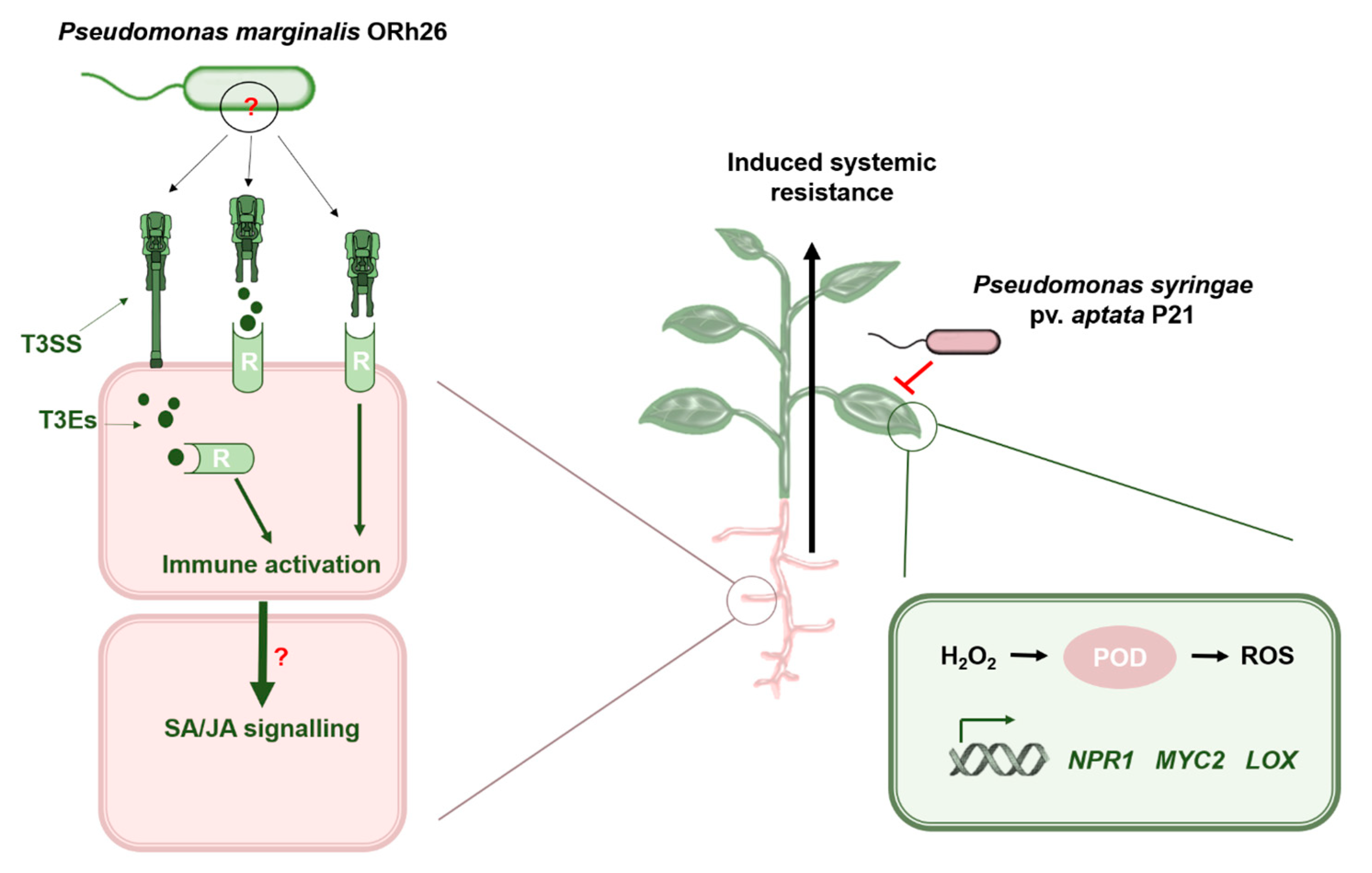
Type III Secretion System-Mediated Induction of Systemic Resistance by Pseudomonas marginalis ORh26 Enhances Sugar Beet Defence Against Pseudomonas syringae pv. aptata
Abstract
1. Introduction
2. Results
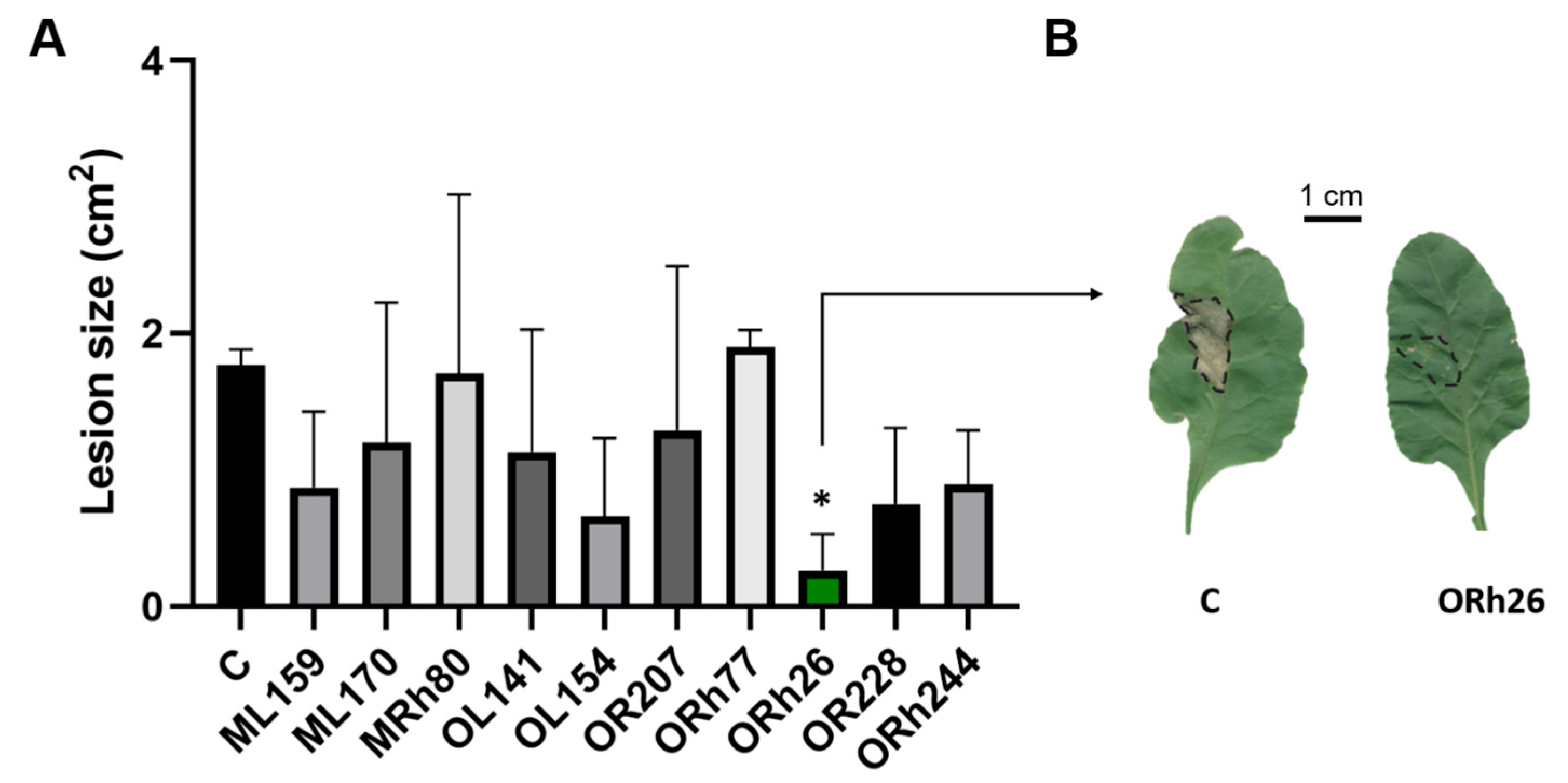
2.1. Effects of T3SS-Positive Pseudomonas Isolates on Sugar Beet Resistance to Pseudomonas syringae pv. aptata P21
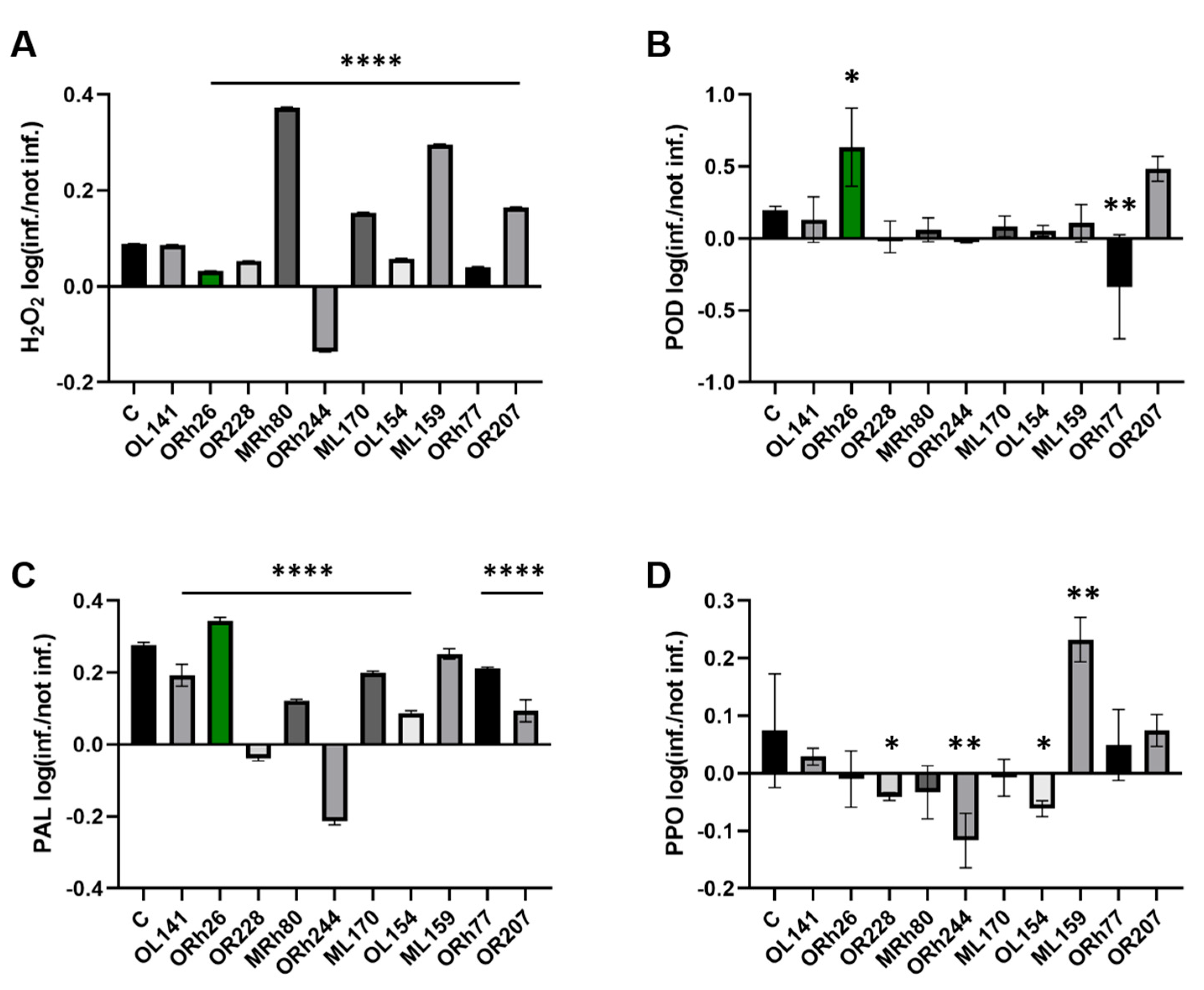
2.2. Differential Immune Responses Across Treatment Conditions
2.3. Characterisation of T3SS Genes in the Pseudomonas marginalis Orh26 Genome
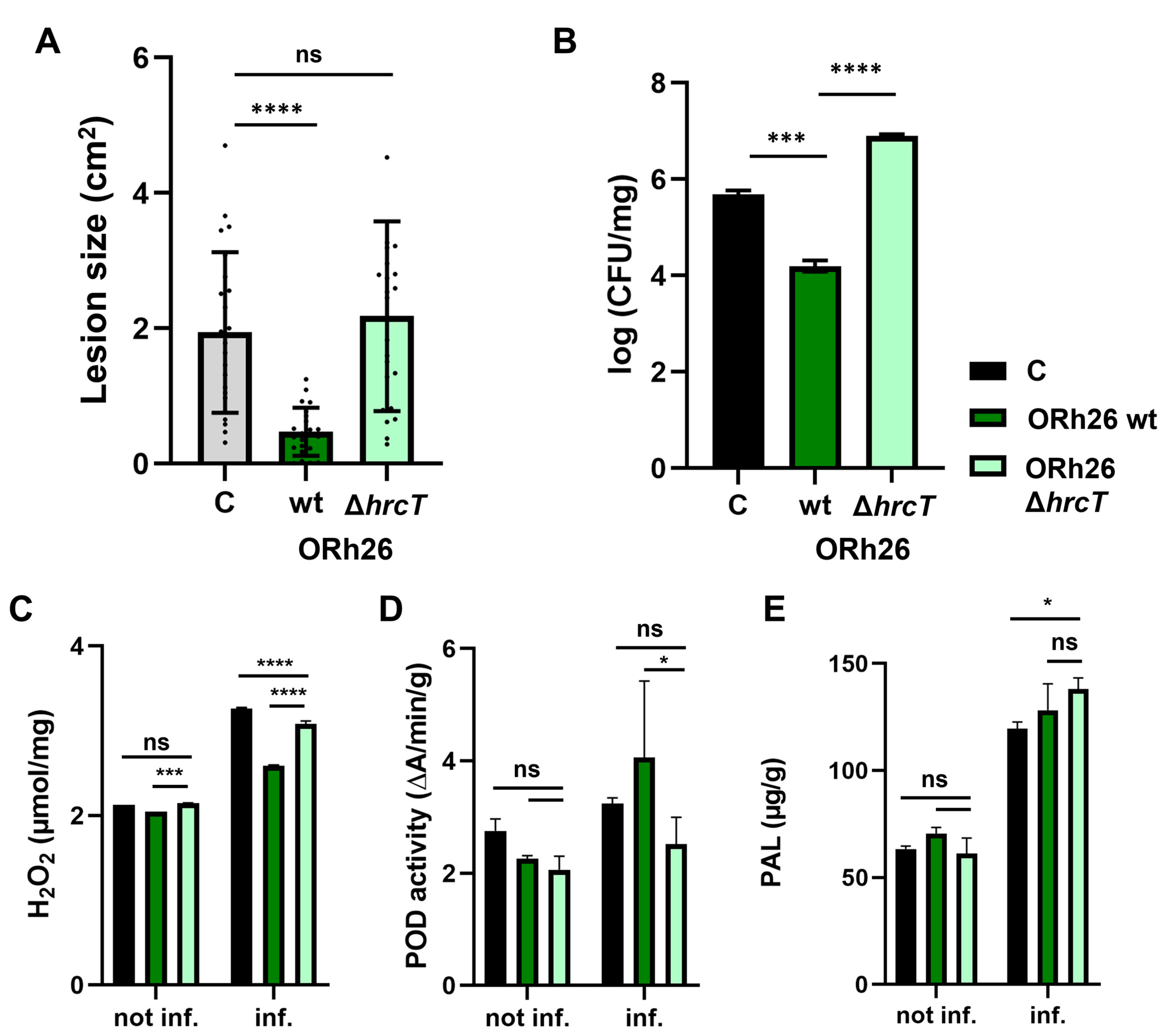
2.4. Induction of ISR and Peroxidase Activity Are T3SS-Dependent
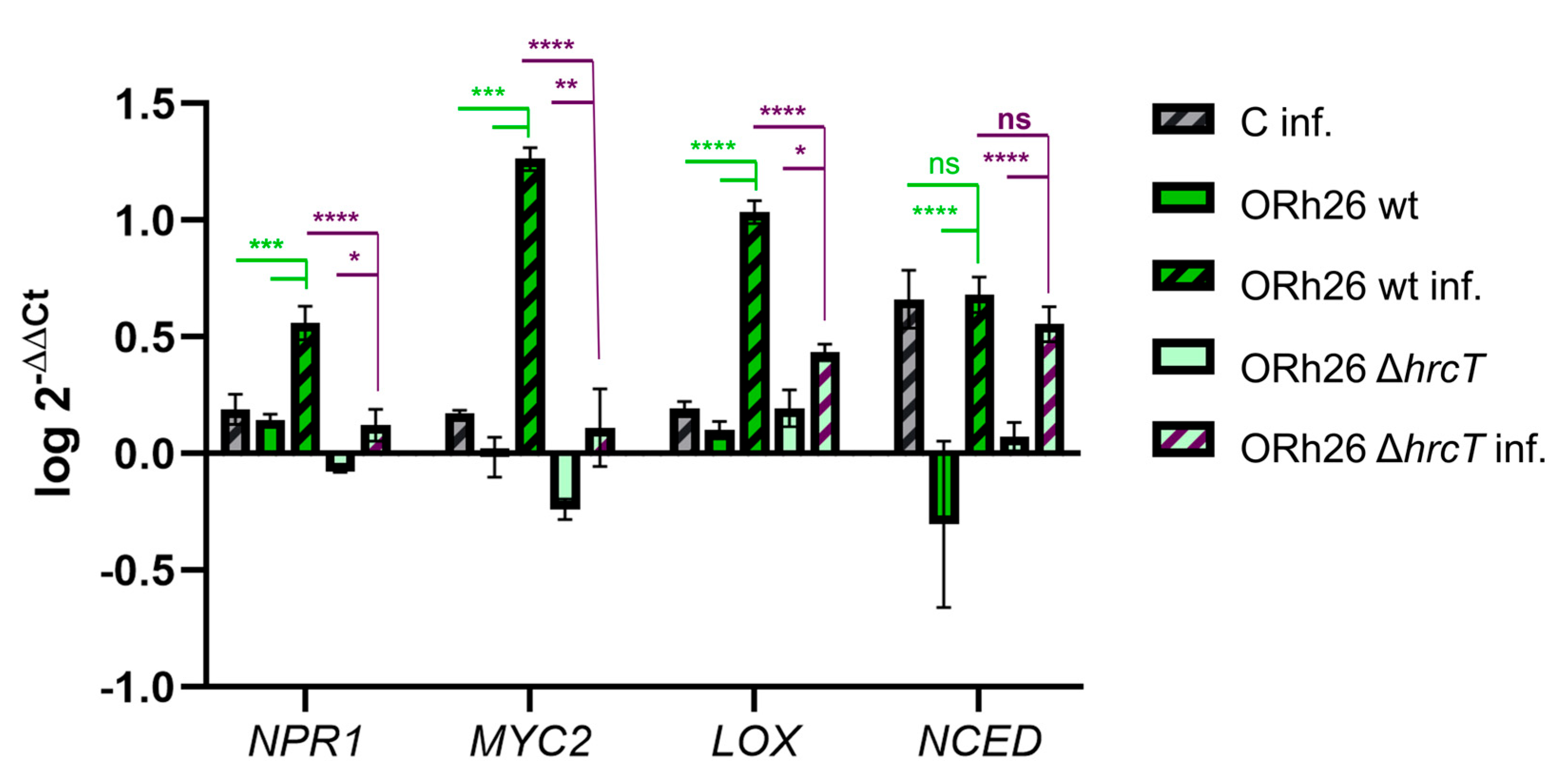
2.5. T3SS-Dependent and Independent Pathways of Systemic Resistance Gene Regulation in Sugar Beet
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Media
4.2. Bacterial Treatment and Infection Susceptibility Assay
4.3. Root Colonisation Assay
4.4. Quantification of Hydrogen Peroxide in Sugar Beet Leaves
4.5. Preparation of Enzyme Extracts
4.6. Peroxidase (POD) Activity
4.7. Polyphenol Oxidase (PPO) Activity
4.8. Phenylalanine Ammonia-Lyase (PAL) Activity
4.9. Genome Sequencing and Annotation of Pseudomonas marginalis Orh26
4.10. Construction of the T3SS Deletion Mutant
4.11. Quantification of Relative Gene Expression Levels
4.12. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| FLS2 | Flagelin Sensitive 2 |
| ISR | Induced Systemic Resistance |
| JA | Jasmonic Acid |
| LOX | LIPOXYGENASE |
| MAMP | Microorganism-Associated Molecular Pattern |
| MYC2 | MYELO-CYTOMATOSIS 2 |
| NCED | NINE-CIS-EPOXYCAROTENOID DIOXYGENASE |
| NPR1 | NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 |
| PAL | Phenylalanine Ammonium Lyase |
| POD | Peroxidase |
| PPO | Polyphenol Oxidase |
| PRR | Pattern Recognition Receptor |
| ROS | Reactive Oxygen Species |
| SA | Salicylic Acid |
| T3E | Type 3 Secretion System effector |
| T3SS | Type 3 Secretion System |
| Wt | Wild Type |
References
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386. [Google Scholar] [CrossRef]
- Bakker, P.A.; Pieterse, C.M.; van Loon, L.C. Induced Systemic Resistance by Fluorescent Pseudomonas spp. Phytopathology 2007, 97, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Meziane, H.; Van der Sluis, I.; Van Loon, L.C.; Höfte, M.; Bakker, P.A. Determinants of Pseudomonas putida WCS358 Involved in Inducing Systemic Resistance in Plants. Mol. Plant Pathol. 2005, 6, 177–185. [Google Scholar] [CrossRef]
- Xie, Y.; Shao, X.; Deng, X. Regulation of Type III Secretion System in Pseudomonas syringae. Environ. Microbiol. 2019, 21, 4465–4477. [Google Scholar] [CrossRef] [PubMed]
- Diepold, A.; Armitage, J.P. Type III Secretion Systems: The Bacterial Flagellum and the Injectisome. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150020. [Google Scholar] [CrossRef]
- Zboralski, A.; Biessy, A.; Filion, M. Bridging the Gap: Type III Secretion Systems in Plant-Beneficial Bacteria. Microorganisms 2022, 10, 187. [Google Scholar] [CrossRef]
- Nedeljković, M.; Mesaroš, A.; Rašić, V.; Nikolić, I.; Stanković, S.; Lozo, J.; Atanasković, I. Effects of T3SS-Positive Pseudomonas Isolates on Sugar Beet Growth Stimulation and Pathogen Resistance. Plant Soil 2024, 1–16. [Google Scholar] [CrossRef]
- Stojšin, V.; Balaž, J.; Budakov, D.; Stanković, S.; Nikolić, I.; Ivanović, Ž.; Popović, T. First Report of Pseudomonas syringae pv. aptata Causing Bacterial Leaf Spot on Sugar Beet in Serbia. Plant Dis. 2015, 99, 281. [Google Scholar] [CrossRef]
- Krstić Tomić, T.; Atanasković, I.; Nikolić, I.; Joković, N.; Stević, T.; Stanković, S.; Berić, T.; Lozo, J. Culture-Dependent and Metabarcoding Characterization of the Sugar Beet (Beta vulgaris L.) Microbiome for High-Yield Isolation of Bacteria with Plant Growth-Promoting Traits. Microorganisms 2023, 11, 1538. [Google Scholar] [CrossRef]
- Martel, A.; Lo, T.; Desveaux, D.; Guttman, D.S. A high-throughput, seedling screen for plant immunity. Mol. Plant Microbe Interact. 2020, 33, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Cheng, J.H.; Yoon, T.M.; Song, J.H.; Rajkarnikar, A.; Kim, W.G.; Yoo, I.D.; Yang, Y.Y.; Suh, J.W. Biological control agent of common scab disease by antagonistic strain Bacillus sp. sunhua. J. Appl. Microbiol. 2005, 99, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jourdan, E.; Schäfer, M.; Kech, C.; Budzikiewicz, H.; Luxen, A.; Thonart, P. Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in tomato. Mol. Plant Microbe Interact. 2005, 18, 562–569. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Zhao, L.; Li, S.; Yu, Q.; Zhang, C.; Wang, L.; Jiang, Y.; Wu, Z.; Pi, Z. Vernalization Promotes GA-Mediated Bolting Initiation via the Inhibition of ABA and JA Biosynthesis. Agronomy 2023, 13, 1251. [Google Scholar] [CrossRef]
- Guilbaud, C.; Morris, C.E.; Barakat, M.; Ortet, P.; Berge, O. Isolation and Identification of Pseudomonas syringae Facilitated by a PCR Targeting the Whole P. syringae Group. FEMS Microbiol. Ecol. 2016, 92, fiv146. [Google Scholar] [CrossRef]
- Buttner, D. Protein Export According to Schedule: Architecture, Assembly, and Regulation of Type III Secretion Systems from Plant- and Animal-Pathogenic Bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 262–310. [Google Scholar] [CrossRef]
- Koike, S.T.; Henderson, D.M.; Bull, C.T.; Goldman, P.H.; Lewellen, R.T. First Report of Bacterial Leaf Spot of Swiss Chard Caused by Pseudomonas syringae pv. aptata in California. Plant Dis. 2003, 87, 1397. [Google Scholar] [CrossRef]
- Nikolić, I.; Glatter, T.; Ranković, T.; Berić, T.; Stanković, S.; Diepold, A. Repertoire and Abundance of Secreted Virulence Factors Shape the Pathogenic Capacity of Pseudomonas syringae pv. aptata. Front. Microbiol. 2023, 14, 1205257. [Google Scholar] [CrossRef]
- Begum, N.; Hu, Z.; Cai, Q.; Lou, L. Influence of PGPB Inoculation on HSP70 and HMA3 Gene Expression in Switchgrass Under Cadmium Stress. Plants 2019, 8, 504. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Joe, A.; Mavrodi, O.V.; Hassan, K.A.; Weller, D.M.; Paulsen, I.T.; Loper, J.E.; Alfano, J.R.; Thomashow, L.S. Structural and Functional Analysis of the Type III Secretion System from Pseudomonas fluorescens Q8r1-96. J. Bacteriol. 2011, 193, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.; Franco, O.L. Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants 2023, 12, 2226. [Google Scholar] [CrossRef] [PubMed]
- Bashan, Y.; Okon, Y.; Henis, Y. Peroxidase, Polyphenoloxidase, and Phenols in Relation to Resistance Against Pseudomonas syringae pv. tomato in Tomato Plants. Can. J. Bot. 1987, 65, 366–372. [Google Scholar] [CrossRef]
- Albert, F.; Anderson, A.J. The Effect of Pseudomonas putida Colonization on Root Surface Peroxidase. Plant Physiol. 1987, 85, 537–541. [Google Scholar] [CrossRef]
- Saikia, R.; Kumar, R.; Singh, T.; Srivastava, A.K.; Arora, D.K.; Lee, M.-W. Induction of Defense-Related Enzymes and Pathogenesis-Related Proteins in Pseudomonas fluorescens-Treated Chickpea in Response to Infection by Fusarium oxysporum f. sp. ciceri. Mycobiology 2004, 32, 47–53. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An Important Role of the Pepper Phenylalanine Ammonia-Lyase Gene (PAL1) in Salicylic Acid-Dependent Signalling of the Defence Response to Microbial Pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef]
- Shine, M.B.; Yang, J.W.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.Q.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative Functioning Between Phenylalanine Ammonia Lyase and Isochorismate Synthase Activities Contributes to Salicylic Acid Biosynthesis in Soybean. New Phytol. 2016, 212, 627–636. [Google Scholar] [CrossRef]
- Ameer Basha, S.; Chatterjee, S.C. Effect of PGPR on Sclerotinia sclerotiorum Infection Through Elicitation of Phenylalanine Ammonia Lyase in Chickpea. Indian Phytopathol. 2007, 60, 313–316. [Google Scholar]
- Huang, Y.; McBeath, J.H. Bacterial-Induced Activation of an Arabidopsis Phenylalanine Ammonia-Lyase Promoter in Transgenic Tobacco Plants. Plant Sci. 1994, 98, 25–35. [Google Scholar] [CrossRef]
- Chen, H.; Chen, J.; Li, M.; Chang, M.; Xu, K.; Shang, Z.; Zhao, Y.; Palmer, I.; Zhang, Y.; McGill, J.; et al. A Bacterial Type III Effector Targets the Master Regulator of Salicylic Acid Signaling, NPR1, to Subvert Plant Immunity. Cell Host Microbe 2017, 22, 777–788.e7. [Google Scholar] [CrossRef]
- Iavicoli, A.; Boutet, E.; Buchala, A.; Métraux, J.P. Induced Systemic Resistance in Arabidopsis thaliana in Response to Root Inoculation with Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 2003, 16, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Van Der Ent, S.; Van Loon, L.C.; Pieterse, C.M.J. Transcription Factor MYC2 Is Involved in Priming for Enhanced Defense During Rhizobacteria-Induced Systemic Resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef]
- Hou, S.; Thiergart, T.; Vannier, N.; Mesny, F.; Ziegler, J.; Pickel, B.; Hacquard, S. A Microbiota-Root-Shoot Circuit Favours Arabidopsis Growth Over Defence Under Suboptimal Light. Nat. Plants 2021, 7, 1078–1092. [Google Scholar] [CrossRef] [PubMed]
- Mariutto, M.; Duby, F.; Adam, A.; Bureau, C.; Fauconnier, M.L.; Ongena, M.; Thonart, P.; Dommes, J. The Elicitation of a Systemic Resistance by Pseudomonas putida BTP1 in Tomato Involves the Stimulation of Two Lipoxygenase Isoforms. BMC Plant Biol. 2011, 11, 29. [Google Scholar] [CrossRef]
- Vanitha, S.C.; Umesha, S. Pseudomonas fluorescens-Mediated Systemic Resistance in Tomato Is Driven Through an Elevated Synthesis of Defense Enzymes. Biol. Plant. 2011, 55, 317–322. [Google Scholar] [CrossRef]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought Induction of Arabidopsis 9-cis-Epoxycarotenoid Dioxygenase Occurs in Vascular Parenchyma Cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef]
- Wang, D.; Wei, L.; Ma, J.; Wan, Y.; Huang, K.; Sun, Y.; Wen, H.; Chen, Z.; Li, Z.; Yu, D.; et al. Bacillus cereus NJ01 Induces SA- and ABA-Mediated Immunity against Bacterial Pathogens through the EDS1-WRKY18 Module. Cell Rep. 2024, 43, 113985. [Google Scholar] [CrossRef]
- Mazurier, S.; Lemunier, M.; Hartmann, A.; Siblot, S.; Lemanceau, P. Conservation of Type III Secretion System Genes in Bradyrhizobium Isolated from Soybean. FEMS Microbiol. Lett. 2006, 259, 317–325. [Google Scholar] [CrossRef][Green Version]
- Liao, C.H.; McCallus, D.E.; Fett, W.F.; Kang, Y. Identification of Gene Loci Controlling Pectate Lyase Production and Soft-Rot Pathogenicity in Pseudomonas marginalis. Can. J. Microbiol. 1997, 43, 425–431. [Google Scholar] [CrossRef]
- Fauvart, M.; Verstraeten, N.; Dombrecht, B.; Venmans, R.; Beullens, S.; Heusdens, C.; Michiels, J. Rhizobium etli HrpW Is a Pectin-Degrading Enzyme and Differs from Phytopathogenic Homologues in Enzymically Crucial Tryptophan and Glycine Residues. Microbiology 2009, 155, 3045–3054. [Google Scholar] [CrossRef]
- Kvitko, B.H.; Ramos, A.R.; Morello, J.E.; Oh, H.S.; Collmer, A. Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 2007, 189, 8059–8072. [Google Scholar] [CrossRef] [PubMed]
- Pineau, C.; Guschinskaya, N.; Gonçalves, I.R.; Ruaudel, F.; Robert, X.; Gouet, P.; Ballut, L.; Shevchik, V.E. Structure–function analysis of pectate lyase Pel3 reveals essential facets of protein recognition by the bacterial type 2 secretion system. J. Biol. Chem. 2021, 296, 100305. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of Leguminous Nodulation Signaling by the Rhizobial Type III Secretion System. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, W.; Zhang, L.Q.; Liu, X.; Wei, H.L. Supramolecular Structure and Functional Analysis of the Type III Secretion System in Pseudomonas fluorescens 2P24. Front. Plant Sci. 2015, 6, 1190. [Google Scholar] [CrossRef]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Targeting of Plant Pattern Recognition Receptor-Triggered Immunity by Bacterial Type-III Secretion System Effectors. Curr. Opin. Microbiol. 2015, 23, 14–22. [Google Scholar] [CrossRef]
- Mammarella, N.D.; Cheng, Z.; Fu, Z.Q.; Daudi, A.; Bolwell, G.P.; Dong, X.; Ausubel, F.M. Apoplastic Peroxidases Are Required for Salicylic Acid-Mediated Defense Against Pseudomonas syringae. Phytochemistry 2015, 112, 110–121. [Google Scholar] [CrossRef]
- Puttilli, M.R.; Danzi, D.; Correia, C.; Brandi, J.; Cecconi, D.; Manfredi, M.; Marengo, E.; Santos, C.; Spinelli, F.; Polverari, A.; et al. Plant Signals Anticipate the Induction of the Type III Secretion System in Pseudomonas syringae pv. actinidiae, Facilitating Efficient Temperature-Dependent Effector Translocation. Microbiol. Spectr. 2022, 10, e0207322. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two Simple Media for the Demonstration of Pyocyanin and Fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ross, A.; Somssich, I.E. A DNA-Based Real-Time PCR Assay for Robust Growth Quantification of the Bacterial Pathogen Pseudomonas syringae on Arabidopsis thaliana. Plant Methods 2016, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.L.; Pelaez, C.A.; Shank, E.A. Monitoring bacterial colonization and maintenance on Arabidopsis thaliana roots in a floating hydroponic system. JoVE-J. Vis. Exp. 2019, e59517. [Google Scholar] [CrossRef]
- Lozo, J.; Danojević, D.; Jovanović, Ž.; Nenadović, Đ.; Fira, S.; Stanković, S.; Radović, S. Genotype-Dependent Antioxidative Response of Four Sweet Pepper Cultivars to Water Deficiency as Affected by Drought-Tolerant Bacillus safensis SS-2.7 and Bacillus thuringiensis SS-29.2 Strains. Horticulturae 2022, 8, 236. [Google Scholar] [CrossRef]
- Gijzen, M.; Van Huystee, R.; Buzzell, R.I. Soybean Seed Coat Peroxidase (A Comparison of High-Activity and Low-Activity Genotypes). Plant Physiol. 1993, 103, 1061–1066. [Google Scholar] [CrossRef][Green Version]
- Cheema, S.; Sommerhalter, M. Characterization of Polyphenol Oxidase Activity in Ataulfo Mango. Food Chem. 2015, 171, 382–387. [Google Scholar] [CrossRef]
- Kleinhofs, A.; Haskins, F.A.; Gorz, H.J. Relationship of Phenylalanine Ammonia-Lyase Activity to o-Hydroxycinnamic Acid Content in Melilotus alba. Plant Physiol. 1966, 41, 1276–1279. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://github.com/s-andrews/FastQC (accessed on 5 December 2024).
- Martin, M. CUTADAPT Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline Reach with Protein Family Model Curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Néron, B.; Denise, R.; Coluzzi, C.; Touchon, M.; Rocha, E.P.; Abby, S.S. MacSyFinder v2: Improved modelling and search engine to identify molecular systems in genomes. Peer Community J. 2023, 3, e28. [Google Scholar] [CrossRef]
- Wagner, N.; Avram, O.; Gold-Binshtok, D.; Zerah, B.; Teper, D.; Pupko, T. Effectidor: An Automated Machine-Learning-Based Web Server for the Prediction of Type-III Secretion System Effectors. Bioinformatics 2022, 38, 2341–2343. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Alexeyev, M.F. The pKNOCK Series of Broad-Host-Range Mobilizable Suicide Vectors for Gene Knockout and Targeted DNA Insertion into the Chromosome of Gram-Negative Bacteria. Biotechniques 1999, 26, 824–826. [Google Scholar] [CrossRef]
- Figurski, D.H.; Helinski, D.R. Replication of an Origin-Containing Derivative of Plasmid RK2 Dependent on a Plasmid Function Provided in Trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef]

| Strain | Species | Sample Source and Month |
|---|---|---|
| ML159 | P. lurida | Phyllosphere in May |
| ML170 | P. marginalis | |
| MRh80 | P. corrugata | Rhizosphere in May |
| OL141 | P. marginalis | Phyllosphere in October |
| OL154 | P. marginalis | |
| OR207 | P. brassicacearum | Rhizosphere in October |
| ORh77 | P. arenae | |
| ORh26 | P. marginalis | |
| OR228 | P. brassicacearum | |
| ORh244 | P. kilonensis |
| Locus Tag | Family | Effector Probability Score | SMART/PFAM Domain Accession |
|---|---|---|---|
| ACKCUY_RS25290 | Polysaccharide lyase family 1 | 1 | SM000656 SM000710 |
| ACKCUY_RS09560 | HopJ type III effector | 1 | PF08888 |
| ACKCUY_RS17770 | Hypothetical protein | 0.99 | SM000354 PF16422 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nedeljković, M.; Mesaroš, A.; Radosavljević, M.; Đorđević, N.; Stanković, S.; Lozo, J.; Atanasković, I. Type III Secretion System-Mediated Induction of Systemic Resistance by Pseudomonas marginalis ORh26 Enhances Sugar Beet Defence Against Pseudomonas syringae pv. aptata. Plants 2025, 14, 1621. https://doi.org/10.3390/plants14111621
Nedeljković M, Mesaroš A, Radosavljević M, Đorđević N, Stanković S, Lozo J, Atanasković I. Type III Secretion System-Mediated Induction of Systemic Resistance by Pseudomonas marginalis ORh26 Enhances Sugar Beet Defence Against Pseudomonas syringae pv. aptata. Plants. 2025; 14(11):1621. https://doi.org/10.3390/plants14111621
Chicago/Turabian StyleNedeljković, Marija, Aleksandra Mesaroš, Marija Radosavljević, Nikola Đorđević, Slaviša Stanković, Jelena Lozo, and Iva Atanasković. 2025. "Type III Secretion System-Mediated Induction of Systemic Resistance by Pseudomonas marginalis ORh26 Enhances Sugar Beet Defence Against Pseudomonas syringae pv. aptata" Plants 14, no. 11: 1621. https://doi.org/10.3390/plants14111621
APA StyleNedeljković, M., Mesaroš, A., Radosavljević, M., Đorđević, N., Stanković, S., Lozo, J., & Atanasković, I. (2025). Type III Secretion System-Mediated Induction of Systemic Resistance by Pseudomonas marginalis ORh26 Enhances Sugar Beet Defence Against Pseudomonas syringae pv. aptata. Plants, 14(11), 1621. https://doi.org/10.3390/plants14111621












