A SUPERMAN-like Gene Controls the Locule Number of Tomato Fruit
Abstract
1. Introduction
2. Results
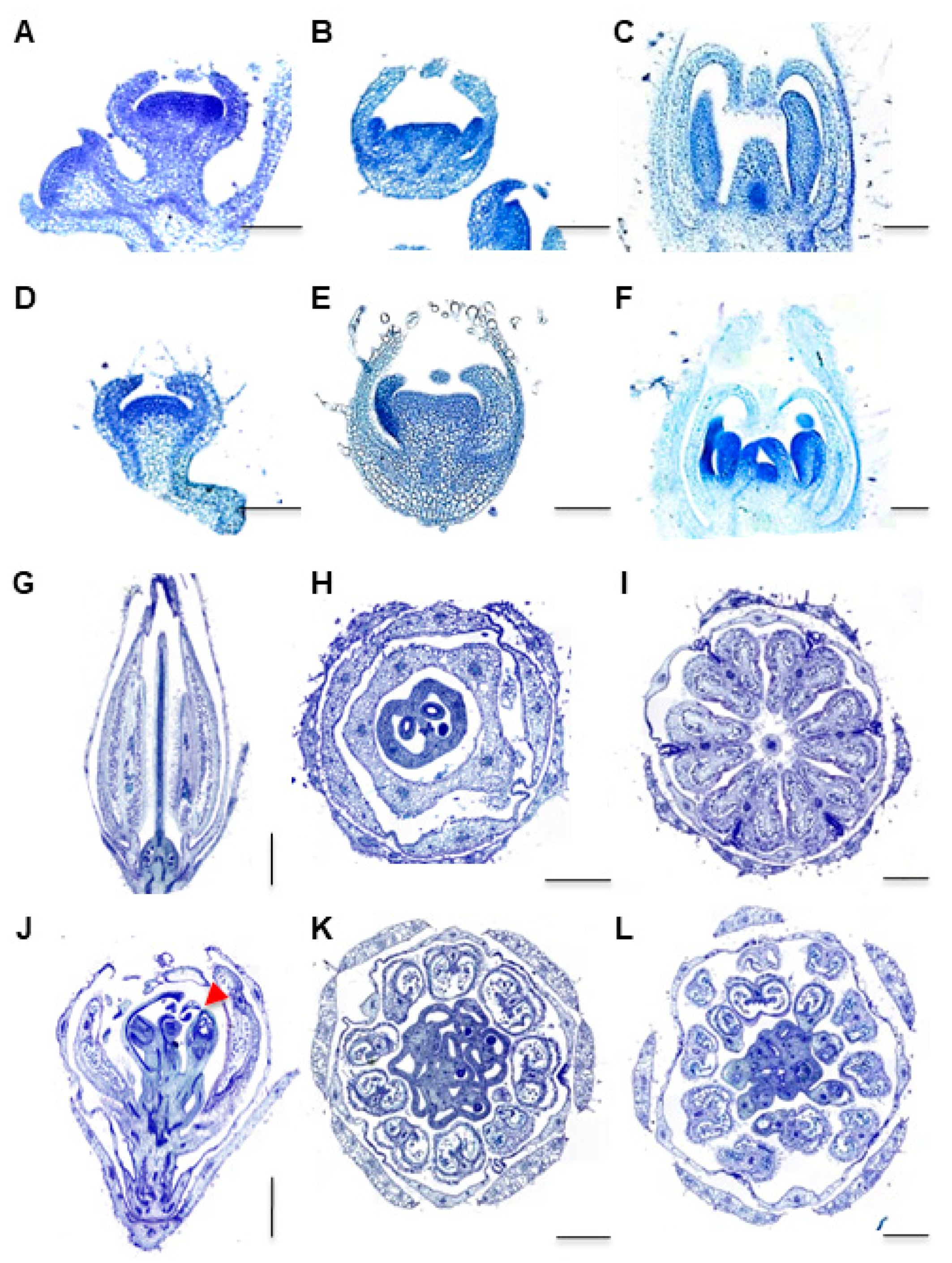
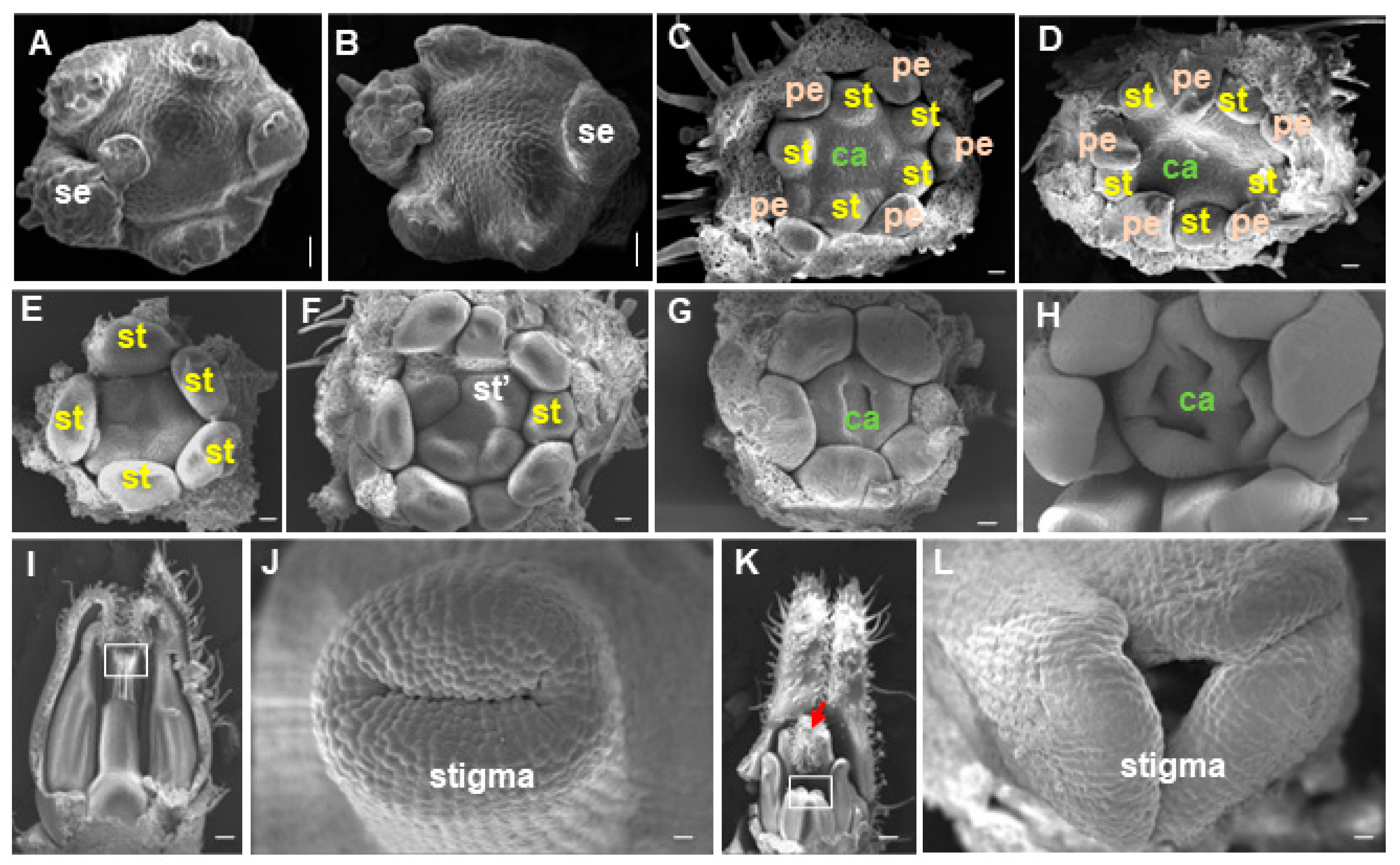
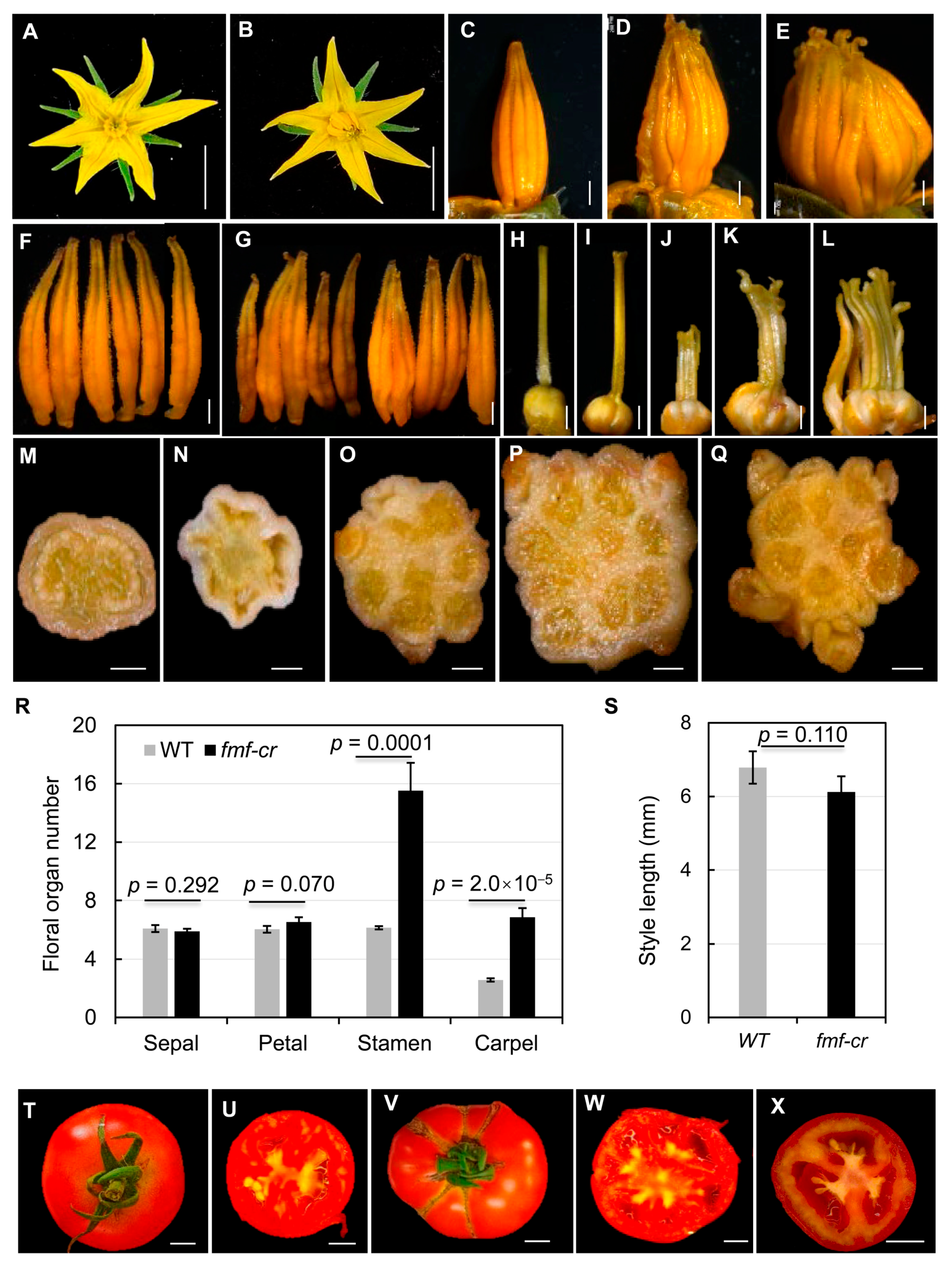
2.1. Phenotypic Analysis of the Fmf Mutant
2.2. Molecular Cloning of the FMF Gene
2.3. Creation of New fmf Alleles by CRISPR-Cas9
2.4. FMF Expression during Flower Development
3. Discussion
4. Materials and Methods
4.1. Materials and Plant Growth Conditions
4.2. Bulked-Segregant Analysis-Seq (BSA-Seq) and Map-Based Cloning
4.3. Generation of fmf-cr Mutants by CRISPR-Cas9
4.4. Phylogenetic Analysis of FMF Homologs
4.5. Microscopy
4.6. In Situ Hybridization
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; Van Der Knaap, E. A Retrotransposon-Mediated Gene Duplication Underlies Morphological Variation of Tomato Fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef]
- Chu, Y.; Jang, J.; Huang, Z.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 2019, 3, e00142. [Google Scholar] [CrossRef]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 406, 800–804. [Google Scholar] [CrossRef]
- Munos, S.; Ranc, N.; Botton, E.; Bérard, A.; Rolland, S.; Duffé, P.; Carretero, Y.; Le Paslier, M.C.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Xiao, H.; Radovich, C.; Welty, N.; Hsu, J.; Li, D.; Meulia, T.; van der Knaap, E. Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 2009, 9, 49. [Google Scholar] [CrossRef]
- Breuil-Broyer, S.; Trehin, C.; Morel, P.; Boltz, V.; Sun, B.; Chambrier, P.; Ito, T.; Negrutiu, I. Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male-female boundary, flower meristem termination and carpel compartmentalization. Ann. Bot. 2016, 117, 905–923. [Google Scholar] [CrossRef]
- Monniaux, M.; Vandenbussche, M. How to Evolve a Perianth: A Review of Cadastral Mechanisms for Perianth Identity. Front. Plant Sci. 2018, 9, 1573. [Google Scholar] [CrossRef]
- Bowman, J.L.; Sakai, H.; Jack, T.; Weigel, D.; Mayer, U.; Meyerowitz, E.M. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 1992, 114, 599–615. [Google Scholar] [CrossRef]
- Sakai, H.; Medrano, L.J.; Meyerowitz, E.M. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 1995, 378, 199–203. [Google Scholar] [CrossRef]
- Prunet, N.; Yang, W.; Das, P.; Meyerowitz, E.M.; Jack, T.P. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl. Acad. Sci. USA 2017, 114, 7166–7171. [Google Scholar] [CrossRef]
- Hiratsu, K.; Mitsuda, N.; Matsui, K.; Ohme-Takagi, M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 2004, 321, 172–178. [Google Scholar] [CrossRef]
- Hiratsu, K.; Ohta, M.; Matsui, K.; Ohme-Takagi, M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 2002, 514, 351–354. [Google Scholar] [CrossRef]
- Nakagawa, H.; Ferrario, S.; Angenent, G.C.; Kobayashi, A.; Takatsuji, H. The Petunia Ortholog of Arabidopsis SUPERMAN Plays a Distinct Role in Floral Organ Morphogenesis. Plant Cell 2004, 16, 920–932. [Google Scholar] [CrossRef]
- Xu, W.; Zhu, W.; Yang, L.; Liang, W.; Li, H.; Chen, M.; Luo, Z.; Huang, G.; Duan, L.; Dreni, L.; et al. SMALL REPRODUCTIVE ORGANS, a SUPERMAN-like transcription factor, regulates stamen and pistil growth in rice. New Phytol. 2021, 233, 1701–1718. [Google Scholar] [CrossRef]
- Rodas, A.L.; Roque, E.; Hamza, R.; Gómez-Mena, C.; Minguet, E.G.; Wen, J.; Mysore, K.S.; Beltrán, J.P.; Cañas, L.A. MtSUPERMAN plays a key role in compound inflorescence and flower development in Medicago truncatula. Plant J. 2020, 105, 816–830. [Google Scholar] [CrossRef]
- Rodas, A.L.; Roque, E.; Hamza, R.; Gómez-Mena, C.; Beltrán, J.P.; Cañas, L.A. SUPERMAN strikes again in legumes. Front. Plant Sci. 2023, 14, 1120342. [Google Scholar] [CrossRef]
- Maurya, D.; Mukherjee, A.; Akhtar, S.; Chattopadhyay, T. Development and validation of the OVATE gene-based functional marker to assist fruit shape selection in tomato. 3 Biotech 2021, 11, 474. [Google Scholar] [CrossRef]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Gardener, B.B.; Francis, D.; van Der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef]
- Sierra-Orozco, E.; Shekasteband, R.; Illa-Berenguer, E.; Snouffer, A.; van der Knaap, E.; Lee, T.G.; Hutton, S.F. Identification and characterization of GLOBE, a major gene controlling fruit shape and impacting fruit size and marketability in tomato. Hortic. Res. 2021, 8, 138. [Google Scholar] [CrossRef]
- van der Knaap, E.; Chakrabarti, M.; Chu, Y.H.; Clevenger, J.P.; Illa-Berenguer, E.; Huang, Z.; Keyhaninejad, N.; Mu, Q.; Sun, L.; Wang, Y.; et al. What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 2014, 5, 227. [Google Scholar] [CrossRef]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato Transcription Factor SlWUS Plays an Important Role in Tomato Flower and Locule Development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef]
- Fletcher, J.C. The CLV-WUS Stem Cell Signaling Pathway: A Roadmap to Crop Yield Optimization. Plants 2018, 7, 87. [Google Scholar] [CrossRef]
- Bondada, R.; Somasundaram, S.; Marimuthu, M.P.; Badarudeen, M.A.; Puthiyaveedu, V.K.; Maruthachalam, R. Natural epialleles of Arabidopsis SUPERMAN display superwoman phenotypes. Commun. Biol. 2020, 3, 772. [Google Scholar] [CrossRef]
- Cao, X.; Jacobsen, S.E. Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef]
- Gaiser, J.C.; Robinson-Beers, K.; Gasser, C.S. The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 1995, 7, 333–345. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Meyerowitz, E.M. Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 1997, 277, 1100–1103. [Google Scholar] [CrossRef]
- Bereterbide, A.; Hernould, M.; Farbos, I.; Glimelius, K.; Mouras, A. Restoration of stamen development and production of functional pollen in an alloplasmic CMS tobacco line by ectopic expression of the Arabidopsis thaliana SUPERMAN gene. Plant J. 2002, 29, 607–615. [Google Scholar] [CrossRef]
- Dinkins, R.; Pflipsen, C.; Thompson, A.; Collins, G.B. Ectopic Expression of an Arabidopsis Single Zinc Finger Gene in Tobacco Results in Dwarf Plants. Plant Cell Physiol. 2002, 43, 743–750. [Google Scholar] [CrossRef]
- Huang, H.; Ma, H. FON1, an Arabidopsis Gene That Terminates Floral Meristem Activity and Controls Flower Organ Number. Plant Cell 1997, 9, 115. [Google Scholar] [CrossRef]
- Ito, T.; Sakai, H.; Meyerowitz, E.M. Whorl-Specific Expression of the SUPERMAN Gene of Arabidopsis Is Mediated by cis Elements in the Transcribed Region. Curr. Biol. 2003, 13, 1524–1530. [Google Scholar] [CrossRef]
- Kater, M.M.; Franken, J.; Van Aelst, A.; Angenent, G.C. Suppression of cell expansion by ectopic expression of the Arabidopsis SUPERMAN gene in transgenic petunia and tobacco. Plant J. 2000, 2, 407–413. [Google Scholar] [CrossRef]
- Kazama, Y.; Fujiwara, M.T.; Koizumi, A.; Nishihara, K.; Nishiyama, R.; Kifune, E.; Abe, T.; Kawano, S. A SUPERMAN-like Gene is Exclusively Expressed in Female Flowers of the Dioecious Plant Silene latifolia. Plant Cell Physiol. 2009, 50, 1127–1141. [Google Scholar] [CrossRef]
- Meister, R.J.; Kotow, L.M.; Gasser, C.S. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 2002, 129, 4281–4289. [Google Scholar] [CrossRef]
- Nandi, A.K.; Kushalappa, K.; Prasad, K.; Vijayraghavan, U. A conserved function for Arabidopsis SUPERMAN in regulating floral-whorl cell proliferation in rice, a monocotyledonous plant. Curr. Biol. 2000, 10, 215–218. [Google Scholar] [CrossRef]
- Nibau, C.; Di Stilio, V.S.; Wu, H.-M.; Cheung, A.Y. Arabidopsis and Tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. J. Exp. Bot. 2010, 62, 949–961. [Google Scholar] [CrossRef]
- Rohde, A.; Grunau, C.; Beck, D.L.; Montagu, M.V.; Rosenthal, A.; Boerjan, W. carpel, a New Arabidopsis Epi-Mutant of the SUPERMAN Gene: Phenotypic Analysis and DNA Methylation Status. Plant Cell Physiol. 1999, 40, 961–972. [Google Scholar] [CrossRef]
- Sakai, H.; Krizek, B.A.; Jacobsen, S.E.; Meyerowitz, E.M. Regulation of SUP Expression Identifies Multiple Regulators Involved in Arabidopsis Floral Meristem Development. Plant Cell 2000, 12, 1607. [Google Scholar] [CrossRef]
- Xu, K.; Wang, L.; Liu, N.; Xie, X.; Zhu, Y. Characterization of a SUPERMAN-like Gene, MdSUP11, in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2018, 125, 136–142. [Google Scholar] [CrossRef]
- Yun, J.-Y.; Weigel, D.; Lee, I. Ectopic Expression of SUPERMAN Suppresses Development of Petals and Stamens. Plant Cell Physiol. 2002, 43, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, M.; Jiang, L.; Ding, L.; Yan, S.S.; Zhang, J.; Dong, Z.; Ren, H.; Zhang, X. Cucumber SUPERMAN Has Conserved Function in Stamen and Fruit Development and a Distinct Role in Floral Patterning. PLoS ONE 2014, 9, e86192. [Google Scholar] [CrossRef] [PubMed]
- Laux, T.; Mayer, K.F.X.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Xu, Q.; Li, R.; Weng, L.; Sun, Y.; Li, M.; Xiao, H. Domain-specific expression of meristematic genes is defined by the LITTLE ZIPPER protein DTM in tomato. Commun. Biol. 2019, 2, 134. [Google Scholar] [CrossRef]
- Lei, Y.; Lu, L.; Liu, H.-Y.; Li, S.; Xing, F.; Chen, L.-L. CRISPR-P: A Web Tool for Synthetic Single-Guide RNA Design of CRISPR-System in Plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Liu, W.; Zhu, X.; Lei, M.; Xia, Q.; Botella, J.R.; Zhu, J.-K.; Mao, Y. A detailed procedure for CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana. Sci. Bull. 2015, 60, 1332–1347. [Google Scholar] [CrossRef][Green Version]
- McCormick, S.; Niedermeyer, J.; Fry, J.; Barnason, A.; Horsch, R.; Fraley, R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986, 5, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Zhao, F.; Li, R.; Xu, C.; Chen, K.; Xiao, H. The Zinc Finger Transcription Factor SlZFP2 Negatively Regulates Abscisic Acid Biosynthesis and Fruit Ripening in Tomato. Plant Physiol. 2015, 167, 931–949. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Zhou, E.; Li, M.; Tian, S.; Xiao, H. A SUPERMAN-like Gene Controls the Locule Number of Tomato Fruit. Plants 2023, 12, 3341. https://doi.org/10.3390/plants12183341
Zhang M, Zhou E, Li M, Tian S, Xiao H. A SUPERMAN-like Gene Controls the Locule Number of Tomato Fruit. Plants. 2023; 12(18):3341. https://doi.org/10.3390/plants12183341
Chicago/Turabian StyleZhang, Mi, Enbai Zhou, Meng Li, Shenglan Tian, and Han Xiao. 2023. "A SUPERMAN-like Gene Controls the Locule Number of Tomato Fruit" Plants 12, no. 18: 3341. https://doi.org/10.3390/plants12183341
APA StyleZhang, M., Zhou, E., Li, M., Tian, S., & Xiao, H. (2023). A SUPERMAN-like Gene Controls the Locule Number of Tomato Fruit. Plants, 12(18), 3341. https://doi.org/10.3390/plants12183341





