonucleic acid |
| OTUs | Observed operational taxonomic units |
| AM, Amen | Organic amended |
| t-test | Student’s t-test |
Appendix A
| Accession | Protein Name | Unique Peptides | Localization | Function | Abundance Ratio | Abundance Ratio (log2) | p-Value |
|---|---|---|---|---|---|---|---|
| A0A078H9F9 | Reticulon-like protein | 1 | Endoplasmic reticulum | ER tubular network organization | 3.06 | 1.61 | 2.64 × 10−15 |
| A0A078GHF0 | Reticulon-like protein | 3 | Endoplasmic reticulum | ER tubular network organization | 2.547 | 1.35 | 2.64 × 10−15 |
| A0A078FBI2 | BnaCnng02600D protein | 3 | - | Response to stress | 2.403 | 1.27 | 2.64 × 10−15 |
| A0A078IVR4 | FACT complex subunit | 1 | Nucleus | DNA repair and replication | 2.414 | 1.27 | 2.17 × 10−12 |
| A0A078I4R4 | Reticulon-like protein | 3 | Endoplasmic reticulum | ER tubular network organization | 2.318 | 1.21 | 2.64 × 10−15 |
| A0A078GVS3 | Imidazole glycerol phosphate synthase hisHF | 1 | Chloroplast | Amino acid metabolism | 2.272 | 1.18 | 3.57 × 10−11 |
| A0A078IXL1 | BnaC08g47240D protein | 1 | Nucleus | Transcription | 2.185 | 1.13 | 1.19 × 10−10 |
| A0A078ILU3 | (rape) hypothetical protein | 1 | - | Defense response | 2.12 | 1.08 | 8.22 × 10−10 |
| A0A078FR89 | BnaA01g34270D protein | 3 | Mitochondria | Aerobic respiration | 2.096 | 1.07 | 2.64 × 10−15 |
| A0A078ICU7 | (rape) hypothetical protein | 3 | Nucleus | Transcription | 2.067 | 1.05 | 2.64 × 10−15 |
| A0A078IB52 | (rape) hypothetical protein | 3 | - | Unknown | 2.072 | 1.05 | 2.64 × 10−15 |
| A0A078IZU4 | (rape) hypothetical protein | 3 | - | Protein folding | 2.055 | 1.04 | 2.64 × 10−15 |
| A0A078G2L0 | NAD(P)H-quinone oxidoreductase subunit 5 | 6 | Chloroplast | Photosynthetic light reactions | 1.981 | 0.99 | 2.64 × 10−15 |
| A0A078G3V0 | BnaA09g01870D protein | 1 | Vacuole | Protein transport | 1.975 | 0.98 | 2.78 × 10−12 |
| A0A078J4S6 | BnaA09g54590D protein | 1 | Nucleus | Transcription | 1.976 | 0.98 | 1.51 × 10−9 |
| A0A078IQJ7 | (rape) hypothetical protein | 1 | - | Protein folding | 1.964 | 0.97 | 2.64 × 10−15 |
| A0A078I462 | (rape) hypothetical protein | 2 | - | Protein folding | 1.932 | 0.95 | 2.64 × 10−15 |
| A0A078FPS6 | (rape) hypothetical protein | 3 | - | Unknown | 1.938 | 0.95 | 7.82 × 10−12 |
| A0A078GHF6 | (rape) hypothetical protein | 2 | Nucleus | Transcription | 1.889 | 0.92 | 2.64 × 10−15 |
| A0A078FU32 | BnaA08g02930D protein | 3 | Nucleus | Transcription | 1.882 | 0.91 | 2.64 × 10−15 |
| A0A078G143 | (rape) hypothetical protein | 1 | Chloroplast | Amino acid metabolism | 1.852 | 0.89 | 2.64 × 10−15 |
| A0A078G4P9 | BnaC09g29190D protein | 6 | - | Aerobic respiration | 1.816 | 0.86 | 2.64 × 10−15 |
| A0A078FCG8 | (rape) hypothetical protein | 15 | Cell membrane | Ion transport | 1.818 | 0.86 | 2.64 × 10−15 |
| A0A078J890 | (rape) hypothetical protein | 8 | Nucleus | Unknown | 1.773 | 0.83 | 2.64 × 10−15 |
| A0A078JLP0 | (rape) hypothetical protein | 13 | Cytosol | Response to stress | 1.766 | 0.82 | 2.64 × 10−15 |
| A0A078HZ14 | BnaC08g10640D protein | 8 | - | Protein transport | 1.737 | 0.8 | 2.64 × 10−15 |
| A0A078H4V5 | (rape) hypothetical protein | 5 | Cytosol | Response to stress | 1.713 | 0.78 | 2.64 × 10−15 |
| A0A078I5S5 | (rape) hypothetical protein | 1 | - | Response to stress | 1.715 | 0.78 | 3.20 × 10−5 |
| A0A078H252 | (rape) hypothetical protein | 4 | Nucleus | Chromatin organization | 1.696 | 0.76 | 2.64 × 10−15 |
| A0A078GEQ7 | BnaC04g01660D protein | 1 | - | Unknown | 1.679 | 0.75 | 5.21 × 10−4 |
| A0A078EZB5 | (rape) hypothetical protein | 1 | - | Unknown | 1.668 | 0.74 | 4.50 × 10−9 |
| A0A078FNQ6 | BnaC08g40680D protein | 2 | - | Unknown | 1.664 | 0.73 | 2.44 × 10−5 |
| Q69BP4 | (rape) hypothetical protein | 13 | Cytosol | Response to stress | 1.646 | 0.72 | 2.64 × 10−15 |
| A0A078IXP0 | (rape) hypothetical protein | 3 | - | Amino acid metabolism | 1.653 | 0.72 | 2.25 × 10−12 |
| A0A078JHC6 | BnaCnng49050D protein | 1 | Nucleus | Chromatin organization | 1.649 | 0.72 | 2.34 × 10−8 |
| A0A078J5L0 | (rape) hypothetical protein | 1 | Cytosol | Unknown | 1.635 | 0.71 | 1.52 × 10−4 |
| A0A078IX55 | (rape) hypothetical protein | 4 | Chloroplast | Malate transport | 1.625 | 0.7 | 2.64 × 10−15 |
| A0A078FBT9 | (rape) hypothetical protein | 3 | - | Unknown | 1.625 | 0.7 | 2.97 × 10−9 |
| A0A078H492 | BnaA06g36180D protein | 5 | Chloroplast | Unknown | 1.615 | 0.69 | 2.64 × 10−15 |
| A0A078FIZ4 | CDP-diacylglycerol--inositol 3-phosphatidyltransferase | 2 | Golgi apparatus | Phospholipid metabolism | 1.614 | 0.69 | 2.94 × 10−5 |
| A0A078F8Q4 | (rape) hypothetical protein | 2 | Nucleus | Transcription | 1.601 | 0.68 | 5.55 × 10−14 |
| A0A078I5B1 | (rape) hypothetical protein | 2 | Endoplasmic reticulum | Lipid metabolism | 1.606 | 0.68 | 6.55 × 10−11 |
| A0A078JJ05 | (rape) hypothetical protein | 1 | - | Unknown | 1.603 | 0.68 | 6.55 × 10−11 |
| A0A078JNA0 | BnaCnng57110D protein (Fragment) | 1 | Cell membrane | Ion transport | 1.603 | 0.68 | 8.62 × 10−4 |
| A0A078G3S7 | BnaC09g29130D protein | 25 | Chloroplast | Protein transport | 1.595 | 0.67 | 2.64 × 10−15 |
| A0A078FUQ6 | (rape) hypothetical protein | 9 | Nucleus | Chromatin organization | 1.583 | 0.66 | 2.64 × 10−15 |
| A0A078G765 | BnaC02g28790D protein | 2 | Nucleus | Transcription | 1.584 | 0.66 | 4.93 × 10−8 |
| A0A078FQZ2 | (rape) hypothetical protein | 2 | Chloroplast | Unknown | 1.575 | 0.66 | 2.96 × 10−7 |
| A0A078H3B2 | (rape) hypothetical protein | 1 | Chloroplast | Malate transport | 1.582 | 0.66 | 1.42 × 10−5 |
| A0A078FCS9 | BnaC06g23930D protein | 1 | - | Response to stress | 1.556 | 0.64 | 7.04 × 10−7 |
| A0A078IBS7 | (rape) hypothetical protein | 5 | - | Unknown | 1.553 | 0.64 | 8.37 × 10−7 |
| A0A078FLH2 | (rape) hypothetical protein | 1 | - | Lipid metabolism | 1.559 | 0.64 | 1.75 × 10−3 |
| A0A078I8F7 | (rape) hypothetical protein | 3 | - | Response to stress | 1.551 | 0.63 | 1.07 × 10−13 |
| A0A078I275 | BnaC08g04910D protein | 1 | - | Lipid metabolism | 1.543 | 0.63 | 2.59 × 10−13 |
| A0A078HYY2 | (rape) hypothetical protein | 3 | Chloroplast | Malate transport | 1.548 | 0.63 | 2.59 × 10−13 |
| A0A078HNK9 | Protein-serine/threonine phosphatase | 3 | - | Signaling | 1.543 | 0.63 | 3.46 × 10−10 |
| A0A078FGL5 | (rape) hypothetical protein | 3 | Endoplasmic reticulum | Protein transport | 1.551 | 0.63 | 3.39 × 10−6 |
| A0A078HQG5 | (rape) hypothetical protein | 4 | Nucleus | Transcription | 1.534 | 0.62 | 1.71 × 10−8 |
| A0A078J925 | BnaC07g48010D protein | 3 | Apoplast | Unknown | 1.537 | 0.62 | 2.62 × 10−8 |
| A0A078H561 | BnaA10g12120D protein | 7 | - | Glucose transport | 1.525 | 0.61 | 1.30 × 10−12 |
| A0A078GMS3 | (rape) hypothetical protein | 4 | - | Carbohydrate metabolism | 1.522 | 0.61 | 3.08 × 10−6 |
| A0A078HE96 | BnaC02g38300D protein | 3 | Cell membrane | Unknown | 1.516 | 0.6 | 5.67 × 10−6 |
| A0A078GWK5 | BnaC07g32090D protein | 1 | Chloroplast | Unknown | 1.513 | 0.6 | 3.11 × 10−5 |
| A0A078HCV4 | (rape) hypothetical protein | 1 | Cell membrane | Ion transport | 1.514 | 0.6 | 5.85 × 10−3 |
| A0A078GAB1 | BnaC09g00530D protein | 1 | - | Signaling | 1.513 | 0.6 | 7.06 × 10−3 |
| A0A078GHL0 | (rape) hypothetical protein | 1 | Vacuole | Ion transport | 1.493 | 0.58 | 1.61 × 10−10 |
| A0A078G686 | BnaC09g00810D protein | 2 | Vacuole | Ion transport | 1.498 | 0.58 | 2.05 × 10−4 |
| A0A078GKT1 | (rape) hypothetical protein | 1 | Vacuole | Ion transport | 1.494 | 0.58 | 2.06 × 10−4 |
| A0A078IXK4 | Potassium transporter | 3 | Cell membrane | Ion transport | 1.493 | 0.58 | 1.09 × 10−3 |
| A0A078FPE7 | (rape) hypothetical protein | 10 | Vacuole | Unknown | 1.489 | 0.57 | 2.85 × 10−11 |
| A0A078JC11 | BnaCnng45920D protein | 3 | Nucleus | Unknown | 1.481 | 0.57 | 5.48 × 10−8 |
| A0A078GWJ7 | BnaA06g14580D protein | 1 | Nucleus | Chromatin organization | 1.475 | 0.56 | 2.08 × 10−5 |
| A0A078F9L1 | BnaA05g27040D protein | 1 | - | Unknown | 1.478 | 0.56 | 2.40 × 10−5 |
| A0A078JHT5 | (rape) hypothetical protein | 3 | Nucleus | Transcription | 1.475 | 0.56 | 2.48 × 10−5 |
| A0A078IXW2 | (rape) hypothetical protein | 2 | - | Unknown | 1.476 | 0.56 | 6.58 × 10−5 |
| A0A078FI66 | (rape) hypothetical protein | 5 | - | Unknown | 1.469 | 0.55 | 1.60 × 10−10 |
| A0A078JGM8 | (rape) hypothetical protein | 2 | Vacuole | Sugar transport | 1.467 | 0.55 | 2.05 × 10−8 |
| A0A078HFX4 | BnaC02g31330D protein | 1 | Mitochondria | Aerobic respiration | 1.468 | 0.55 | 2.05 × 10−4 |
| F8K8N9 | (rape) hypothetical protein | 1 | Mitochondria | Ribosome biogenesis | 1.464 | 0.55 | 2.43 × 10−2 |
| A0A078HDB7 | BnaC02g09720D protein | 4 | - | Unknown | 1.449 | 0.54 | 2.23 × 10−9 |
| A0A078JBL3 | BnaC03g78270D protein | 6 | - | Response to stress | 1.454 | 0.54 | 3.09 × 10−8 |
| A0A078G8E9 | (rape) hypothetical protein | 10 | Cell membrane | Ion transport | 1.452 | 0.54 | 3.78 × 10−8 |
| A0A078F7I4 | (rape) hypothetical protein | 2 | - | Unknown | 1.455 | 0.54 | 1.05 × 10−4 |
| A0A078H8B0 | (rape) hypothetical protein | 1 | Nucleus | Transcription | 1.452 | 0.54 | 2.35 × 10−4 |
| A0A078I8I3 | (rape) hypothetical protein | 4 | - | Ion transport | 1.44 | 0.53 | 7.31 × 10−8 |
| A0A078JPD9 | Thioglucosidase (Fragment) | 2 | Vacuole | Glucosinolate metabollism | 1.444 | 0.53 | 3.50 × 10−7 |
| A0A078FS08 | (rape) hypothetical protein | 1 | - | Unknown | 1.448 | 0.53 | 6.46 × 10−3 |
| A0A078FHQ0 | BnaC09g38510D protein | 1 | - | DNA repair and replication | 1.449 | 0.53 | 1.93 × 10−2 |
| A0A078F8K3 | BnaA02g26510D protein | 4 | Nucleus | Proteolysis | 1.436 | 0.52 | 2.23 × 10−9 |
| A0A078J441 | BnaA09g52620D protein | 5 | - | Transcription | 1.43 | 0.52 | 1.46 × 10−6 |
| A0A078GNT1 | Thioglucosidase | 8 | Vacuole | Glucosinolate metabollism | 1.423 | 0.51 | 6.03 × 10−9 |
| A0A078F5Z9 | BnaA05g27150D protein | 5 | Chloroplast | Unknown | 1.422 | 0.51 | 8.02 × 10−8 |
| A0A078GQ28 | (rape) hypothetical protein | 6 | Nucleus | Chromatin organization | 1.428 | 0.51 | 1.40 × 10−6 |
| A0A078JC75 | BnaAnng17860D protein | 3 | - | Unknown | 1.42 | 0.51 | 3.65 × 10−4 |
| A0A078FS29 | (rape) hypothetical protein | 2 | - | Unknown | 1.423 | 0.51 | 1.75 × 10−3 |
| A0A078FTF1 | BnaA08g28520D protein | 1 | - | Unknown | 1.423 | 0.51 | 9.96 × 10−3 |
| A0A078J084 | (rape) hypothetical protein | 2 | - | Unknown | 1.429 | 0.51 | 1.83 × 10−2 |
| A0A078HV25 | BnaC04g00810D protein | 4 | Nucleus | Transcription | 1.416 | 0.5 | 1.59 × 10−4 |
| A0A078DH63 | (rape) hypothetical protein | 2 | Endosome | Vesicle mediated transport | 1.41 | 0.5 | 2.86 × 10−4 |
| A0A078IVG4 | (rape) hypothetical protein | 2 | - | Unknown | 1.407 | 0.49 | 4.72 × 10−6 |
| A0A078FPB0 | Non-specific lipid-transfer protein | 2 | - | Lipid transport | 1.402 | 0.49 | 5.61 × 10−6 |
| A0A078FTJ8 | Chloride channel protein | 4 | - | Ion transport | 1.405 | 0.49 | 5.61 × 10−6 |
| A0A078FQK8 | (rape) hypothetical protein | 4 | - | Unknown | 1.398 | 0.48 | 3.91 × 10−8 |
| A0A078HHL6 | Thioglucosidase | 13 | Vacuole | Glucosinolate metabollism | 1.397 | 0.48 | 4.35 × 10−8 |
| A0A078G2E4 | BnaC05g13830D protein | 8 | - | Unknown | 1.395 | 0.48 | 6.17 × 10−8 |
| A0A078IY82 | (rape) hypothetical protein | 5 | - | Response to stress | 1.392 | 0.48 | 8.37 × 10−7 |
| A0A078IZ88 | (rape) hypothetical protein | 2 | Nucleus | Transcription | 1.39 | 0.48 | 1.92 × 10−6 |
| A0A078IJX2 | (rape) hypothetical protein | 4 | - | Unknown | 1.393 | 0.48 | 3.16 × 10−6 |
| A0A078ITH8 | BnaA09g15670D protein | 1 | - | Unknown | 1.398 | 0.48 | 4.46 × 10−3 |
| A0A078I5M0 | BnaA06g34100D protein | 1 | - | Proteolysis | 1.391 | 0.48 | 2.36 × 10−2 |
| A0A078G4P5 | (rape) hypothetical protein | 10 | Chloroplast | Protein transport | 1.382 | 0.47 | 8.88 × 10−7 |
| A0A078HST0 | BnaC05g43420D protein | 1 | - | Translation | 1.387 | 0.47 | 3.40 × 10−3 |
| A0A078IF26 | 1,3-beta-glucan synthase | 4 | Cell membrane | Cell wall organization | 1.373 | 0.46 | 4.68 × 10−4 |
| A0A078FZE9 | BnaA06g35320D protein | 13 | Nucleus | DNA repair and replication | 1.367 | 0.45 | 2.52 × 10−7 |
| A0A078IID2 | BnaA02g19360D protein | 6 | - | Transcription | 1.368 | 0.45 | 3.93 × 10−5 |
| A0A078I4Y5 | Germin-like protein | 4 | Apoplast | Unknown | 1.371 | 0.45 | 9.36 × 10−4 |
| A0A078GN58 | BnaA06g13580D protein | 2 | Vacuole | Sugar transport | 1.365 | 0.45 | 1.49 × 10−3 |
| A0A078HR75 | (rape) hypothetical protein | 3 | Chloroplast | Unknown | 1.368 | 0.45 | 1.75 × 10−3 |
| A0A078GQ62 | V-type proton ATPase subunit G | 1 | Vacuole | Ion transport | 1.365 | 0.45 | 2.56 × 10−2 |
| A0A078JMC4 | BnaA09g54410D protein | 5 | - | Unknown | 1.36 | 0.44 | 2.52 × 10−7 |
| A0A078G5T7 | (rape) hypothetical protein | 13 | Nucleus | Chromatin organization | 1.359 | 0.44 | 7.62 × 10−7 |
| A0A078FZ88 | T-complex protein 1 subunit delta | 1 | Cytosol | Protein folding | 1.356 | 0.44 | 2.00 × 10−4 |
| A0A078GQS7 | (rape) hypothetical protein | 2 | - | Unknown | 1.359 | 0.44 | 9.40 × 10−3 |
| A0A078HF75 | (rape) hypothetical protein | 1 | Nucleus | Unknown | 1.352 | 0.44 | 1.50 × 10−2 |
| A0A078HWR7 | Endoplasmic reticulum transmembrane protein | 1 | Endoplasmic reticulum | Protein transport | 1.359 | 0.44 | 3.88 × 10−2 |
| A0A078FU98 | Copper transport protein | 1 | Cell membrane | Ion transport | 1.343 | 0.43 | 3.18 × 10−3 |
| A0A078IX79 | (rape) hypothetical protein | 1 | - | Unknown | 1.346 | 0.43 | 9.86 × 10−3 |
| A0A078HX16 | L-ascorbate peroxidase | 2 | Chloroplast | Redox homeostasis | 1.348 | 0.43 | 1.49 × 10−2 |
| A0A078FKD7 | BnaA03g51800D protein | 6 | Nucleus | DNA repair and replication | 1.337 | 0.42 | 1.08 × 10−5 |
| A0A078ID05 | BnaA02g24310D protein | 3 | - | Unknown | 1.34 | 0.42 | 4.34 × 10−5 |
| A0A078G149 | (rape) hypothetical protein | 2 | - | Lipid metabolism | 1.339 | 0.42 | 4.14 × 10−3 |
| A0A078HUF2 | Reticulon-like protein | 3 | Endoplasmic reticulum | Unknown | 1.339 | 0.42 | 2.21 × 10−2 |
| A0A078IPP8 | Glutathione transferase | 3 | Cytosol | Redox homeostasis | 1.326 | 0.41 | 1.44 × 10−4 |
| A0A078I263 | Chlorophyll a-b binding protein | 2 | Chloroplast | Photosynthetic light reactions | 1.329 | 0.41 | 3.56 × 10−4 |
| A0A078GI82 | Thioglucosidase | 2 | - | Glucosinolate metabollism | 1.325 | 0.41 | 6.70 × 10−3 |
| A0A078IZG5 | BnaA06g38150D protein | 2 | - | Ion transport | 1.325 | 0.41 | 1.07 × 10−2 |
| A0A078IBE6 | BnaCnng16060D protein | 1 | Vacuole | Ion transport | 1.324 | 0.41 | 2.32 × 10−2 |
| A0A078G1M4 | BnaC04g05890D protein | 1 | Nucleus | Unknown | 1.325 | 0.41 | 2.62 × 10−2 |
| A0A078ICP6 | (rape) hypothetical protein | 3 | - | Unknown | 1.316 | 0.4 | 2.05 × 10−5 |
| A0A078JP84 | BnaCnng59680D protein (Fragment) | 9 | - | Unknown | 1.316 | 0.4 | 5.80 × 10−5 |
| A0A078G9H2 | (rape) hypothetical protein | 2 | - | Response to stress | 1.319 | 0.4 | 1.08 × 10−2 |
| A0A078FDJ2 | (rape) hypothetical protein | 2 | Cytosol | Unknown | 1.315 | 0.4 | 4.08 × 10−2 |
| A0A078JIV5 | (rape) hypothetical protein | 6 | Cell membrane | Response to stress | 1.311 | 0.39 | 1.85 × 10−5 |
| A0A078GU87 | Carbonic anhydrase | 7 | - | Inorganic carbon utilization | 1.312 | 0.39 | 2.00 × 10−5 |
| A0A078J666 | (rape) hypothetical protein | 3 | - | Unknown | 1.314 | 0.39 | 1.20 × 10−4 |
| A0A078IZX1 | BnaA02g20200D protein | 2 | - | Unknown | 1.313 | 0.39 | 1.59 × 10−4 |
| A0A078JI91 | (rape) hypothetical protein | 2 | - | Unknown | 1.315 | 0.39 | 7.85 × 10−3 |
| A0A078GJA8 | BnaA05g30820D protein | 3 | Golgi apparatus | Cell wall organization | 1.306 | 0.39 | 2.16 × 10−2 |
| A0A078IGY8 | Allene-oxide cyclase | 3 | Chloroplast | Jasmonic acid metabolism | 1.305 | 0.38 | 4.49 × 10−3 |
| A0A078HP42 | (rape) hypothetical protein | 2 | Chloroplast | Unknown | 1.305 | 0.38 | 1.13 × 10−2 |
| A0A078HSH2 | (rape) hypothetical protein | 3 | - | Unknown | 1.293 | 0.37 | 3.11 × 10−3 |
| A0A078G6G4 | Dihydrolipoyllysine-residue succinyltransferase | 1 | Mitochondria | TCA cycle | 1.296 | 0.37 | 4.83 × 10−2 |
| A0A078FAM6 | ABC-type xenobiotic transporter | 19 | - | Unknown | 1.28 | 0.36 | 1.57 × 10−4 |
| A0A078IWZ8 | Transmembrane 9 superfamily member | 1 | Golgi apparatus | Protein transport | 1.285 | 0.36 | 6.96 × 10−4 |
| A0A078GN01 | (rape) hypothetical protein | 1 | Vacuole | Ion transport | 1.287 | 0.36 | 1.61 × 10−2 |
| A0A078GHK2 | BnaA06g00960D protein | 3 | - | Unknown | 1.283 | 0.36 | 1.72 × 10−2 |
| A0A078G4K4 | Co-chaperone protein p23 | 4 | Nucleus | Protein folding | 1.288 | 0.36 | 2.09 × 10−2 |
| Q9ZSL7 | Non-specific lipid-transfer protein | 2 | - | Lipid transport | 1.274 | 0.35 | 2.73 × 10−4 |
| A0A078I6I1 | BnaCnng12500D protein | 1 | - | Unknown | 1.275 | 0.35 | 2.36 × 10−2 |
| A0A078I056 | (rape) hypothetical protein | 3 | - | Unknown | 1.273 | 0.35 | 2.97 × 10−2 |
| A0A078JND8 | (rape) hypothetical protein | 1 | Cytosol | Redox homeostasis | 1.278 | 0.35 | 3.14 × 10−2 |
| A0A078J2 × 0 | BnaA07g38390D protein | 2 | - | Unknown | 1.27 | 0.35 | 3.58 × 10−2 |
| A0A078J4Y4 | ADP, ATP carrier protein | 13 | Chloroplast | ATP transport | 1.262 | 0.34 | 4.40 × 10−4 |
| A0A078GX94 | (rape) hypothetical protein | 5 | - | Unknown | 1.263 | 0.34 | 6.25 × 10−4 |
| A0A078HK20 | Non-specific lipid-transfer protein | 2 | - | Lipid transport | 1.263 | 0.34 | 1.67 × 10−3 |
| A0A078FC69 | BnaA03g46100D protein | 4 | - | Unknown | 1.267 | 0.34 | 3.43 × 10−3 |
| A0A078JJB9 | (rape) hypothetical protein | 1 | - | Unknown | 1.262 | 0.34 | 4.54 × 10−2 |
| A0A078IVF6 | (rape) hypothetical protein | 4 | Mitochondria | Translation | 1.261 | 0.34 | 4.87 × 10−2 |
| Q84 × 96 | 3-oxoacyl-[acyl-carrier-protein] reductase | 8 | Chloroplast | Lipid metabolism | 1.253 | 0.33 | 1.19 × 10−3 |
| A0A078FIN1 | (rape) hypothetical protein | 7 | Vacuole | Ion transport | 1.254 | 0.33 | 2.95 × 10−3 |
| A0A078HLQ1 | (rape) hypothetical protein | 2 | - | Unknown | 1.254 | 0.33 | 2.18 × 10−2 |
| A0A078GE42 | (rape) hypothetical protein | 3 | Nucleus | Transcription | 1.256 | 0.33 | 2.84 × 10−2 |
| A0A078HAX8 | BnaA05g16460D protein | 4 | - | Unknown | 1.251 | 0.32 | 7.37 × 10−4 |
| A0A078H280 | (rape) hypothetical protein | 7 | Vacuole | Unknown | 1.249 | 0.32 | 3.26 × 10−3 |
| A0A078GNY7 | (rape) hypothetical protein | 3 | - | Unknown | 1.25 | 0.32 | 4.48 × 10−3 |
| A0A078J719 | (rape) hypothetical protein | 4 | - | Unknown | 1.248 | 0.32 | 4.98 × 10−3 |
| A0A078GZX0 | (rape) hypothetical protein | 15 | Nucleus | Chromatin organization | 1.243 | 0.31 | 1.31 × 10−3 |
| A0A078I5D5 | Sucrose-phosphate synthase | 8 | - | Sucrose biosynthesis | 1.242 | 0.31 | 1.40 × 10−3 |
| A0A078G056 | (rape) hypothetical protein | 4 | Chloroplast | Unknown | 1.238 | 0.31 | 3.48 × 10−3 |
| A0A078FZM8 | Zeaxanthin epoxidase | 5 | Chloroplast | ABA biosynthesis | 1.24 | 0.31 | 4.24 × 10−2 |
| A0A078J1V7 | (rape) hypothetical protein | 6 | Cytosol | Unknown | 1.23 | 0.3 | 2.81 × 10−3 |
| A0A078GPH3 | Eukaryotic translation initiation factor 3 subunit C | 1 | Cytosol | Translation | 1.229 | 0.3 | 2.83 × 10−3 |
| A0A078FYW6 | Non-specific lipid-transfer protein | 1 | - | Lipid transport | 1.232 | 0.3 | 8.50 × 10−3 |
| A0A078I7K6 | Chalcone-flavonone isomerase family protein | 5 | Chloroplast | Lipid metabolism | 1.235 | 0.3 | 1.75 × 10−2 |
| A0A078INI4 | BnaC01g44250D protein | 4 | - | Amino acid metabolism | 1.232 | 0.3 | 1.92 × 10−2 |
| A0A078HHQ4 | (rape) hypothetical protein | 5 | Nucleus | Chromatin organization | 1.229 | 0.3 | 2.16 × 10−2 |
| A0A078HPA3 | (rape) hypothetical protein | 15 | - | Unknown | 1.221 | 0.29 | 4.32 × 10−3 |
| A0A078JG83 | (rape) hypothetical protein | 7 | - | Unknown | 1.226 | 0.29 | 4.42 × 10−3 |
| A0A078J0C6 | BnaC07g50320D protein | 4 | - | Unknown | 1.226 | 0.29 | 4.46 × 10−3 |
| A0A078FUY5 | Dihydroorotate dehydrogenase (quinone) | 9 | Mitochondria | Pyrimidine biosynthesis | 1.225 | 0.29 | 4.62 × 10−3 |
| A0A078I3D0 | (rape) hypothetical protein | 6 | - | Unknown | 1.221 | 0.29 | 8.26 × 10−3 |
| A0A078G3T1 | Chlorophyll a-b binding protein | 2 | Chloroplast | Photosynthetic light reactions | 1.226 | 0.29 | 1.07 × 10−2 |
| A0A078GPI5 | Glycosyltransferase | 1 | Cytosol | Unknown | 1.222 | 0.29 | 2.74 × 10−2 |
| A0A078I909 | Xyloglucan endotransglucosylase/hydrolase | 5 | Apoplast | Cell wall organization | 1.22 | 0.29 | 2.97 × 10−2 |
| A0A078J5T4 | (rape) hypothetical protein | 13 | - | Unknown | 1.218 | 0.28 | 3.66 × 10−3 |
| A0A078JW32 | (rape) hypothetical protein | 11 | - | Unknown | 1.217 | 0.28 | 7.49 × 10−3 |
| A0A078JR63 | (rape) hypothetical protein | 1 | Cytosol | Protein folding | 1.212 | 0.28 | 1.77 × 10−2 |
| A0A078I708 | GrpE protein homolog | 5 | Mitochondria | Protein transport | 1.211 | 0.28 | 2.00 × 10−2 |
| A0A078ISW8 | (rape) hypothetical protein | 4 | - | Lignin biosynthesis | 1.215 | 0.28 | 4.50 × 10−2 |
| A0A078EQE4 | Non-specific lipid-transfer protein | 3 | - | Lipid transport | 1.21 | 0.27 | 4.62 × 10−3 |
| A0A078H8M2 | 1,3-beta-glucan synthase | 13 | Cell membrane | Cell wall organization | 1.205 | 0.27 | 2.14 × 10−2 |
| A0A078GSR8 | BnaA01g06570D protein | 3 | Vacuole | Vacuolar protein processing | 1.209 | 0.27 | 2.51 × 10−2 |
| A0A078F8D8 | ABC-type xenobiotic transporter | 6 | - | Xenobiotics transport | 1.204 | 0.27 | 2.84 × 10−2 |
| A0A078IDI4 | BnaA02g29390D protein | 7 | - | Unknown | 1.208 | 0.27 | 3.40 × 10−2 |
| A0A078JCY9 | BnaC06g42330D protein | 4 | Nucleus | Proteolysis | 1.207 | 0.27 | 4.61 × 10−2 |
| A0A078HR88 | (rape) hypothetical protein | 6 | - | Lipid metabolism | 1.199 | 0.26 | 1.41 × 10−2 |
| A0A078HYE9 | (rape) hypothetical protein | 10 | Vacuole | Unknown | 1.194 | 0.26 | 2.10 × 10−2 |
| A0A078I7F1 | Peroxidase | 6 | - | Redox homeostasis | 1.194 | 0.26 | 2.10 × 10−2 |
| A0A078I415 | BnaC02g00800D protein | 5 | - | Protein folding | 1.194 | 0.26 | 2.43 × 10−2 |
| A0A078JE52 | (rape) hypothetical protein | 7 | Chloroplast | Unknown | 1.199 | 0.26 | 2.67 × 10−2 |
| A0A078G6 × 1 | BnaA02g08820D protein | 1 | Cytosol | Protein folding | 1.197 | 0.26 | 3.84 × 10−2 |
| A0A078GSE8 | (rape) hypothetical protein | 1 | - | Redox homeostasis | 1.192 | 0.25 | 1.82 × 10−2 |
| P93063 | (rape) hypothetical protein | 6 | - | Unknown | 1.187 | 0.25 | 2.14 × 10−2 |
| A0A078HWD9 | (rape) hypothetical protein | 9 | - | Unknown | 1.187 | 0.25 | 2.19 × 10−2 |
| A0A078JHI7 | (rape) hypothetical protein | 1 | Chloroplast | Photosynthetic light reactions | 1.188 | 0.25 | 4.43 × 10−2 |
| A0A078J7I5 | Lipoxygenase | 4 | - | Lipid metabolism | 1.181 | 0.24 | 2.21 × 10−2 |
| A0A078GP42 | BnaC09g36360D protein | 1 | Cytosol | Unknown | 1.183 | 0.24 | 2.35 × 10−2 |
| A0A078FL44 | Chlorophyll a-b binding protein | 3 | Chloroplast | Photosynthetic light reactions | 1.179 | 0.24 | 2.82 × 10−2 |
| A0A078FPX0 | BnaC05g00300D protein | 3 | Chloroplast | Ion transport | 1.18 | 0.24 | 3.51 × 10−2 |
| A0A078GHK3 | 1,3-beta-glucan synthase | 4 | Cell membrane | Cell wall organization | 1.181 | 0.24 | 3.72 × 10−2 |
| A0A078FGY6 | BnaA03g01740D protein | 6 | - | Proteolysis | 1.183 | 0.24 | 3.95 × 10−2 |
| A0A078FTS5 | Lipoxygenase | 9 | - | Lipid metabolism | 1.176 | 0.23 | 3.26 × 10−2 |
| A0A078JJT8 | BnaC02g46730D protein | 9 | - | Unknown | 1.173 | 0.23 | 3.58 × 10−2 |
| A0A078FLH0 | Inositol-3-phosphate synthase | 2 | Cytosol | Lipid metabolism | 1.174 | 0.23 | 4.19 × 10−2 |
| A0A078FAW1 | Phospholipase A1 | 2 | - | Lipid metabolism | 1.173 | 0.23 | 4.21 × 10−2 |
| A0A078IWD9 | Branched-chain-amino-acid aminotransferase | 3 | Chloroplast | Amino acid metabolism | 1.175 | 0.23 | 4.62 × 10−2 |
| A0A078HXB3 | BnaA05g32660D protein | 4 | - | Unknown | 0.83 | −0.27 | 4.79 × 10−2 |
| A0A078FTG4 | (rape) hypothetical protein | 13 | - | Unknown | 0.821 | −0.28 | 3.38 × 10−2 |
| A0A078I495 | (rape) hypothetical protein | 16 | - | Unknown | 0.816 | −0.29 | 2.16 × 10−2 |
| A0A078GVN3 | Carboxypeptidase | 9 | Apoplast | Proteolysis | 0.817 | −0.29 | 4.63 × 10−2 |
| A0A078JCF7 | BnaCnng39740D protein | 8 | Cell membrane | Unknown | 0.81 | −0.3 | 1.52 × 10−2 |
| A0A078I1Z2 | BnaA02g13970D protein | 7 | - | Carbohydrate metabolism | 0.812 | −0.3 | 1.70 × 10−2 |
| A0A078GT00 | BnaA05g31420D protein | 15 | - | Unknown | 0.815 | −0.3 | 2.07 × 10−2 |
| A0A078JKB1 | Pectin acetylesterase (Fragment) | 9 | - | Cell wall organization | 0.809 | −0.31 | 2.57 × 10−2 |
| A0A078HZ31 | Glyceraldehyde-3-phosphate dehydrogenase | 4 | - | Calvin Benson cycle | 0.806 | −0.31 | 2.68 × 10−2 |
| A0A078H104 | BnaA08g26240D protein | 3 | - | Unknown | 0.807 | −0.31 | 3.68 × 10−2 |
| A0A078HA31 | Carboxypeptidase | 1 | Apoplast | Proteolysis | 0.8 | −0.32 | 7.63 × 10−3 |
| A0A078JBU5 | (rape) hypothetical protein | 6 | Chloroplast | Chlorophyll metabolism | 0.803 | −0.32 | 8.23 × 10−3 |
| A0A078F5V9 | (rape) hypothetical protein | 3 | - | Unknown | 0.802 | −0.32 | 2.10 × 10−2 |
| A0A078FN47 | BnaA10g07360D protein | 3 | - | Unknown | 0.803 | −0.32 | 2.96 × 10−2 |
| A0A078J6M0 | BnaAnng16600D protein | 10 | - | Unknown | 0.793 | −0.33 | 4.30 × 10−3 |
| A0A078I9C2 | (rape) hypothetical protein | 15 | - | Amino acid metabolism | 0.794 | −0.33 | 4.54 × 10−3 |
| A0A078GA58 | (rape) hypothetical protein | 6 | Apoplast | Unknown | 0.797 | −0.33 | 5.64 × 10−3 |
| A0A078I1U9 | NAD(P)H dehydrogenase (quinone) | 2 | - | Redox homeostasis | 0.794 | −0.33 | 7.90 × 10−3 |
| A0A078G1G3 | BnaC02g24210D protein | 2 | Cytosol | Unknown | 0.796 | −0.33 | 8.66 × 10−3 |
| A0A078FAN3 | (rape) hypothetical protein | 5 | Nucleus | Coenzyme A biosynthesis | 0.793 | −0.33 | 1.37 × 10−2 |
| A0A078J542 | BnaA05g35220D protein | 2 | - | Unknown | 0.795 | −0.33 | 1.58 × 10−2 |
| A0A078GHM5 | BnaA03g59800D protein | 5 | - | Sulfate assimilation | 0.795 | −0.33 | 1.80 × 10−2 |
| A0A078HA55 | Succinate--CoA ligase [ADP-forming] subunit beta | 1 | Mitochondria | TCA cycle | 0.797 | −0.33 | 1.80 × 10−2 |
| A0A078INE2 | BnaA01g30370D protein | 5 | - | Proteolysis | 0.794 | −0.33 | 2.18 × 10−2 |
| A0A078IYC4 | BnaC04g53560D protein | 4 | Cytosol | Unknown | 0.798 | −0.33 | 2.46 × 10−2 |
| A0A078HV59 | BnaA03g53100D protein | 13 | - | Proteolysis | 0.789 | −0.34 | 3.43 × 10−3 |
| A0A078I103 | Starch synthase | 5 | Chloroplast | Starch biosynthesis | 0.788 | −0.34 | 1.93 × 10−2 |
| A0A078IMS4 | (rape) hypothetical protein | 4 | - | Unknown | 0.792 | −0.34 | 3.21 × 10−2 |
| A0A078GXJ1 | Glutathione hydrolase | 7 | Cell membrane | Redox homeostasis | 0.785 | −0.35 | 4.10 × 10−3 |
| A0A078F6Z3 | BnaA02g05770D protein | 6 | Cytosol | Unknown | 0.783 | −0.35 | 7.57 × 10−3 |
| A0A078FRX3 | (rape) hypothetical protein | 2 | Chloroplast | Photosynthetic light reactions | 0.785 | −0.35 | 8.26 × 10−3 |
| Q42625 | Glutamine synthetase | 7 | Cytosol | Amino acid metabolism | 0.78 | −0.36 | 2.72 × 10−3 |
| A0A078J1J0 | (rape) hypothetical protein | 3 | Plasmodesmata | Defense response | 0.778 | −0.36 | 4.22 × 10−2 |
| A0A078GXM3 | (rape) hypothetical protein | 1 | Chloroplast | Unknown | 0.777 | −0.36 | 4.50 × 10−2 |
| A0A078HMR7 | Ferredoxin--NADP reductase | 10 | Chloroplast | Photosynthetic light reactions | 0.776 | −0.37 | 9.90 × 10−4 |
| A0A078HXA1 | NAD(P)H dehydrogenase (quinone) | 4 | - | Redox homeostasis | 0.776 | −0.37 | 1.75 × 10−3 |
| A0A078GBH5 | (rape) hypothetical protein | 2 | Cell membrane | Unknown | 0.773 | −0.37 | 3.32 × 10−2 |
| A0A078IJ48 | Apyrase | 3 | - | Nucleoside diphosphate metabolism | 0.772 | −0.37 | 3.34 × 10−2 |
| A0A078JFA9 | (rape) hypothetical protein | 2 | Apoplast | Unknown | 0.769 | −0.38 | 2.51 × 10−3 |
| A0A078FBH3 | Histone H2A | 1 | Nucleus | Chromatin organization | 0.768 | −0.38 | 1.31 × 10−2 |
| A0A078IUE0 | Histone H2A | 1 | Nucleus | Chromatin organization | 0.771 | −0.38 | 3.58 × 10−2 |
| A0A078GPW5 | (rape) hypothetical protein | 2 | - | Unknown | 0.763 | −0.39 | 2.21 × 10−2 |
| A0A078JIB2 | BnaC08g46420D protein | 1 | - | Unknown | 0.762 | −0.39 | 2.32 × 10−2 |
| A0A078J451 | (rape) hypothetical protein | 3 | Cytosol | Unknown | 0.764 | −0.39 | 2.84 × 10−2 |
| A0A078F9R8 | Ferritin | 6 | Chloroplast | Ion transport | 0.755 | −0.4 | 3.02 × 10−4 |
| A0A078GVU9 | (rape) hypothetical protein | 5 | - | Unknown | 0.76 | −0.4 | 5.84 × 10−4 |
| A0A078GH45 | Alanine transaminase | 1 | - | Photorespiration | 0.76 | −0.4 | 2.78 × 10−2 |
| A0A078F8F7 | (rape) hypothetical protein | 6 | Nucleus | Chromosome condensation | 0.75 | −0.41 | 8.26 × 10−5 |
| A0A078F4V7 | Pectinesterase | 3 | - | Cell wall organization | 0.754 | −0.41 | 2.10 × 10−2 |
| A0A078HKL0 | BnaA07g02120D protein | 2 | Cytosol | Translation | 0.746 | −0.42 | 7.23 × 10−4 |
| A0A078FQN7 | (rape) hypothetical protein | 1 | - | Unknown | 0.747 | −0.42 | 3.01 × 10−2 |
| A0A078JI07 | (rape) hypothetical protein | 1 | Chloroplast | Rubisco assembly | 0.746 | −0.42 | 4.76 × 10−2 |
| A0A078IZV0 | (rape) hypothetical protein | 8 | Apoplast | Unknown | 0.744 | −0.43 | 9.07 × 10−5 |
| A0A078J6D0 | BnaC04g53030D protein | 2 | - | Cell wall organization | 0.737 | −0.44 | 5.14 × 10−3 |
| A0A078FS35 | (rape) hypothetical protein | 1 | Cytosol | Translation | 0.738 | −0.44 | 6.81 × 10−3 |
| A0A078IRE8 | (rape) hypothetical protein | 2 | - | Unknown | 0.726 | −0.46 | 4.78 × 10−6 |
| A0A078G019 | (rape) hypothetical protein | 1 | Chloroplast | Photosynthetic light reactions | 0.726 | −0.46 | 1.27 × 10−5 |
| A0A078HZN5 | Expansin | 2 | - | Cell wall organization | 0.724 | −0.47 | 5.78 × 10−4 |
| A0A078I7K4 | BnaC02g22590D protein | 2 | Vacuole | Unknown | 0.724 | −0.47 | 1.87 × 10−3 |
| A0A078I0P0 | BnaC05g36680D protein | 2 | Plasmodesmata | Signaling | 0.717 | −0.48 | 1.75 × 10−3 |
| A0A078HMK9 | (rape) hypothetical protein | 1 | - | Unknown | 0.711 | −0.49 | 1.44 × 10−2 |
| A0A078FB79 | BnaA02g05290D protein | 8 | Nucleus | Defense response | 0.706 | −0.5 | 6.87 × 10−7 |
| A0A078GTF2 | Non-specific serine/threonine protein kinase | 2 | - | Signaling | 0.7 | −0.52 | 6.41 × 10−4 |
| A0A078HAP6 | BnaA02g30740D protein | 3 | - | Cell wall organization | 0.691 | −0.53 | 9.09 × 10−7 |
| A0A078GNZ7 | Glycine cleavage system H protein | 2 | Mitochondria | Photorespiration | 0.694 | −0.53 | 1.35 × 10−6 |
| A0A078F776 | Histone H2B | 1 | Nucleus | Chromatin organization | 0.694 | −0.53 | 1.93 × 10−2 |
| A0A078JLL8 | BnaC01g44910D protein | 1 | - | Lipid transport | 0.691 | −0.53 | 2.63 × 10−2 |
| A0A078I390 | (rape) hypothetical protein | 1 | - | Unknown | 0.69 | −0.54 | 8.04 × 10−3 |
| A0A078HLM5 | (rape) hypothetical protein | 2 | - | Unknown | 0.666 | −0.59 | 6.50 × 10−5 |
| A0A078F5L4 | (rape) hypothetical protein | 2 | Plasmodesmata | Signaling | 0.659 | −0.6 | 9.65 × 10−6 |
| A0A078JG76 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase | 1 | Chloroplast | Lipid metabolism | 0.661 | −0.6 | 1.45 × 10−2 |
| A0A078G4U7 | (rape) hypothetical protein | 1 | - | Unknown | 0.652 | −0.62 | 4.49 × 10−3 |
| A0A078I2L1 | BnaC02g41300D protein | 2 | Plasmodesmata | defense response | 0.631 | −0.66 | 2.53 × 10−6 |
| A0A078I8 × 8 | (rape) hypothetical protein | 1 | Chloroplast | Translation | 0.625 | −0.68 | 8.50 × 10−9 |
| A0A078G384 | DNA polymerase | 1 | Nucleus | DNA repair and replication | 0.61 | −0.71 | 1.15 × 10−7 |
| A0A078HN54 | (rape) hypothetical protein | 1 | - | Unknown | 0.611 | −0.71 | 1.42 × 10−4 |
| A0A078GRB9 | Pectinesterase | 2 | - | Cell wall organization | 0.568 | −0.82 | 1.19 × 10−9 |
| A0A078HF73 | BnaC03g41590D protein | 2 | - | Unknown | 0.561 | −0.83 | 1.36 × 10−10 |
| A0A078F748 | BnaC08g11470D protein | 1 | - | Unknown | 0.56 | −0.84 | 1.29 × 10−5 |
| A0A078GMM9 | (rape) hypothetical protein | 1 | - | Unknown | 0.364 | −1.46 | 2.64 × 10−15 |
| A0A078HA23 | (rape) hypothetical protein | 1 | - | Protein transport | 0.275 | −1.86 | 2.64 × 10−15 |
| Control | AM | |
|---|---|---|
| Richness (nº OTUs) | 283 ± 52 a | 317 ± 65 a |
| Actinobacteriota | 0.057 ± 0.016 b | 0.121 ± 0.009 a |
| Bacteroidota | 0.068 ± 0.002 a | 0.077 ± 0.004 a |
| Bdellovibrionota | 0.010 ± 0.004 a | 0.005 ± 0.003 a |
| Chloroflexi | 0.068 ± 0.014 a | 0.083 ± 0.004 a |
| Cyanobacteria | 0.042 ± 0.012 a | 0.027 ± 0.014 a |
| Fibrobacterota | 0.002 ± 0.001 a | 0.001 ± 0.001 a |
| Firmicutes | 0.008 ± 0.001 a | 0.018 ± 0.005 a |
| Gemmatimonadota | 0.024 ± 0.004 a | 0.022 ± 0.004 a |
| Myxococcota | 0.014 ± 0.005 a | 0.017 ± 0.004 a |
| Nitrospirota | 0.013 ± 0.001 a | 0.010 ± 0.001 a |
| Patescibacteria | 0.107 ± 0.031 a | 0.033 ± 0.009 a |
| Planctomycetota | 0.162 ± 0.017 a | 0.198 ± 0.017 a |
| Proteobacteria | 0.178 ± 0.024 a | 0.164 ± 0.012 a |
| Verrucomicrobiota | 0.087 ± 0.003 a | 0.078 ± 0.004 a |

References
- John, D.A.; Babu, G.R. Lessons From the Aftermaths of Green Revolution on Food System and Health. Front. Sustain. Food Syst. 2021, 5, 644559. [Google Scholar] [CrossRef] [PubMed]
- Soria-Lopez, A.; Garcia-Perez, P.; Carpena, M.; Garcia-Oliveira, P.; Otero, P.; Fraga-Corral, M.; Cao, H.; Prieto, M.A.; Simal-Gandara, J. Challenges for Future Food Systems: From the Green Revolution to Food Supply Chains with a Special Focus on Sustainability. Food Front. 2023, 4, 9–20. [Google Scholar] [CrossRef]
- FAO. The Future of Food and Agriculture: Trends and Challenges; FAO: Rome, Italy, 2017; ISBN 978-92-5-109551-5.
- Khan, N.; Ray, R.L.; Sargani, G.R.; Ihtisham, M.; Khayyam, M.; Ismail, S. Current Progress and Future Prospects of Agriculture Technology: Gateway to Sustainable Agriculture. Sustainability 2021, 13, 4883. [Google Scholar] [CrossRef]
- Liu, Y.; Pan, X.; Li, J. Current Agricultural Practices Threaten Future Global Food Production. J. Agric. Environ. Ethics 2014, 28, 203–216. [Google Scholar] [CrossRef]
- Seneviratne, S.I.; Zhang, X.; Adnan, M.; Badi, W.; Dereczynski, C.; Luca, A.D.; Ghosh, S.; Iskandar, I.; Kossin, J.; Lewis, S.; et al. Weather and Climate Extreme Events in a Changing Climate. In Climate Change 2021—The Physical Science Basis: Working Group I Contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC, Ed.; Cambridge University Press: Cambridge, UK, 2023; pp. 1513–1766. ISBN 978-1-00-915788-9. [Google Scholar]
- Akanmu, A.O.; Babalola, O.O.; Venturi, V.; Ayilara, M.S.; Adeleke, B.S.; Amoo, A.E.; Sobowale, A.A.; Fadiji, A.E.; Glick, B.R. Plant Disease Management: Leveraging on the Plant-Microbe-Soil Interface in the Biorational Use of Organic Amendments. Front. Plant Sci. 2021, 12, 700507. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lin, J.Y.; Sayre, J.M.; Schmidt, R.; Fonte, S.J.; Rodrigues, J.L.M.; Scow, K.M. Compost Amendment Maintains Soil Structure and Carbon Storage by Increasing Available Carbon and Microbial Biomass in Agricultural Soil—A Six-Year Field Study. Geoderma 2022, 427, 116117. [Google Scholar] [CrossRef]
- Matisic, M.; Dugan, I.; Bogunovic, I. Challenges in Sustainable Agriculture—The Role of Organic Amendments. Agriculture 2024, 14, 643. [Google Scholar] [CrossRef]
- Iqbal, S.; Yahya Khan, M.; Asghar, H.; Akhtar, M. Combined Use of Phosphate Solubilizing Bacteria and Poultry Manure to Enhance the Growth and Yield of Mung Bean in Calcareous Soil. Soil Environ. 2016, 35, 146–154. [Google Scholar]
- Amadou, A.; Song, X.; Huang, S.; Song, A.; Tang, Z.; Dong, W.; Zhao, S.; Zhang, B.; Yi, K.; Fan, F. Effects of Long-Term Organic Amendment on the Fertility of Soil, Nodulation, Yield, and Seed Quality of Soybean in a Soybean-Wheat Rotation System. J. Soils Sediments 2021, 21, 1385–1394. [Google Scholar] [CrossRef]
- Amanullah; Khan, S.-T.; Iqbal, A.; Fahad, S. Growth and Productivity Response of Hybrid Rice to Application of Animal Manures, Plant Residues and Phosphorus. Front. Plant Sci. 2016, 7, 1440. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Siddique, K.H.M.; Ahmad, P. Understanding Drought Tolerance in Plants. Physiol. Plant. 2021, 172, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Anee, T.; Islam, M.; Hassan, M.; Masud, A.; Alam, M.; Hasanuzzaman, M. Organic Amendments Improve Plant Morpho-Physiology and Antioxidant Metabolism in Mitigating Drought Stress in Bread Wheat (Triticum aestivum L.). Phyton-Int. J. Exp. Bot. 2022, 91, 1959–1972. [Google Scholar] [CrossRef]
- Shi, L.; Guo, Z.; Liu, S.; Xiao, X.; Peng, C.; Feng, W.; Ran, H.; Zeng, P. Effects of Combined Soil Amendments on Cd Accumulation, Translocation and Food Safety in Rice: A Field Study in Southern China. Environ. Geochem. Health 2022, 44, 2451–2463. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, H.; Dai, Y.; Tian, H.; Zhou, W.; Lv, J. Soil Organic Carbon Transformation and Dynamics of Microorganisms under Different Organic Amendments. Sci. Total Environ. 2021, 750, 141719. [Google Scholar] [CrossRef]
- Vida, C.; de Vicente, A.; Cazorla, F.M. The Role of Organic Amendments to Soil for Crop Protection: Induction of Suppression of Soilborne Pathogens. Ann. Appl. Biol. 2020, 176, 1–15. [Google Scholar] [CrossRef]
- Yadav, V.; Karak, T.; Singh, S.; Singh, A.K.; Khare, P. Benefits of Biochar over Other Organic Amendments: Responses for Plant Productivity (Pelargonium graveolens L.) and Nitrogen and Phosphorus Losses. Ind. Crops Prod. 2019, 131, 96–105. [Google Scholar] [CrossRef]
- Akanmu, A.O.; Sobowale, A.A.; Abiala, M.A.; Olawuyi, O.J.; Odebode, A.C. Efficacy of Biochar in the Management of Fusarium Verticillioides Sacc. Causing Ear Rot in Zea mays L. Biotechnol. Rep. 2020, 26, e00474. [Google Scholar] [CrossRef]
- Mohamed, I.A.A.; Shalby, N.; El-Badri, A.M.; Awad-Allah, E.F.A.; Batool, M.; Saleem, M.H.; Wang, Z.; Wen, J.; Ge, X.; Xu, Z.; et al. Multipurpose Uses of Rapeseed (Brassica napus L.) Crop (Food, Feed, Industrial, Medicinal, and Environmental Conservation Uses) and Improvement Strategies in China. J. Agric. Food Res. 2025, 20, 101794. [Google Scholar] [CrossRef]
- Raza, A. Eco-Physiological and Biochemical Responses of Rapeseed (Brassica napus L.) to Abiotic Stresses: Consequences and Mitigation Strategies. J. Plant Growth Regul. 2021, 40, 1368–1388. [Google Scholar] [CrossRef]
- Amanullah, P.; Khalid, S. Integrated Use of Phosphorus, Animal Manures and Biofertilizers Improve Maize Productivity under Semiarid Condition. In Organic Fertilizers—From Basic Concepts to Applied Outcomes; InTechOpen: London, UK, 2016; pp. 137–155. ISBN 978-953-51-2449-8. [Google Scholar]
- Gräf, M.; Immitzer, M.; Hietz, P.; Stangl, R. Water-Stressed Plants Do Not Cool: Leaf Surface Temperature of Living Wall Plants under Drought Stress. Sustainability 2021, 13, 3910. [Google Scholar] [CrossRef]
- Petrík, P.; Petek-Petrik, A.; Mukarram, M.; Schuldt, B.; Lamarque, L.J. Leaf Physiological and Morphological Constraints of Water-Use Efficiency in C3 Plants. AoB Plants 2023, 15, plad047. [Google Scholar] [CrossRef] [PubMed]
- Gu, L. Optimizing the Electron Transport Chain to Sustainably Improve Photosynthesis. Plant Physiol. 2023, 193, 2398–2412. [Google Scholar] [CrossRef] [PubMed]
- Kierans, S.J.; Taylor, C.T. Glycolysis: A Multifaceted Metabolic Pathway and Signaling Hub. J. Biol. Chem. 2024, 300, 107906. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Luo, X.; Wen, M.; Dong, X.; Sharifi, S.; Xie, D.; He, X. Funneliformis mosseae Improves Growth and Nutrient Accumulation in Wheat by Facilitating Soil Nutrient Uptake under Elevated CO2 at Daytime, Not Nighttime. J. Fungi 2021, 7, 458. [Google Scholar] [CrossRef]
- Xu, Z.; Mahmood, K.; Rothstein, S.J. ROS Induces Anthocyanin Production via Late Biosynthetic Genes and Anthocyanin Deficiency Confers the Hypersensitivity to ROS-Generating Stresses in Arabidopsis. Plant Cell Physiol. 2017, 58, 1364–1377. [Google Scholar] [CrossRef]
- Singh, P.; Choudhary, K.K.; Chaudhary, N.; Gupta, S.; Sahu, M.; Tejaswini, B.; Sarkar, S. Salt Stress Resilience in Plants Mediated through Osmolyte Accumulation and Its Crosstalk Mechanism with Phytohormones. Front. Plant Sci. 2022, 13, 1006617. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes Benefaction Role in Soil and Plant Health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Mitra, D.; Mondal, R.; Khoshru, B.; Senapati, A.; Radha, T.; Mahakur, B.; Uniyal, N.; Myo, E.M.; Boutaj, H.; Guerra Sierra, B.E.; et al. Actinobacteria-Enhanced Plant Growth, Nutrient Acquisition, and Crop Protection: Advances in Soil, Plant, and Microbial Multifactorial Interactions. Pedosphere 2022, 32, 149–170. [Google Scholar] [CrossRef]
- von Caemmerer, S.; Farquhar, G.D. Some Relationships between the Biochemistry of Photosynthesis and the Gas Exchange of Leaves. Planta 1981, 153, 376–387. [Google Scholar] [CrossRef]
- Harley, P.C.; Loreto, F.; Di Marco, G.; Sharkey, T.D. Theoretical Considerations When Estimating the Mesophyll Conductance to CO2 Flux by Analysis of the Response of Photosynthesis to CO2. Plant Physiol. 1992, 98, 1429–1436. [Google Scholar] [CrossRef]
- López-Ferrer, D.; Martínez-Bartolomé, S.; Villar, M.; Campillos, M.; Martín-Maroto, F.; Vázquez, J. Statistical Model for Large-Scale Peptide Identification in Databases from Tandem Mass Spectra Using SEQUEST. Anal. Chem. 2004, 76, 6853–6860. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An Open Source Document Editor for Creating and Analyzing Targeted Proteomics Experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles Instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Seo, M.; Jikumaru, Y.; Kamiya, Y. Profiling of Hormones and Related Metabolites in Seed Dormancy and Germination Studies. In Seed Dormancy: Methods and Protocols; Kermode, A.R., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 99–111. ISBN 978-1-61779-231-1. [Google Scholar]

| Treatment | Biomass (g plant−1) | SW (g plant−1) | LW (g plant−1) | RW (g plant−1) | Yield (g plant−1) | N (%) |
|---|---|---|---|---|---|---|
| Control | 10.7 ± 0.52 b | 9.84 ± 0.63 b | 0.83 ± 0.27 b | 4.24 ± 1.05 a | 276 ± 25.2 a | 2.3 ± 0.40 b |
| AM | 19.4 ± 0.83 a | 16.4 ± 0.26 a | 3.01 ± 0.62 a | 7.34 ± 1.55 a | 748 ± 245 a | 4.0 ± 0.24 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picazo, P.J.; Ancín, M.; Gakière, B.; Gilard, F.; Soba, D.; Gámez, A.L.; Houdusse, D.; Aranjuelo, I. Advancing Sustainable Agriculture: Molecular and Physiological Insights into Rapeseed Responsiveness to Organic Amendment Fertilization. Plants 2025, 14, 2937. https://doi.org/10.3390/plants14182937
Picazo PJ, Ancín M, Gakière B, Gilard F, Soba D, Gámez AL, Houdusse D, Aranjuelo I. Advancing Sustainable Agriculture: Molecular and Physiological Insights into Rapeseed Responsiveness to Organic Amendment Fertilization. Plants. 2025; 14(18):2937. https://doi.org/10.3390/plants14182937
Chicago/Turabian StylePicazo, Pedro J., María Ancín, Bertrand Gakière, Françoise Gilard, David Soba, Angie L. Gámez, Diane Houdusse, and Iker Aranjuelo. 2025. "Advancing Sustainable Agriculture: Molecular and Physiological Insights into Rapeseed Responsiveness to Organic Amendment Fertilization" Plants 14, no. 18: 2937. https://doi.org/10.3390/plants14182937
APA StylePicazo, P. J., Ancín, M., Gakière, B., Gilard, F., Soba, D., Gámez, A. L., Houdusse, D., & Aranjuelo, I. (2025). Advancing Sustainable Agriculture: Molecular and Physiological Insights into Rapeseed Responsiveness to Organic Amendment Fertilization. Plants, 14(18), 2937. https://doi.org/10.3390/plants14182937








