Multicellularity and the Need for Communication—A Systematic Overview on (Algal) Plasmodesmata and Other Types of Symplasmic Cell Connections †
Abstract
1. Multicellularity and the Need of Communication
2. Please Contact—Various Types of Non-Plasmodesmatal Cell Connections
2.1. Animals
2.2. Fungi
2.3. Red Algae
2.4. Green Algae—Ulvophyceae and Chlorophyta with Uncertain Taxonomic Position
2.5. Green Algae—Volvocaceae
3. Conclusions I
- (1)
- Intercellular bridges of animals, fungal septal pores, and pit plugs of red algae show striking similarities pertaining to their general structure and to their mechanisms of formation although they have evolved independently from each other [43]. Suppression of abscission, leaving an open, membrane-lined cytoplasmic pore in the center of an ingressive cleavage furrow, might be a simple mode to form symplasmic cell connections.The core molecular machinery of abscission appears to be highly conserved across eukaryotes [43] and slight molecular changes may be sufficient to convert full into incomplete abscission as shown for Chlamydomonas reinhardtii [99]. Repeated independent evolutionary origins of (simple) multicellularity are, therefore, highly probable.Cell connections retain symplasmic communication between daughter cells, but also serve cell cohesion [96] which might be relevant in early phases of the transition from uni- to multicellular body plans. Such a mechanical function may also pertain to the cell connections interconnecting non-clonally related cells across lateral walls in pseudoparenchymatous thalli of Rhodophyta.
- (2)
- Plugs observed with fungal, red algal, Ulvophyceae, and permanent volvocaceaen cell connections also occur in PD at particular interfaces of streptophyte algae [59] and between undifferentiated cells of plant calluses [39]. Targeted occlusion has likely evolved multiple times to exert systematic size restriction on symplasmic exchange [81] and eventually creates partially or fully isolated symplasmic domains [31,59,69] which subdivide the multicellular organisms into heterogenous regions with different developmental fates and/or metabolic functions to allow division of labor.
- (3)
- We suggest the term ‘conjunctor’ pit connections to replace the term ‘secondary’ pit connections in Rhodophyta, since they develop primarily in the course of a cell division [31] and connect clonally non-related cells only after cell fusion events.
- (4)
- Cytoplasmic bridges formed temporarily or permanently in developing Volvocaceae colonies are slightly wider than canonical PD and do not always possess an ER-derived substructure. At least in V. carteri, they develop among fusing vesicles in a cellplate-like structure. In this respect, they resemble canonical land-plant PD [31] and might be regarded as PD-like connections (PDLCs) in contrast to all other types of cell connection discussed in this section (Figure 2).
4. Get in Touch—PD and Plasmodesmata-Like Cell Connections (PDLCs)

4.1. PD/PDLCs in Fungi
4.2. PD/PDLCs in Brown Algae—Phaeophyceae
4.3. PD/PDLCs in the Green Lineage—Viridiplantae (Prasinodermophyta and Unicellular Core Chlorophyta)
4.4. PD/PDLCs in the Green Lineage—Viridiplantae (Multicellular Chlorophyta)
4.4.1. Trebouxiophyceae
| Class | Order | Family | Genus | n (Species Examined) | n (Publications) | |
|---|---|---|---|---|---|---|
| Total | with PD(LC) | |||||
| Trebouxiophyceae | Prasiolales | Stichococcaceae | Deuterostichococcus | 1 | 0 | 1 |
| Stichococcus | 1 | 0 | 3 | |||
| +6 | ||||||
| Koliellaceae | Koliella | 1 | 0 | 1 | ||
| +4 | ||||||
| Prasiolaceae | Prasiola | 1 | 0 | 1 | ||
| +8 | ||||||
| +1 | ||||||
| Microthamniales | Microthamniaceae | Microthamnion | 1 | 0 | 1 | |
| +2 | ||||||
| ordo i.s. | familia i.s. | Leptosira | 1 | 0 | 2 | |
| +4 | ||||||
| Ulvophyceae | Ulotrichales | Ulotrichaceae | Ulothrix | 11 | 1 a | 6 + 2 |
| +20 | ||||||
| Sarcinofilaceae | Sarcinofilum | 1 | 0 | 2 | ||
| +1 | ||||||
| Helicodictyaceae | Rhexinema | 1 | 0 | 1 | ||
| +2 | ||||||
| Tupiellaceae | Vischerioclonium | 1 | 0 | 2 | ||
| +1 | ||||||
| +11 | ||||||
| Ulvales | Ctenocladaceae | Ctenocladus | 1 | 0 | 3 + 1 | |
| +3 | ||||||
| Ulvaceae | Ulva | 4 | 0 | 6 | ||
| Percursaria | 1 | 0 | 1 | |||
| +11 | ||||||
| Kornmanniaceae | Lithotrichon | 1 | 0 | 1 | ||
| +7 | ||||||
| Ulvellaceae | Ulvella | 1 | 0 | 1 | ||
| +6 | ||||||
| +3 | ||||||
| Trentepohliales | Trentepohliaceae | Trentepohlia | 7 + 4 | 7 + 4 | 9 | |
| Cephaleuros | 2 + 2 | 2 + 2 | 3 | |||
| Phycopeltis | 1 | 1 | 1 | |||
| +6 | ||||||
| +10 | ||||||
| Chlorophyceae | Chaetophorales | Aphanochaetaceae | Aphanochaete | 2 | 2 | 2 |
| +3 | ||||||
| Barrancaceae | Barranca | 1 | 0 | 1 | ||
| Chaetophoraceae | Stigeoclonium | 6 + 1 | 6 + 1 | 6 | ||
| Caespitella | 1 | 1 | 1 | |||
| Chaetophora | 1 | 1 | 1 | |||
| Draparnaldia | 1 + 1 | 1 + 1 | 1 + 1 | |||
| Gongrosira b | 1 | 0 | 2 | |||
| Pleurococcus c | 1 | 0 | 1 | |||
| Sporocladopsis | 1 | 1 | 1 | |||
| Leptosiropsis | 1 | 0 | 1 | |||
| +42 | ||||||
| Fritschiellaceae | Fritschiella | 2 | 1 d | 2 | ||
| Schizomeridaceae | Schizomeris | 1 | 1 | 1 + 1 | ||
| Uronemataceae | Uronema | 3 + 1 e | 3 + 1 e | 2 | ||
| Chaetopeltidales | Chaetopeltidaceae | Pseudulvella | 1 | 0 | 1 | |
| Koshicola | 1 | 0 | 1 | |||
| Oncosaccus | 1 | 0 | 1 | |||
| +7 | ||||||
| +1 | ||||||
| Oedogoniales | Oedogoniaceae | Oedogonium | 3 + 2 | 1 + 2 | 5 | |
| Bulbochaete | 1 | 1 | 5 | |||
| +2 | ||||||
| Sphaeropleales | Microsporaceae | Microspora | 1 | 0 | 2 | |
| Flintia | 1 | 0 | 1 | |||
| Cylindrocapsaceae | Cylindrocapsa | 2 | 0 | 3 | ||
| +2 | ||||||
| +20 | ||||||
| +1 | ||||||
4.4.2. Ulvophyceae
4.4.3. Chlorophyceae
4.5. PD/PDLCs Structures in the Green Lineage—Viridiplantae (Streptophyte Algae)
| Class | Order | Family | Genus | n (Species Examined) | n (Publications) | |
|---|---|---|---|---|---|---|
| Total | with PD (LC) | |||||
| Chlorokybophyceae | Chlorokybales | Chlorokybaceae | Chlorokybus | 1 | 0 | 1 |
| Klebsormidiophyceae | Klebsormidiales | Klebsormidiaceae | Klebsormidium | 10 + 2 | 0 | 6 |
| Interfilum a | 3 + 3 | 0 | 2 | |||
| +4 | ||||||
| +1 | ||||||
| Charophyceae | Charales | Characeae | Chara | 6 + 2 | 6 + 2 | 13 |
| Nitella | 2 | 2 | 3 | |||
| +48 | ||||||
| +5 | ||||||
| +3 | ||||||
| Coleochaetophyceae | Coleochaetales | Coleochaetaceae | Coleochaete | 5 | 5 | 5 |
| +4 | ||||||
| +1 | ||||||
| Zygnematophyceae | Spirogyrales | Spirogyraceae | Spirogyra | 1 | 0 | 1 |
| +1 | ||||||
| Zygnematales | Zygnemataceae | Zygnema | 0 + 3 | 0 | 1 | |
| Mougeotia | 2 + 1 | 0 | 2 | |||
| +24 | ||||||
| +1 | ||||||
| +3 | ||||||
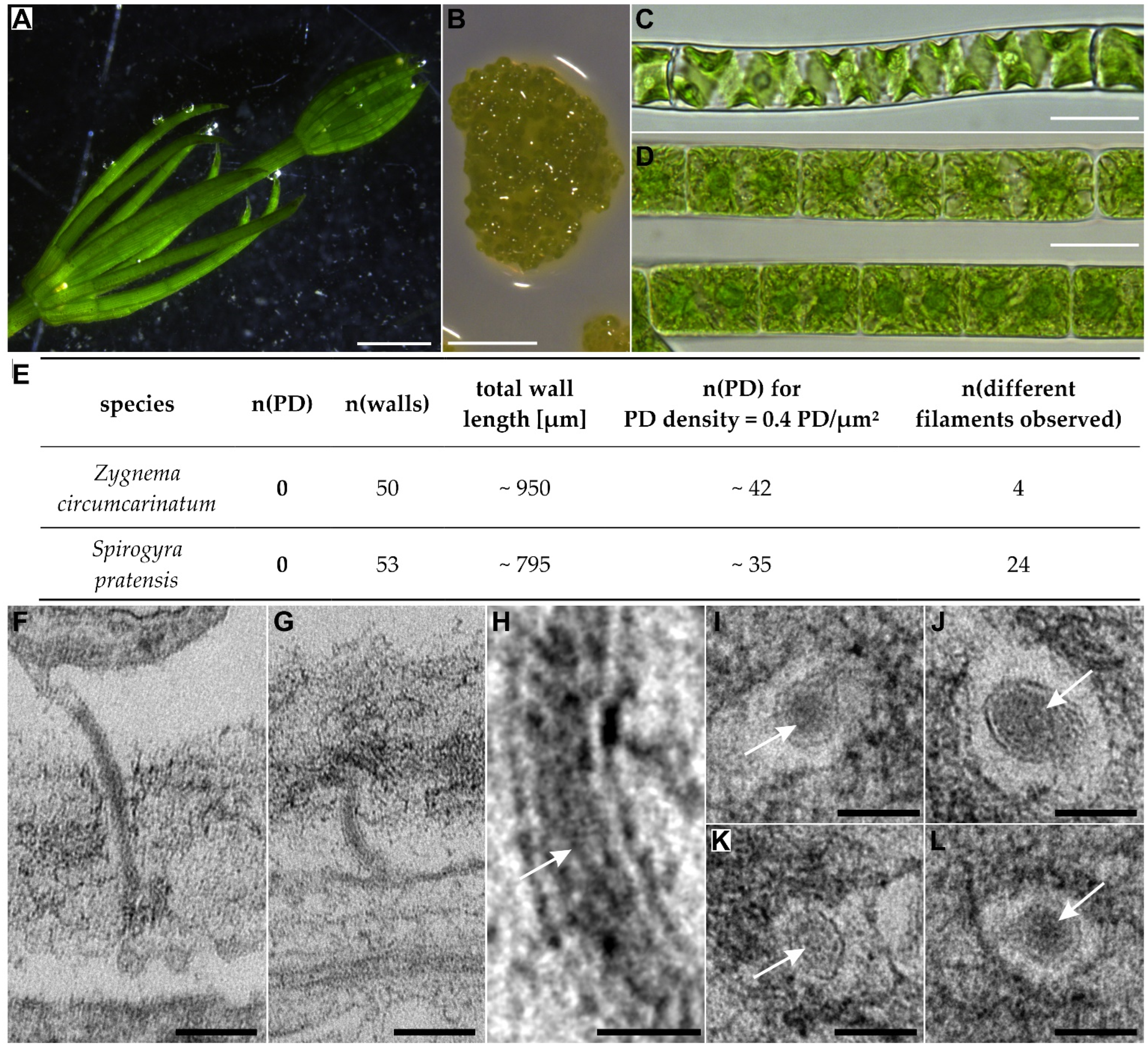
5. Conclusions II
- (1)
- The data gathered from the literature to map the present-day distribution of PD/PDLCs on today’s taxonomic classification of extant algal lineages (Figure 2, Figures S1 and S2) show that there are several taxa which have not or not sufficiently been investigated, although they have an interesting phylogenetic position; e.g., the Discosporangiales at the base of the radiation of Phaeophyceae, the newly described Barrancaceae within the Chaetophorales, the Acrosiphoniales and Cladophorales within the Ulvophyceae, the genus Oedocladium in the Oedogoniales, and, of course, the streptophyte algae Coleochaetales and Charales.
- (2)
- According to the available data, we would conclude, however, that there have been at least six independent origins of PD/PDLCs:
- -
- at least one in the Phaeophyceae, probably before the start of their radiation;
- -
- an extraordinary one in the Trentepohliales, which, after the proposed reclassification of ‘Ulothrix fimbriata’ as ‘Uronema fimbriata’, would be the only PDLC-bearing taxon in the Ulvophyceae. The evolution of PDLCs in the Trentepohliales might be a consequence of a special selection pressure in their terrestrial habitats or a prerequisite for the survival in their ecological niche (see Section 4.4.2);
- -
- most likely three in the Chlorophyceae:
- (i)
- Volvophyceae (Chlamydomonadales, CS clade), since their ‘cytoplasmic bridges’ resemble PDLCs rather than non-PD cell connections if the mechanism of their formation is considered;
- (ii)
- Oedogoniales whose PDLCs differ structurally from all other algal PDLCs;
- (iii)
- Chaetophorales (possibly with a very special situation in Sporocladopsis; see Section 4.4.3);
- -
- at least one in the Phragmoplastophyta, including the land plants. Homology of all PD in this taxon would be supported by the presented hints on the occurrence of a permanent ER substructure in their PD. Yet, two or three independent origins of PD/PDLCs in the Phragmoplastophyta can still not be ruled out.
- (3)
- Data must be considered with care, though, since the present-day occurrence of PD/PDLCs in extant algal taxa might not mirror the complete picture of PD evolution. In view of the enormous age of the green lineage, it can be expected that several independent transitions of unicellular green algae to coenocytic, siphonous, and multicellular body plans with increasing complexity have taken place. However, reductions of algal body plans have also occurred in other lineages [7] so that complexity and even multicellularity have been lost at several evolutionary points. This interplay between gain and loss of traits hampers inferences about the evolution of particular characteristics, like the occurrence of PD.Moreover, as discussed in Section 4.1 (fungi), Section 4.4.2 (Ulvales, Trentepohliales), and Section 4.4.3 (Sporocladopsis novae-zelandiae), it should be taken into account that other mechanisms like horizontal gene transfer and cross-kingdom interactions might have influenced the final picture [157,217,218]. It will be a challenging task for future studies to decipher the impact of symbiotic interactions between eukaryotes and bacteria or viruses, as well as between different eukaryotic lineages on the evolution of multicellularity and symplasmic cell connections.
- (4)
- According to the data presented, PD/PDLCs have evolved in parallel with complex multicellularity rather than with simple multicellularity [19]. Supposedly, direct symplasmic communication was selected for as a group-level trait [23,24] correlated with the evolution of division of labor. It might be a prerequisite for—or a direct consequence of—coordinated development and cooperative functioning of distinct cell types within an algal thallus, in particular with those who had probably undergone the transition from unicellular—via colonial—to multicellular body plans [4,5]. Throughout the phyla, simple multicellular algal organisms were found to lack both cell-type diversification and cell connectivity (i.e., Xanthophyceae, Trebouxiouphyceae, Ulotrichales, Chaetopeltidales, Klebsormidiophyceae, Zygnematophyceae; Figure 2 and Figure S2), while complex multicellular body plans with cell-type specification often coincide with the occurrence of PD/PDLCs (PD-bearing Phaeophyceae, Trentepohliales, Volvocaceae, Oedogoniales, Chaetophorales, Charophyceae, Coleochaetophyceae, and Embryophyta; Figure 2 and Figure S2).The only exceptions would be the following:
- (i)
- the Ulvales whose complex body plans presumably have evolved in the absence of PD/PDLCs via a distinct pathway dependent on cross-kingdom interaction with bacteria;
- (ii)
- the siphonocladous Cladophorales and the (partially siphonocladous) Sphaeropleales which also show complex multicellularity, but lack PD/PDLCs. Independent loss of PD/PDLCs in the latter two taxa might be a conceivable explanation for this finding, but, as speculated in Section 4.4.2, it is also possible that these taxa have achieved complex multicellularity via the ‘siphonous–multicellular’ transformation [4,5] and presence of specialized structures for symplasmic communication might not be a decisive factor in this evolutionary pathway.
- (5)
- We suggest to differentiate clearly between distinct types of cell connections, which can be assumed to have distinct functional capacities (Figure 1):
- -
- Membrane-lined cytosolic channels, enabling plasma-membrane connectivity besides cytosolic connectivity, should be distinguished from other types of cell connections including the proteinaceous gap junctions of animals.
- -
- Cytoplasmic pores developing as single, large channels after incomplete abscission in a furrowing process (and functionally regulated by plugging) should be distinguished from multiple smaller PD/PDLCs which develop during cytokinesis in a cell plate-like structure (either among the fusing vesicles or by protrusion) or which are formed secondarily in a rigid cell wall.
- -
- PDLCs representing the dominant type(s) of symplasmic connections throughout the algal taxa should be distinguished from canonical PD which contain a permanent ER component and, thus, provide connectivity of the endomembrane systems of neighboring cells.
- (6)
- Based on the presented data, we suggest a four-step process towards the evolution of canonical PD:
- (i)
- Establishment of specific plasma-membrane domains [219,220,221] is most likely necessary to control the membrane stability/flexibility required to form small cytosolic channels with relatively constant inner diameters of about 20 nm (reminiscent of Type I PD [34]) in a cell-plate(-like) structure during cytokinesis. This may happen either by leaving open pores between the fusing vesicles or by membrane invaginations (Phaeophyceae) into the developing wall. The latter would pave the way for secondary PD/PDLC formation as found with Phaeophyceae and angiosperms. It is worth noting that PDLCs may develop in centripetally (many algae) or centrifugally growing cell plates in which vesicle fusion occurs either simultaneously (many algae) or successively. The ER is obviously not necessarily required for this process, but a cell plate alone would probably not be sufficient to produce PDLCs as shown with the Ulotrichales (see Section 4.4.2). Membrane domains with proper lipid and protein equipment and, possibly, the presence of a phycoplast or a phragmoplast during cytokinesis might be indispensable for PD/PDLC formation.
- (ii)
- Recruitment of (proteinaceous) substructures attached to the inner face of the plasma-membrane lining the PDLC can be assumed to stabilize the PDLC, as shown with Oedogoniales and with Volvocaceae (Figure 1O,P,U).
- (iii)
- Targeted establishment of barriers within the cytosolic channel of the PDLCs as shown for Phaeophyceae, Oedogoniales, and some Chaetophorales presumably limits and controls the symplasmic transport capacity. Alternatively or in parallel, specialized cell-wall areas form collars around the PDLC (Phaeophyceae) or neck constrictions at the orifices (Oedogoniales) which may control the transport pathways functionally. Specialization of the cell-wall collars around PDLCs might also be a prerequisite for the development of PD twinning as a mode to form PD secondarily in existing walls (Phaeophyceae) and also for branching as a secondary modification of PD/PDLCs (Chara).
- (iv)
- Recruitment of the ER to the PDLC and a permanent fixation of the ER as indelible substructure of the cell connections gives rise to canonical PD enabling an additional endomembrane transport pathway. This process is likely mediated by special membrane-tether proteins and modifications of the ER membrane [124,125,126]. A temporary attachment of the ER to PDLCs, as suggested for some Chaetophorales and some Chara species, might represent an intermediate stage in this process.
6. Outlook
- (1)
- Functional investigations on the symplasmic connectivity mediated by algal PD/PDLCs have only rarely been performed, and, thus, we know hardly anything about the size and biochemical nature of molecules which are transported through PD/PDLCs in Phaeophyceae, Chlorophyta, and streptophyte algae. We need to answer the following questions before we can conclude which role PD/PDLCs might play in the coordination of developmental processes of the algal thalli or in their cell- or non-cell-autonomous responses to environmental changes and (a) biotic stresses.
- (i)
- What is the size-exclusion limit for diffusional transport through algal PD/PDLCs?
- (ii)
- Is there also a targeted transport of macromolecules, like transcription factors, mRNA, and small RNA species with regulatory functions, as was reported for angiosperm PD?
- (iii)
- Is there a membrane flow through algal PD/PDLCs or is the intercellular exchange of lipids restricted by plasma-membrane domains like in angiosperm PD [221]?
- (iv)
- Are there differences in the numbers and/or functional capacities of PD/PDLCs at the diverse cell interfaces of the algal thalli which might establish distinct symplasmic domains like in angiosperm tissues?
- (v)
- Do algae alter their PD/PDLC numbers and/or the functional capacities of their symplasmic cell connections temporarily or permanently during development or in response to physiological stresses like angiosperms do?
- (vi)
- How are putative functional changes of algal PD/PDLC permeability and possible alterations of algal PD/PDLC frequencies achieved?
- (2)
- Furthermore, we know almost nothing about the molecular composition of the algal PD/PDLCs in distinct taxa, except for the fact that typical molecular components of angiosperm PD lack in the streptophyte algae and only single potentially orthologous genes can be identified [222]. Proteomics on algal PDLCs has not been performed, except for a single initial study on Chara corallina [213]. Whether morphological similarities of PD/PDLC in closely or distantly related taxa reflect any homologies on the molecular level is unclear. Do PDLCs in distinct taxa differ with respect to their molecular PDLC equipment, and can the (respective) ‘molecular evolution’ of (analogous) PDLC be traced back with the help of recent species? Final conclusions on the evolution of PD/PDLCs cannot not be drawn without molecular data.
7. Material and Methods
7.1. Ulothrix fimbriata 18S rRNA Gene Phylogeny
7.2. Algae Cultures
7.3. Transmission Electron Microscopy (TEM)
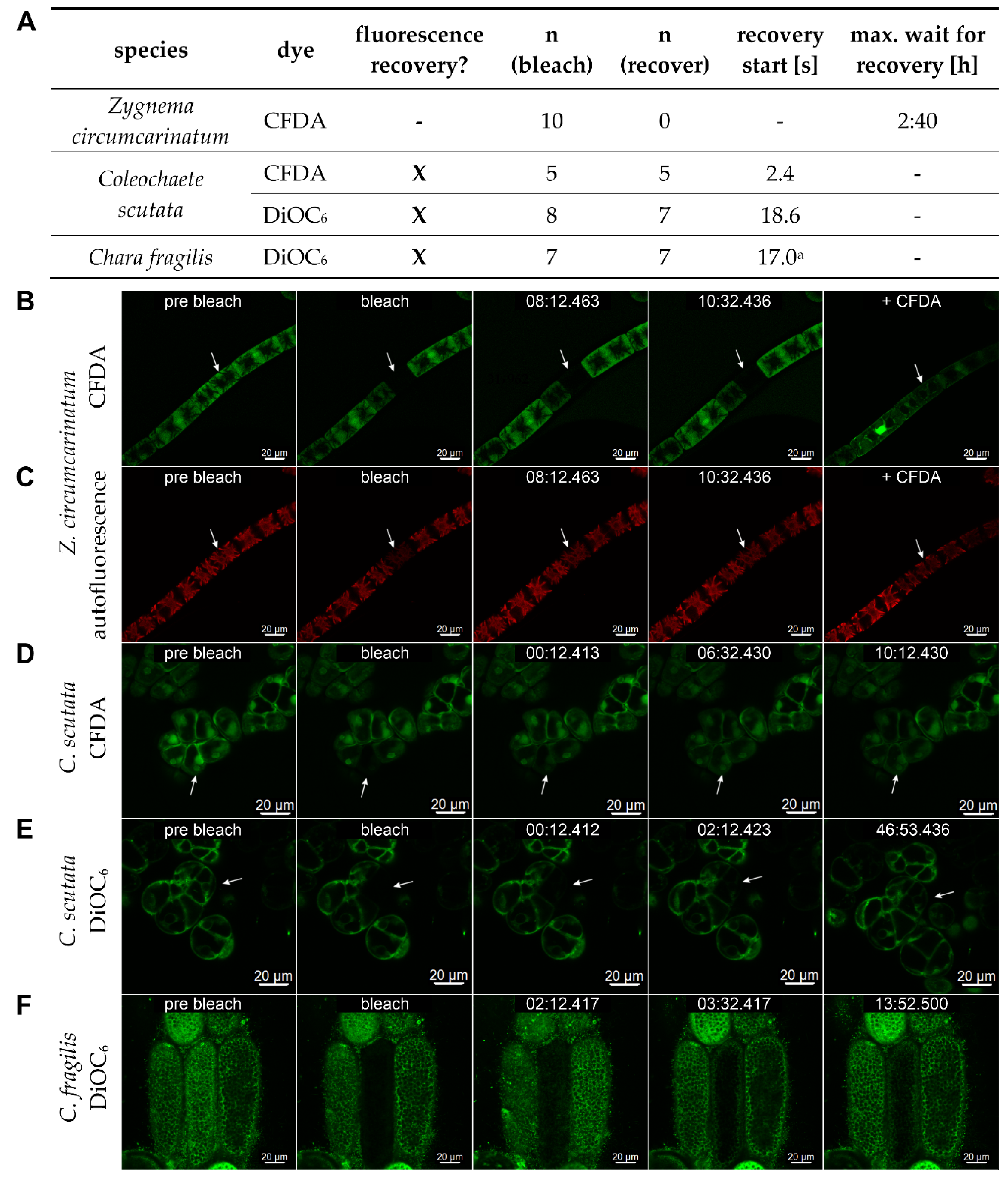
7.4. Fluorescence Recovery after Photobleaching (FRAP)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Umen, J.; Herron, M.D. Green Algal Models for Multicellularity. Annu. Rev. Genet. 2021, 55, 603–632. [Google Scholar] [CrossRef] [PubMed]
- Niklas, K.J.; Newman, S.A. The many roads to and from multicellularity. J. Exp. Bot. 2020, 71, 3247–3253. [Google Scholar] [CrossRef] [PubMed]
- Umen, J.G. Green algae and the origins of multicellularity in the plant kingdom. Cold Spring Harb. Perspect. Biol. 2014, 6, a016170. [Google Scholar] [CrossRef] [PubMed]
- Niklas, K.J. The evolutionary-developmental origins of multicellularity. Am. J. Bot. 2014, 101, 6–25. [Google Scholar] [CrossRef] [PubMed]
- Niklas, K.J.; Newman, S.A. The origins of multicellular organisms. Evol. Dev. 2013, 15, 41–52. [Google Scholar] [CrossRef]
- Lamża, Ł. Diversity of ‘simple’multicellular eukaryotes: 45 independent cases and six types of multicellularity. Biol. Rev. 2023; online version of record before inclusion in an issue. [Google Scholar]
- Hess, S.; Williams, S.; Busch, A.; Irisarri, I.; Delwiche, C.; De Vries, S.; Tatyana, D.; Roger, A.; Archibald, J.; Buschmann, H.; et al. A phylogenomically informed five-order system for the closest relatives of land plants. Curr. Biol. 2022, 32, 4473–4482.e7. [Google Scholar] [CrossRef]
- Day, T.C.; Márquez-Zacarías, P.; Bravo, P.; Pokhrel, A.R.; MacGillivray, K.A.; Ratcliff, W.C.; Yunker, P.J. Varied solutions to multicellularity: The biophysical and evolutionary consequences of diverse intercellular bonds. Biophys. Rev. 2022, 3, 021305. [Google Scholar] [CrossRef]
- Niklas, K.J.; Cobb, E.D.; Crawford, D.R. The evo-devo of multinucleate cells, tissues, and organisms, and an alternative route to multicellularity. Evol. Dev. 2013, 15, 466–474. [Google Scholar] [CrossRef]
- Kirk, D.L. A twelve-step program for evolving multicellularity and a division of labor. Bioessays 2005, 27, 299–310. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Herron, M.D.; Howell, K.; Pentz, J.T.; Rosenzweig, F.; Travisano, M. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat. Commun. 2013, 4, 2742. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. USA 2012, 109, 1595–1600. [Google Scholar] [CrossRef]
- Cope, D. Real-time Evolution of Multicellularity with Artificial Gene Regulation. arXiv 2023, arXiv:2305.12249. [Google Scholar]
- Colizzi, E.S.; Vroomans, R.M.A.; Merks, R.M.H. Evolution of multicellularity by collective integration of spatial information. eLife 2020, 9, e56349. [Google Scholar] [CrossRef] [PubMed]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.C.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De novo origins of multicellularity in response to predation. Sci. Rep. 2019, 9, 2328. [Google Scholar] [CrossRef] [PubMed]
- Boraas, M.E.; Seale, D.B.; Boxhorn, J.E. Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol. Ecol. 1998, 12, 153–164. [Google Scholar] [CrossRef]
- Koschwanez, J.H.; Foster, K.R.; Murray, A.W. Sucrose Utilization in Budding Yeast as a Model for the Origin of Undifferentiated Multicellularity. PLoS Biol. 2011, 9, e1001122. [Google Scholar] [CrossRef]
- Cheloni, G.; Slaveykova, V.I. Morphological plasticity in Chlamydomonas reinhardtii and acclimation to micropollutant stress. Aquat. Toxicol. 2021, 231, 105711. [Google Scholar] [CrossRef]
- Knoll, A.H. The Multiple Origins of Complex Multicellularity. Annu. Rev. Earth Planet. Sci. 2011, 39, 217–239. [Google Scholar] [CrossRef]
- Buschmann, H. Into another dimension: How streptophyte algae gained morphological complexity. J. Exp. Bot. 2020, 71, 3279–3286. [Google Scholar] [CrossRef]
- Márquez-Zacarías, P.; Pineau, R.M.; Gomez, M.; Veliz-Cuba, A.; Murrugarra, D.; Ratcliff, W.C.; Niklas, K.J. Evolution of Cellular Differentiation: From Hypotheses to Models. Trends Ecol. Evol. 2020, 36, 49–60. [Google Scholar] [CrossRef]
- West, S.A.; Fisher, R.M.; Gardner, A.; Kiers, E.T. Major evolutionary transitions in individuality. Proc. Natl. Acad. Sci. USA 2015, 112, 10112–10119. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.J.; Hammerschmidt, K. What Do We Mean by Multicellularity? The Evolutionary Transitions Framework Provides Answers. Front. Ecol. Evol. 2021, 9, 730714. [Google Scholar] [CrossRef]
- Bourrat, P.; Doulcier, G.; Rose, C.J.; Rainey, P.B.; Hammerschmidt, K. Tradeoff breaking as a model of evolutionary transitions in individuality and limits of the fitness-decoupling metaphor. eLife 2022, 11, e73715. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.A.; Rutter, J. Metabolites as signalling molecules. Nat. Rev. Mol. Cell Biol. 2023, 24, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Ullo, M.F.; Case, L.B. How cells sense and integrate information from different sources. WIREs Mech. Dis. 2023, 15, e1604. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Gao, J.; He, Y.; Jiang, L. Plant extracellular vesicles. Protoplasma 2020, 257, 3–12. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, Z.; Liu, X.; Tyler, B.M. Biogenesis and Biological Functions of Extracellular Vesicles in Cellular and Organismal Communication With Microbes. Front. Microbiol. 2022, 13, 817844. [Google Scholar] [CrossRef]
- Borniego, M.L.; Innes, R.W. Extracellular RNA: Mechanisms of secretion and potential functions. J. Exp. Bot. 2023, 74, 2389–2404. [Google Scholar] [CrossRef]
- Ehlers, K.; Kollmann, R. Primary and secondary plasmodesmata: Structure, origin, and functioning. Protoplasma 2001, 216, 1–30. [Google Scholar] [CrossRef]
- Ehlers, K.; Große Westerloh, M. Developmental Control of Plasmodesmata Frequency, Structure, and Function. In Symplasmic Transport in Vascular Plants; Sokołowska, K., Sowiński, P., Eds.; Springer: New York, NY, USA, 2013; pp. 41–82. [Google Scholar]
- Sager, R.; Lee, J.-Y. Plasmodesmata in integrated cell signalling: Insights from development and environmental signals and stresses. J. Exp. Bot. 2014, 65, 6337–6358. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, W.J.; Grison, M.S.; Trépout, S.; Gaston, A.; Fouché, M.; Cordelières, F.P.; Oparka, K.; Tilsner, J.; Brocard, L.; Bayer, E.M. Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants 2017, 3, 17082. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Zambryski, P.C. Plasmodesmata paradigm shift: Regulation from without versus within. Annu. Rev. Plant Biol. 2012, 63, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.H.; Shine, M.B.; de Lorenzo, L.; Yu, K.; Cui, W.; Navarre, D.; Hunt, A.G.; Lee, J.Y.; Kachroo, A.; Kachroo, P. Plasmodesmata Localizing Proteins Regulate Transport and Signaling during Systemic Acquired Immunity in Plants. Cell Host Microbe 2016, 19, 541–549. [Google Scholar] [CrossRef]
- Li, Z.; Paterlini, A.; Glavier, M.; Bayer, E. Intercellular trafficking via plasmodesmata: Molecular layers of complexity. Cell. Mol. Life Sci. CMLS 2021, 78, 799–816. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Stonebloom, S.; Xu, M.; Zambryski, P.C. Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma 2011, 248, 61–74. [Google Scholar] [CrossRef]
- Ehlers, K.; Binding, H.; Kollmann, R. The formation of symplasmic domains by plugging of plasmodesmata: A general event in plant morphogenesis? Protoplasma 1999, 209, 181–192. [Google Scholar] [CrossRef]
- Bayer, E.; Thomas, C.; Maule, A. Symplastic domains in the Arabidopsis shoot apical meristem correlate with PDLP1 expression patterns. Plant Signal. Behav. 2008, 3, 853–855. [Google Scholar] [CrossRef]
- Shimizu, K.; Stopfer, M. Gap junctions. Curr. Biol. 2013, 23, R1026–R1031. [Google Scholar] [CrossRef]
- Drab, M.; Stopar, D.; Kralj-Iglič, V.; Iglič, A. Inception Mechanisms of Tunneling Nanotubes. Cells 2019, 8, 626. [Google Scholar] [CrossRef]
- Chaigne, A.; Brunet, T. Incomplete abscission and cytoplasmic bridges in the evolution of eukaryotic multicellularity. Curr. Biol. 2022, 32, R385–R397. [Google Scholar] [CrossRef] [PubMed]
- Ziemons, S.; Kück, U. Cell-to-cell communication in plants, animals, and fungi: A comparative review. Naturwissenschaften 2012, 100, 3–19. [Google Scholar] [CrossRef]
- Steinberg, G.; Harmer, N.J.; Schuster, M.; Kilaru, S. Woronin body-based sealing of septal pores. Fungal Genet. Biol. 2017, 109, 53–55. [Google Scholar] [CrossRef]
- Venneman, J.; Pawlick, J.S.; Audenaert, K.; Meyer, E.; Demeyere, K.; Leus, L.; Baert, G.; Kogel, K.-H.; Haesaert, G.; Vereecke, D.; et al. Evaluation of genome size and quantitative features of the dolipore septum as taxonomic predictors for the Serendipita ‘williamsii’ species complex. Fungal Biol. 2020, 124, 781–800. [Google Scholar] [CrossRef] [PubMed]
- Pueschel, C.M. An expanded survey of the ultrastructure of red algal pit plugs. J. Phycol. 1989, 25, 625–636. [Google Scholar] [CrossRef]
- Brawley, S.H.; Sears, J.R. Septal Plugs in a Green Alga. Am. J. Bot. 1982, 69, 455–463. [Google Scholar] [CrossRef]
- Chappell, D.F.; Stewart, K.D.; Mattox, K.R. On Pits and Plasmodesmata of Trentepohlialean Algae (Chlorophyta). Trans. Am. Microsc. Soc. 1978, 97, 88–94. [Google Scholar] [CrossRef]
- Küster, E. Die Plasmodesmen von Codium. Protoplasma 1933, 19, 335–349. [Google Scholar] [CrossRef]
- Green, K.J.; Viamontes, G.I.; Kirk, D.L. Mechanism of formation, ultrastructure, and function of the cytoplasmic bridge system during morphogenesis in Volvox. J. Cell Biol. 1981, 91, 756–769. [Google Scholar] [CrossRef]
- Ikushima, N.; Maruyama, S. The Protoplasmic Connection in Volvox. J. Protozool. 1968, 15, 136–140. [Google Scholar] [CrossRef]
- Floyd, G.L.; Stewart, K.D.; Mattox, K.R. Cytokinesis and plasmodesmata in Ulothrix. J. Phycol. 1971, 7, 306–309. [Google Scholar] [CrossRef]
- Terauchi, M.; Nagasato, C.; Motomura, T. Plasmodesmata of brown algae. J. Plant Res. 2015, 128, 7–15. [Google Scholar] [CrossRef]
- Michetti, K.M.; Leonardi, P.I.; Cáceres, E.J. Morphology, cytology and taxonomic remarks of four species of Stigeoclonium (Chaetophorales, Chlorophyceae) from Argentina. Phycol. Res. 2010, 58, 35–43. [Google Scholar] [CrossRef]
- Katsaros, C.I.; Varvarigos, V.; Gachon, C.M.M.; Brand, J.; Motomura, T.; Nagasato, C.; Küpper, F.C. Comparative Immunofluorescence and Ultrastructural Analysis of Microtubule Organization in Uronema sp., Klebsormidium flaccidum, K. subtilissimum, Stichococcus bacillaris and S. chloranthus (Chlorophyta). Protist 2011, 162, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, V.A.; Beilby, M.J.; Bisson, M.A. When is a cell not a cell? A theory relating coenocytic structure to the unusual electrophysiology of Ventricaria ventricosa (Valonia ventricosa). Protoplasma 2004, 223, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Fraser, T.W.; Gunning, B.E.S. The ultrastructure of plasmodesmata in the filamentous green alga, Bulbochaete hiloensis (Nordst.) tiffany. Planta 1969, 88, 244–254. [Google Scholar] [CrossRef]
- Kwiatkowska, M. Plasmodesmal changes are related to different developmental stages of antheridia of Chara species. Protoplasma 2003, 222, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.L.; Kieninger, A.-K.; Maldener, I.; Forchhammer, K.; Pilhofer, M. Structure and Function of a Bacterial Gap Junction Analog. Cell 2019, 178, 374–384.e315. [Google Scholar] [CrossRef]
- Goodenough, D.A.; Paul, D.L. Gap Junctions. Cold Spring Harb. Perspect. Biol. 2009, 1, a002576. [Google Scholar] [CrossRef]
- Lampe, P.D.; Laird, D.W. Recent advances in connexin gap junction biology. Fac. Rev. 2022, 11, 14. [Google Scholar] [CrossRef]
- Sharma, P.C.; Negi, N.; Thakur, H.; Battu, J.R.; Turnbull, M. Insect Gap Junctions Could Be a Potential Target for Pest Management. Ann. Entomol. Soc. Am. 2022, 115, 449–460. [Google Scholar] [CrossRef]
- Rustom, A.; Saffrich, R.; Markovic, I.; Walther, P.; Gerdes, H.-H. Nanotubular Highways for Intercellular Organelle Transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef]
- Korenkova, O.; Pepe, A.; Zurzolo, C. Fine intercellular connections in development: TNTs, cytonemes, or intercellular bridges? Cell Stress 2020, 4, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Ottonelli, I.; Caraffi, R.; Tosi, G.; Vandelli, M.A.; Duskey, J.T.; Ruozi, B. Tunneling Nanotubes: A New Target for Nanomedicine? Int. J. Mol. Sci. 2022, 23, 2237. [Google Scholar] [CrossRef] [PubMed]
- Gousset, K.; Zurzolo, C. Tunnelling nanotubes. Prion 2009, 3, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Roehlecke, C.; Schmidt, M.H.H. Tunneling Nanotubes and Tumor Microtubes in Cancer. Cancers 2020, 12, 857. [Google Scholar] [CrossRef]
- Steinberg, G.; Peñalva, M.A.; Riquelme, M.; Wösten, H.A.; Harris, S.D. Cell Biology of Hyphal Growth. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- D’Avino, P.P.; Giansanti, M.G.; Petronczki, M. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 2015, 7, a015834. [Google Scholar] [CrossRef]
- Müller, W.H.; Humbel, B.M.; Van Aelst, A.C.; Van der Krift, T.P.; Boekhout, T. The Perforate Septal Pore Cap of Basidiomycetes. In Plasmodesmata: Structure, Function, Role in Cell Communication; van Bel, A.J.E., Van Kesteren, W.J.P., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 119–127. [Google Scholar]
- van Driel, K.G.A.; Humbel, B.M.; Verkleij, A.J.; Stalpers, J.; Müller, W.H.; Boekhout, T. Septal pore complex morphology in the Agaricomycotina (Basidiomycota) with emphasis on the Cantharellales and Hymenochaetales. Mycol. Res. 2009, 113, 559–576. [Google Scholar] [CrossRef]
- Iizuka, T.; Nozawa, M.; Ikeo, K. Direct link between convergent evolution at sequence level and phenotypic level of septal pore cap in Agaricomycotina. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhegunov, G.; Pogozhykh, D. The Unified System. In Life. Death. Immortality.: The Reign of the Genome; Springer International Publishing: Cham, Switzerland, 2023; pp. 57–81. [Google Scholar]
- Markham, P. Occlusions of septal pores in filamentous fungi. Mycol. Res. 1994, 98, 1089–1106. [Google Scholar] [CrossRef]
- Mamun, M.A.A.; Cao, W.; Nakamura, S.; Maruyama, J.-i. Large-scale identification of genes involved in septal pore plugging in multicellular fungi. Nat. Commun. 2023, 14, 1418. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef]
- Ramus, J. Pit connection formation in the red alga Pseudogloiophloea. J. Phycol. 1969, 5, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Pueschel, C.M.; Cole, K.M. Rhodophycean pit plugs: An ultrastructural survey with taxonomic implications. Am. J. Bot. 1982, 69, 703–720. [Google Scholar] [CrossRef]
- Raven, J.A. Evolution of Plasmodesmata. In Plasmodesmata; Oparka, K.J., Ed.; Annual Plant Reviews Series; Wiley: Hoboken, NJ, USA, 2005; Volume 18, pp. 33–52. [Google Scholar]
- Kim, G.H.; Nagasato, C.; Kwak, M.; Lee, J.W.; Hong, C.Y.; Klochkova, T.A.; Motomura, T. Intercellular transport across pit-connections in the filamentous red alga Griffithsia monilis. Algae 2022, 37, 75–84. [Google Scholar] [CrossRef]
- Bauman, R.W., Jr.; Jones, B.R. Electrophysiological investigations of the red alga Griffithsia pagifig akyl. J. Phycol. 1986, 22, 49–56. [Google Scholar] [CrossRef]
- Wetherbee, R.; Quirk, H.M.; Mallett, J.E.; Ricker, R.W. The structure and formation of host-parasite pit connections between the red algal alloparasite Harveyella mirabilis and its red algal host Odonthalia floccosa. Protoplasma 1984, 119, 62–73. [Google Scholar] [CrossRef]
- Goff, L.J.; Coleman, A.W. The role of secondary pit connections in red algal parasitism. J. Phycol. 1985, 21, 483–508. [Google Scholar] [CrossRef]
- Freese, J.M.; Lane, C.E. Parasitism finds many solutions to the same problems in red algae (Florideophyceae, Rhodophyta). Mol. Biochem. Parasitol. 2017, 214, 105–111. [Google Scholar] [CrossRef]
- Pueschel, C.M. Secondary pit connections in Hildenbrandia (Rhodophyta, Hildenbrandiales). Br. Phycol. J. 1988, 23, 25–32. [Google Scholar] [CrossRef]
- Pueschel, C.M. Formation of secondary pit connections by conjunctor cells in a coralline red alga. Phycologia 2021, 60, 644–652. [Google Scholar] [CrossRef]
- Sears, J.R.; Brawley, S.H. Smithsoniella gen. nov., a Possible Evolutionary Link between the Multicellular and Siphonous Habits in the Ulvophyceae, Chlorophyta. Am. J. Bot. 1982, 69, 1450–1461. [Google Scholar] [CrossRef]
- Arino, X.; Hernández Mariné, M.; Saiz-Jimenez, C. Ctenocladus circinnatus (Chlorophyta) in stuccos from archaeological sites of southern Spain. Phycologia 1996, 35, 183–189. [Google Scholar] [CrossRef]
- Ding, L.; Wang, X.; Huang, B.; Chen, W.; Chen, S. The environmental adaptability and reproductive properties of invasive green alga Codium fragile from the Nan’ao Island, South China Sea. Acta Oceanol. Sin. 2022, 41, 70–75. [Google Scholar] [CrossRef]
- Herron, M.D. Origins of multicellular complexity: Volvox and the volvocine algae. Mol. Ecol. 2016, 25, 1213–1223. [Google Scholar] [CrossRef]
- Hallmann, A. Morphogenesis in the Family Volvocaceae: Different Tactics for Turning an Embryo Right-side Out. Protist 2006, 157, 445–461. [Google Scholar] [CrossRef]
- Yamashita, S.; Arakaki, Y.; Kawai-Toyooka, H.; Noga, A.; Hirono, M.; Nozaki, H. Alternative evolution of a spheroidal colony in volvocine algae: Developmental analysis of embryogenesis in Astrephomene (Volvocales, Chlorophyta). BMC Evol. Biol. 2016, 16, 243. [Google Scholar] [CrossRef]
- Herron, M.D.; Hackett, J.D.; Aylward, F.O.; Michod, R.E. Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. USA 2009, 106, 3254–3258. [Google Scholar] [CrossRef]
- Green, K.J.; Kirk, D.L. Cleavage patterns, cell lineages, and development of a cytoplasmic bridge system in Volvox embryos. J. Cell Biol. 1981, 91, 743–755. [Google Scholar] [CrossRef]
- Hoops, H.J.; Nishii, I.; Kirk, D.L. Cytoplasmic Bridges in Volvox and Its Relatives. In Cell-Cell Channels; Springer: New York, NY, USA, 2006; pp. 65–84. [Google Scholar]
- Nishii, I.; Ogihara, S.; Kirk, D.L. A Kinesin, InvA, Plays an Essential Role in Volvox Morphogenesis. Cell 2003, 113, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Iida, H.; Ota, S.; Inouye, I. Cleavage, incomplete inversion, and cytoplasmic bridges in Gonium pectorale (Volvocales, Chlorophyta). J. Plant Res. 2013, 126, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Hanschen, E.R.; Marriage, T.N.; Ferris, P.J.; Hamaji, T.; Toyoda, A.; Fujiyama, A.; Neme, R.; Noguchi, H.; Minakuchi, Y.; Suzuki, M.; et al. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat. Commun. 2016, 7, 11370. [Google Scholar] [CrossRef] [PubMed]
- Marchant, H.J. Plasmodesmata in Algae and Fungi. In Intercellular Communication in Plants: Studies on Plasmodemata; Gunning, B.E.S., Robards, A.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1976; pp. 59–80. [Google Scholar]
- Raven, J.A. Miniview: Multiple origins of plasmodesmata. Eur. J. Phycol. 1997, 32, 95–101. [Google Scholar] [CrossRef]
- Cook, M.E.; Graham, L.E. Evolution of Plasmodesmata. In Plasmodesmata: Structure, Function, Role in Cell Communication; van Bel, A.J.E., Van Kesteren, W.J.P., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 101–117. [Google Scholar]
- Brunkard, J.O.; Zambryski, P.C. Plasmodesmata enable multicellularity: New insights into their evolution, biogenesis, and functions in development and immunity. Curr. Opin. Plant Biol. 2017, 35, 76–83. [Google Scholar] [CrossRef]
- Jill Harrison, C. Development and genetics in the evolution of land plant body plans. Philos. Trans. R Soc. Lond. B Biol. Sci. 2017, 372, 20150490. [Google Scholar] [CrossRef]
- Leebens-Mack, J.H.; Barker, M.S.; Carpenter, E.J.; Deyholos, M.K.; Gitzendanner, M.A.; Graham, S.W.; Grosse, I.; Li, Z.; Melkonian, M.; Mirarab, S.; et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The New Tree of Eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Wang, H.; Sahu, S.K.; Marin, B.; Li, H.; Xu, Y.; Liang, H.; Li, Z.; Cheng, S.; et al. The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants. Nat. Ecol. Evol. 2020, 4, 1220–1231. [Google Scholar] [CrossRef]
- Irisarri, I.; Strassert, J.F.H.; Burki, F. Phylogenomic Insights into the Origin of Primary Plastids. Syst. Biol. 2022, 71, 105–120. [Google Scholar] [CrossRef]
- Bowles, A.M.C.; Williamson, C.J.; Williams, T.A.; Lenton, T.M.; Donoghue, P.C.J. The origin and early evolution of plants. Trends Plant Sci. 2023, 28, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. Available online: https://www.algaebase.org (accessed on 13 July 2023).
- Liu, B.; Hu, Y.; Hu, Z.; Liu, G.; Zhu, H. Taxonomic scheme of the order Chaetophorales (Chlorophyceae, Chlorophyta) based on chloroplast genomes. BMC Genom. 2020, 21, 442. [Google Scholar] [CrossRef] [PubMed]
- Darienko, T.; Rad-Menéndez, C.; Campbell, C.N.; Pröschold, T. Molecular Phylogeny of Unicellular Marine Coccoid Green Algae Revealed New Insights into the Systematics of the Ulvophyceae (Chlorophyta). Microorganisms 2021, 9, 1586. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Ma, X.; Shi, X.; Li, X.; Yang, L.; Xiao, S.; De Clerck, O.; Leliaert, F.; Zhong, B. Phylotranscriptomic insights into a Mesoproterozoic–Neoproterozoic origin and early radiation of green seaweeds (Ulvophyceae). Nat. Commun. 2022, 13, 1610. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and Molecular Evolution of the Green Algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Škaloud, P.; Rindi, F.; Boedeker, C.; Leliaert, F. Freshwater Flora of Central Europe, Vol 13: Chlorophyta: Ulvophyceae (Süßwasserflora von Mitteleuropa, Bd. 13: Chlorophyta: Ulvophyceae); Springer: Berlin/Heidelberg, Germany, 2018; Volume 13. [Google Scholar]
- Hawker, L.E.; Gooday, M.A. Delimitation of the gametangia of Rhizopus sexualis (Smith) Callen: An electron microscope study of septum formation. Microbiology 1967, 49, 371–376. [Google Scholar] [CrossRef]
- Scheuer, C.; Bauer, R.; Lutz, M.; Stabentheiner, E.; Mel’nik, V.A.; Grube, M. Bartheletia paradoxa is a living fossil on Ginkgo leaf litter with a unique septal structure in the Basidiomycota. Mycol. Res. 2008, 112, 1265–1279. [Google Scholar] [CrossRef]
- Lucas, W.J.; Ding, B.; van der Schoot, C. Plasmodesmata and the supracellular nature of plants. New Phytol. 1993, 125, 435–476. [Google Scholar] [CrossRef]
- Bredeweg, E.L.; Baker, S.E. Horizontal Gene Transfer in Fungi. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 317–332. [Google Scholar]
- Wang, S.; Huang, J. Fungal genes in the innovation and evolution of land plants. Plant Signal. Behav. 2021, 16, 1879534. [Google Scholar] [CrossRef]
- Irwin, N.A.T.; Pittis, A.A.; Richards, T.A.; Keeling, P.J. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat. Microbiol. 2022, 7, 327–336. [Google Scholar] [CrossRef]
- Ciach, M.A.; Pawłowska, J.; Górecki, P.; Muszewska, A. The interkingdom horizontal gene transfer in 44 early diverging fungi boosted their metabolic, adaptive and immune capabilities. bioRxiv 2023. [Google Scholar] [CrossRef]
- Knox, K.; Wang, P.; Kriechbaumer, V.; Tilsner, J.; Frigerio, L.; Sparkes, I.; Hawes, C.; Oparka, K. Putting the Squeeze on Plasmodesmata: A Role for Reticulons in Primary Plasmodesmata Formation. Plant Physiol. 2015, 168, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Zheng, J.Y.; Lazarowitz, S.G. Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 2015, 25, 2018–2025. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, W.J.; Grison, M.S.; Bayer, E.M. Shaping intercellular channels of plasmodesmata: The structure-to-function missing link. J. Exp. Bot. 2018, 69, 91–103. [Google Scholar] [CrossRef]
- Brault, M.L.; Petit, J.D.; Immel, F.; Nicolas, W.J.; Glavier, M.; Brocard, L.; Gaston, A.; Fouché, M.; Hawkins, T.J.; Crowet, J.M.; et al. Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep. 2019, 20, e47182. [Google Scholar] [CrossRef] [PubMed]
- Kawai, H.; Hanyuda, T.; Draisma, S.G.A.; Wilce, R.T.; Andersen, R.A. Molecular phylogeny of two unusual brown algae, Phaeostrophion irregulare and Platysiphon glacialis, proposal of the Stschapoviales ord. nov. and Platysiphonaceae fam. nov., and a re-examination of divergence times for brown algal orders. J. Phycol. 2015, 51, 918–928. [Google Scholar] [CrossRef]
- Drobnitch, S.T.; Jensen, K.H.; Prentice, P.; Pittermann, J. Convergent evolution of vascular optimization in kelp (Laminariales). Proc. R. Soc. B Biol. Sci. 2015, 282, 20151667. [Google Scholar] [CrossRef]
- Knoblauch, J.; Tepler Drobnitch, S.; Peters, W.S.; Knoblauch, M. In situ microscopy reveals reversible cell wall swelling in kelp sieve tubes: One mechanism for turgor generation and flow control? Plant Cell Environ. 2016, 39, 1727–1736. [Google Scholar] [CrossRef]
- Bringloe, T.T.; Starko, S.; Wade, R.M.; Vieira, C.; Kawai, H.; De Clerck, O.; Cock, J.M.; Coelho, S.M.; Destombe, C.; Valero, M.; et al. Phylogeny and Evolution of the Brown Algae. Crit. Rev. Plant Sci. 2020, 39, 281–321. [Google Scholar] [CrossRef]
- Silberfeld, T.; Rousseau, F.; Reviers, B.d. An Updated Classification of Brown Algae (Ochrophyta, Phaeophyceae). Cryptogam. Algol. 2014, 35, 117–156. [Google Scholar] [CrossRef]
- Sanders, W.B.; Moe, R.L.; Ascaso, C. Ultrastructural study of the brown alga Petroderma maculiforme (Phaeophyceae) in the free-living state and in lichen symbiosis with the intertidal marine fungus Verrucaria tavaresiae (Ascomycotina). Eur. J. Phycol. 2005, 40, 353–361. [Google Scholar] [CrossRef]
- O’Kelly, C.J. Preservation of cytoplasmic ultrastructure in dried herbarium specimens: The lectotype of Pilinia rimosa (Phaeophyta, formerly Chlorophyta). Phycologia 1989, 28, 369–374. [Google Scholar] [CrossRef]
- Kawai, H.; Hanyuda, T.; Henry, E.C. Transfer of Pilinia from Ectocarpales to Ishigeales (Phaeophyceae) with proposal of Piliniaceae fam. nov., and taxonomy of Porterinema in Ectocarpales. Eur. J. Phycol. 2022, 57, 318–327. [Google Scholar] [CrossRef]
- Kawai, H.; Maeba, S.; Sasaki, H.; Okuda, K.; Henry, E.C. Schizocladia ischiensis: A New Filamentous Marine Chromophyte Belonging to a New Class, Schizocladiophyceae. Protist 2003, 154, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Katsaros, C.; Motomura, T.; Nagasato, C.; Galatis, B. Diaphragm development in cytokinetic vegetative cells of brown algae. Bot. Mar. 2009, 52, 150–161. [Google Scholar] [CrossRef]
- Terauchi, M.; Nagasato, C.; Kajimura, N.; Mineyuki, Y.; Okuda, K.; Katsaros, C.; Motomura, T. Ultrastructural study of plasmodesmata in the brown alga Dictyota dichotoma (Dictyotales, Phaeophyceae). Planta 2012, 236, 1013–1026. [Google Scholar] [CrossRef]
- Katsaros, C.; Panse, S.L.; Milne, G.; Carrano, C.J.; Küpper, F.C. New insights on Laminaria digitata ultrastructure through combined conventional chemical fixation and cryofixation. Bot. Mar. 2021, 64, 177–187. [Google Scholar] [CrossRef]
- Nagasato, C.; Terauchi, M.; Tanaka, A.; Motomura, T. Development and function of plasmodesmata in zygotes of Fucus distichus. Bot. Mar. 2015, 58, 229–238. [Google Scholar] [CrossRef]
- Faulkner, C.; Akman, O.E.; Bell, K.; Jeffree, C.; Oparka, K. Peeking into Pit Fields: A Multiple Twinning Model of Secondary Plasmodesmata Formation in Tobacco. Plant Cell 2008, 20, 1504–1518. [Google Scholar] [CrossRef]
- Chambaud, C.; Cookson, S.J.; Ollat, N.; Bayer, E.; Brocard, L. A correlative light electron microscopy approach reveals plasmodesmata ultrastructure at the graft interface. Plant Physiol. 2022, 188, 44–55. [Google Scholar] [CrossRef]
- Ehlers, K.; van Bel, A.J. Dynamics of plasmodesmal connectivity in successive interfaces of the cambial zone. Planta 2010, 231, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Fitzgibbon, J.; Beck, M.; Zhou, J.; Faulkner, C.; Robatzek, S.; Oparka, K. A Developmental Framework for Complex Plasmodesmata Formation Revealed by Large-Scale Imaging of the Arabidopsis Leaf Epidermis. Plant Cell 2013, 25, 57–70. [Google Scholar] [CrossRef] [PubMed]
- González, A.V.; Beltrán, J.; Flores, V.; Santelices, B. Morphological convergence in the inter-holdfast coalescence process among kelp and kelp-like seaweeds (Lessonia, Macrocystis, Durvillaea). Phycologia 2015, 54, 283–291. [Google Scholar] [CrossRef]
- Nagasato, C.; Tanaka, A.; Ito, T.; Katsaros, C.; Motomura, T. Intercellular translocation of molecules via plasmodesmata in the multiseriate filamentous brown alga, Halopteris congesta (Sphacelariales, Phaeophyceae). J. Phycol. 2017, 53, 333–341. [Google Scholar] [CrossRef]
- Knoll, A.H. Life on a Young Planet: The First Three Billion Years of Evolution on Earth; Princeton University Press: Princeton, NJ, USA, 2003. [Google Scholar]
- De Clerck, O.; Bogaert, K.A.; Leliaert, F. Chapter Two—Diversity and Evolution of Algae: Primary Endosymbiosis. In Advances in Botanical Research; Piganeau, G., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 64, pp. 55–86. [Google Scholar]
- Becker, B.; Marin, B. Streptophyte algae and the origin of embryophytes. Ann. Bot. 2009, 103, 999–1004. [Google Scholar] [CrossRef]
- Lopes dos Santos, A.; Pollina, T.; Gourvil, P.; Corre, E.; Marie, D.; Garrido, J.L.; Rodríguez, F.; Noël, M.-H.; Vaulot, D.; Eikrem, W. Chloropicophyceae, a new class of picophytoplanktonic prasinophytes. Sci. Rep. 2017, 7, 14019. [Google Scholar] [CrossRef]
- Leliaert, F.; Tronholm, A.; Lemieux, C.; Turmel, M.; DePriest, M.S.; Bhattacharya, D.; Karol, K.G.; Fredericq, S.; Zechman, F.W.; Lopez-Bautista, J.M. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep. 2016, 6, 25367. [Google Scholar] [CrossRef]
- Lee, R.E. Phycology, 4th ed.; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Liu, B.; Wang, Q.; Li, S.; Fang, J.; Liu, G.; Hu, Z. Taxonomic transfer of Gongrosira fluminensis Fritsch (Chaetophorales, Chlorophyceae) to Lithotrichon Darienko et Pröschold (Ulvales, Ulvophyceae) based on morphological observation and phylogenetic analyses. Fottea 2019, 19, 25–32. [Google Scholar] [CrossRef]
- McBride, G.E. Cytokinesis and ultrastructure in Fritschiella tuberosa lyengar. Arch. Protist. 1970, 112, 365–375. [Google Scholar]
- McBride, G.E. Cytokinesis in the Green Alga Fritschiella. Nature 1967, 216, 939. [Google Scholar] [CrossRef]
- Cocquyt, E.; Verbruggen, H.; Leliaert, F.; De Clerck, O. Evolution and Cytological Diversification of the Green Seaweeds (Ulvophyceae). Mol. Biol. Evol. 2010, 27, 2052–2061. [Google Scholar] [CrossRef]
- Spoerner, M.; Wichard, T.; Bachhuber, T.; Stratmann, J.; Oertel, W. Growth and Thallus Morphogenesis of Ulva mutabilis (Chlorophyta) Depends on A Combination of Two Bacterial Species Excreting Regulatory Factors. J. Phycol. 2012, 48, 1433–1447. [Google Scholar] [CrossRef]
- Wichard, T. Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta). Front. Plant Sci. 2015, 6, 86. [Google Scholar] [CrossRef]
- De Clerck, O.; Kao, S.-M.; Bogaert, K.A.; Blomme, J.; Foflonker, F.; Kwantes, M.; Vancaester, E.; Vanderstraeten, L.; Aydogdu, E.; Boesger, J.; et al. Insights into the Evolution of Multicellularity from the Sea Lettuce Genome. Curr. Biol. 2018, 28, 2921–2933.e2925. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Townsley, B.T.; Ichihashi, Y.; Sinha, N.R.; Chitwood, D.H. An Intracellular Transcriptomic Atlas of the Giant Coenocyte Caulerpa taxifolia. PLoS Genet. 2015, 11, e1004900. [Google Scholar] [CrossRef] [PubMed]
- Arimoto, A.; Nishitsuji, K.; Narisoko, H.; Shoguchi, E.; Satoh, N. Differential gene expression in fronds and stolons of the siphonous macroalga, Caulerpa lentillifera. Dev. Growth Differ. 2019, 61, 475–484. [Google Scholar] [CrossRef] [PubMed]
- McNaughton, E.E.; Goff, L.J. The role of microtubules in establishing nuclear spatial patterns in multinucleate green algae. Protoplasma 1990, 157, 19–37. [Google Scholar] [CrossRef]
- Woodcock, C.L. The anchoring of nuclei by cytoplasmic microtubules in Acetabularia. J. Cell Sci. 1971, 8, 611–621. [Google Scholar] [CrossRef]
- Baluška, F.; Volkmann, D.; Barlow, P.W. Eukaryotic Cells and their Cell Bodies: Cell Theory Revised. Ann. Bot. 2004, 94, 9–32. [Google Scholar] [CrossRef]
- McDonald, K.L.; Pickett-Heaps, J.D. Ultrastructure and Differentiation in Cladophora glomerata. I. Cell Division. Am. J. Bot. 1976, 63, 592–601. [Google Scholar] [CrossRef]
- Del Cortona, A.; Jackson, C.J.; Bucchini, F.; Van Bel, M.; D’hondt, S.; Škaloud, P.; Delwiche, C.F.; Knoll, A.H.; Raven, J.A.; Verbruggen, H.; et al. Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds. Proc. Natl. Acad. Sci. USA 2020, 117, 2551–2559. [Google Scholar] [CrossRef] [PubMed]
- Lovlie, A.; Bråten, T. On mitosis in the multicellular alga Ulva mutabilis Føyn. J. Cell Sci. 1970, 6, 109–129. [Google Scholar] [CrossRef] [PubMed]
- Mikhailyuk, T.I.; Sluiman, H.J.; Massalski, A.; Mudimu, O.; Demchenko, E.M.; Kondratyuk, S.Y.; Friedl, T. New streptophyte green algae from terrestrial habitats and an assessment of the genus Interfilum (klebsormidiophyceae, streptophyta). J. Phycol. 2008, 44, 1586–1603. [Google Scholar] [CrossRef] [PubMed]
- Rindi, F.; Mikhailyuk, T.I.; Sluiman, H.J.; Friedl, T.; López-Bautista, J.M. Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta). Mol. Phylogenetics Evol. 2011, 58, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Sluiman, H.J.; Roberts, K.R.; Stewart, K.D.; Mattox, K.R. Comparative cytology and taxonomy of the ulvophyceae. IV. mitosis and cytokinesis in Ulothrix (chlorophyta). Acta Bot. Neerl. 1983, 32, 257–269. [Google Scholar] [CrossRef]
- Stewart, K. Classification of the green algae: A concept based on comparative cytology. Syst. Assoc. 1984, 27, 29–72. [Google Scholar]
- Chapman, R.L.; Henk, M.C. Phragmoplasts in cytokinesis of Cephaleuros parasiticus (chlorophyta) vegetative cells J. Phycol. 1986, 22, 83–88. [Google Scholar] [CrossRef]
- Chapman, R.L.; Borkhsenious, O.; Brown, R.C.; Henk, M.C.; Waters, D.A. Phragmoplast-mediated cytokinesis in Trentepohlia: Results of TEM and immunofluorescence cytochemistry. Int. J. Syst. Evol. Microbiol. 2001, 51, 759–765. [Google Scholar] [CrossRef]
- López-Bautista, J.M.; Waters, D.A.; Chapman, R.L. Phragmoplastin, green algae and the evolution of cytokinesis. Int. J. Syst. Evol. Microbiol. 2003, 53, 1715–1718. [Google Scholar] [CrossRef]
- Buschmann, H.; Zachgo, S. The Evolution of Cell Division: From Streptophyte Algae to Land Plants. Trends Plant Sci. 2016, 21, 872–883. [Google Scholar] [CrossRef]
- Chapman, R.L.; Good, B.H. Ultrastructure of plasmodesmata and cross walls in Cephaleuros, Phycopeltis and Trentepohlia (Chroolepidaceae; Chlorophyta). Br. Phycol. J. 1978, 13, 241–246. [Google Scholar] [CrossRef]
- Turmel, M.; Brouard, J.-S.; Gagnon, C.; Otis, C.; Lemieux, C. Deep division in the chlorophyceae (chlorophyta) revealed by chloroplast phylogenomic analyses. J. Phycol. 2008, 44, 739–750. [Google Scholar] [CrossRef]
- Buchheim, M.A.; Sutherland, D.M.; Schleicher, T.; Förster, F.; Wolf, M. Phylogeny of Oedogoniales, Chaetophorales and Chaetopeltidales (Chlorophyceae): Inferences from sequence-structure analysis of ITS2. Ann. Bot. 2012, 109, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Sluiman, H.J. Mitosis and cell division in Cylindrocapsa geminella (chlorophyceae). J. Phycol. 1985, 21, 523–532. [Google Scholar] [CrossRef]
- Mikhailyuk, T.; Holzinger, A.; Massalski, A.; Karsten, U. Morphology and ultrastructure of Interfilum and Klebsormidium (Klebsormidiales, Streptophyta) with special reference to cell division and thallus formation. Eur. J. Phycol. 2014, 49, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Pickett-Heaps, J.; Fowke, L. Cell Division in Oedogonium I. Mitosis, Cytokinesis, and Cell Elongation. Aust. J. Biol. Sci. 1969, 22, 857–894. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D. Cell division in Bulbochaete. I. Divisions utilizing the wall ring. J. Phycol. 1973, 9, 408–420. [Google Scholar] [CrossRef]
- O’Kelly, C.J.; Watanabe, S.; Floyd, G.L. Ultrastructure and Phylogenetic Relationships of Chaetopeltidales Ord. Nov.(Chlorophyta, Chlorophyceae). J. Phycol. 1994, 30, 118–128. [Google Scholar] [CrossRef]
- Caisová, L.; Reyes, C.P.; Álamo, V.C.; Quintana, A.M.; Surek, B.; Melkonian, M. Barrancaceae: A new green algal lineage with structural and behavioral adaptations to a fluctuating environment. Am. J. Bot. 2015, 102, 1482–1492. [Google Scholar] [CrossRef]
- Egorova, I.N.; Kulakova, N.V.; Boldina, O.N.; Tupikova, G.S. Barranca variabilis sp. nov.—A New Terrestrial Alga of the Genus Barranca (Chaetophorales, Chlorophyta) from the Baikal Region (Russia). Diversity 2023, 15, 583. [Google Scholar] [CrossRef]
- Stewart, K.D.; Mattox, K.R.; Floyd, G.L. Mitosis, cytokinesis, the distribution of plasmodesmata, and other cytological characteristics in the ulotrichales, ulvales, and chaetophorales: Phylogenetic and taxonomic considerations. J. Phycol. 1973, 9, 128–141. [Google Scholar] [CrossRef]
- Leonardi, P.I.; Cáceres, E.J.; Correa, J.A. Ultrastructure and Taxonomy of Sporocladopsis novae-zelandiae (Ulvophyceae, Chlorophyta). Bot. Mar. 2002, 45, 324–330. [Google Scholar] [CrossRef]
- Correa, J.; Martínez, E. Factors associated with host specificity in Sporocladopsis novae-zelandiae (Chlorophyta). J. Phycol. 1996, 32, 22–27. [Google Scholar] [CrossRef]
- Rensing, S.A. Great moments in evolution: The conquest of land by plants. Curr. Opin. Plant Biol. 2018, 42, 49–54. [Google Scholar] [CrossRef]
- Marin, B.; Melkonian, M. Mesostigmatophyceae, a new class of streptophyte green algae revealed by SSU rRNA sequence comparisons. Protist 1999, 150, 399–417. [Google Scholar] [CrossRef]
- Lokhorst, G.M.; Sluiman, H.J.; Star, W. The ultrastructure of mitosis and cytokinesis in the sarcinoid Chlorokybus atmophyticus (chlorophyta, charophyceae) revealed by rapid freeze fixation and freeze substitution. J. Phycol. 1988, 24, 237–248. [Google Scholar] [CrossRef]
- Lokhorst, G.M.; Star, W. Ultrastructure of mitosis and cytokinesis in Klebsormidium mucosum nov. comb., formerly Ulothrix verrucosa (chlorophyta). J. Phycol. 1985, 21, 466–476. [Google Scholar] [CrossRef]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef]
- Puttick, M.N.; Morris, J.L.; Williams, T.A.; Cox, C.J.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Schneider, H.; Pisani, D.; et al. The Interrelationships of Land Plants and the Nature of the Ancestral Embryophyte. Curr. Biol. 2018, 28, 733–745.e732. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara Genome: Secondary Complexity and Implications for Plant Terrestrialization. Cell 2018, 174, 448–464.e424. [Google Scholar] [CrossRef]
- Graham, L.E. The occurrence, evolution, and phylogenetic significance of parenchyma in Coleochaete bréb. (chlorophyta). Am. J. Bot. 1982, 69, 447–454. [Google Scholar] [CrossRef]
- Delwiche, C.F.; Karol, K.G.; Cimino, M.T.; Sytsma, K.J. Phylogeny of the genus Coleochaete (coleochaetales, charophyta) and related taxa inferred by analysis of the chloroplast gene rbcL1. J. Phycol. 2002, 38, 394–403. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D.; Wetherbee, R. Spindle function in the green alga Mougeotia: Absence of anaphase A correlates with postmitotic nuclear migration. Cell Motil. 1987, 7, 68–77. [Google Scholar] [CrossRef]
- Holzinger, A.; Albert, A.; Aigner, S.; Uhl, J.; Schmitt-Kopplin, P.; Trumhová, K.; Pichrtová, M. Arctic, Antarctic, and temperate green algae Zygnema spp. under UV-B stress: Vegetative cells perform better than pre-akinetes. Protoplasma 2018, 255, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
- Porth, M.L. Cell Connections in Algae: Studies on the Evolutionary Development of Plasmodesmata. Master’s Thesis, Justus-Liebig University, Giessen, Germany, 2023. [Google Scholar]
- Imaichi, R.; Hiratsuka, R. Evolution of shoot apical meristem structures in vascular plants with respect to plasmodesmatal network. Am J Bot 2007, 94, 1911–1921. [Google Scholar] [CrossRef]
- Marchant, H.J.; Pickett-Heaps, J.D. Mitosis and cytokinesis in Coleochaete scutata. J. Phycol. 1973, 9, 461–471. [Google Scholar] [CrossRef]
- Brown, R.C.; Lemmon, B.E.; Graham, L.E. Morphogenetic Plastid Migration and Microtubule Arrays in Mitosis and Cytokinesis in the Green Alga Coleochaete orbicularis. Am. J. Bot. 1994, 81, 127–133. [Google Scholar] [CrossRef]
- Cook, M.E. Cytokinesis in Coleochaete orbicularis (Charophyceae): An Ancestral Mechanism Inherited by Plants. Am. J. Bot. 2004, 91, 313–320. [Google Scholar] [CrossRef]
- Cook, M.; Graham, L.; Botha, C.; Lavin, C. Comparative ultrastructure of plasmodesmata of Chara and selected bryophytes: Toward an elucidation of the evolutionary origin of plant plasmodesmata. Am. J. Bot. 1997, 84, 1169. [Google Scholar] [CrossRef]
- Franceschi, V.R.; Ding, B.; Lucas, W.J. Mechanism of plasmodesmata formation in characean algae in relation to evolution of intercellular communication in higher plants. Planta 1994, 192, 347–358. [Google Scholar] [CrossRef]
- Pickett-Heaps, J. Ultrastructure and differentiation in Chara sp. I. Vegetative cells. Aust. J. Biol. Sci. 1967, 20, 539–552. [Google Scholar] [CrossRef]
- Pickett-Heaps, J. Ultrastructure and differentiation in Chara sp. II. Mitosis. Aust. J. Biol. Sci. 1967, 20, 883–894. [Google Scholar] [CrossRef]
- Kikuyama, M.; Hara, Y.; Shimada, K.; Yamamoto, K.; Hiramoto, Y. Intercellular transport of macromolecules in Nitella. Plant Cell Physiol. 1992, 33, 413–417. [Google Scholar]
- Spanswick, R.M.; Costerton, J.W.F. Plasmodesmata in Nitella translucens: Structure and electrical resistance. J. Cell Sci. 1967, 2, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Gunning, B.E.S. Age-related and origin-related control of the numbers of plasmodesmata in cell walls of developing Azolla roots. Planta 1978, 143, 181–190. [Google Scholar] [CrossRef]
- Bowman, J.L. The origin of a land flora. Nat. Plants 2022, 8, 1352–1369. [Google Scholar] [CrossRef]
- Paterlini, A. Uncharted routes: Exploring the relevance of auxin movement via plasmodesmata. Biol. Open 2020, 9, bio055541. [Google Scholar] [CrossRef]
- Faulkner, C.R.; Blackman, L.M.; Cordwell, S.J.; Overall, R.L. Proteomic identification of putative plasmodesmatal proteins from Chara corallina. Proteomics 2005, 5, 2866–2875. [Google Scholar] [CrossRef]
- Vaattovaara, A.; Brandt, B.; Rajaraman, S.; Safronov, O.; Veidenberg, A.; Luklová, M.; Kangasjärvi, J.; Löytynoja, A.; Hothorn, M.; Salojärvi, J.; et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019, 2, 56. [Google Scholar] [CrossRef]
- Terasaki, M.; Song, J.; Wong, J.R.; Weiss, M.J.; Chen, L.B. Localization of endoplasmic reticulum in living and glutaraldehyde-fixed cells with fluorescent dyes. Cell 1984, 38, 101–108. [Google Scholar] [CrossRef]
- Yuan, S.; Heath, I.B. A comparison of fluorescent membrane probes in hyphal tips of Saprolegnia ferax. Exp. Mycol. 1991, 15, 103–115. [Google Scholar] [CrossRef]
- Monier, A.; Pagarete, A.; de Vargas, C.; Allen, M.J.; Claverie, J.-M.; Ogata, H. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009, 19, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, A.; Oppermann, J.; Wietek, J.; Fernandez Lahore, R.G.; Sandaa, R.-A.; Bratbak, G.; Hegemann, P.; Béjà, O. Lateral Gene Transfer of Anion-Conducting Channelrhodopsins between Green Algae and Giant Viruses. Curr. Biol. 2020, 30, 4910–4920.e4915. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.M.; Mongrand, S.; Tilsner, J. Specialized membrane domains of plasmodesmata, plant intercellular nanopores. Front. Plant Sci. 2014, 5, 507. [Google Scholar] [CrossRef]
- Grison, M.S.; Brocard, L.; Fouillen, L.; Nicolas, W.; Wewer, V.; Dörmann, P.; Nacir, H.; Benitez-Alfonso, Y.; Claverol, S.; Germain, V.; et al. Specific Membrane Lipid Composition Is Important for Plasmodesmata Function in Arabidopsis. Plant Cell 2015, 27, 1228–1250. [Google Scholar] [CrossRef]
- Béziat, C.; Jaillais, Y. Should I stay or should I go: The functional importance and regulation of lipid diffusion in biological membranes. J. Exp. Bot. 2023, 74, 2479–2488. [Google Scholar] [CrossRef]
- Timme, R.E.; Delwiche, C.F. Uncovering the evolutionary origin of plant molecular processes: Comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010, 10, 96. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- van de Weyer, K. Bestimmungsschlüssel. In Armleuchteralgen: Die Characeen Deutschlands; Springer: Berlin/Heidelberg, Germany, 2016; pp. 193–208. [Google Scholar]
- Hoesch, A. Einfacher Bestimmungsschlüssel für die häufigsten Characeae-Arten in Seen Deutschlands. Lauterbornia 2003, 48, 15–24. [Google Scholar]
- Althoff, F.; Wegner, L.; Ehlers, K.; Buschmann, H.; Zachgo, S. Developmental Plasticity of the Amphibious Liverwort Riccia fluitans. Front. Plant Sci. 2022, 13, 909327. [Google Scholar] [CrossRef] [PubMed]
- Permann, C.; Pierangelini, M.; Remias, D.; Lewis, L.A.; Holzinger, A. Photophysiological investigations of the temperature stress responses of Zygnema spp (Zygnematophyceae) from subpolar and polar habitats (Iceland, Svalbard). Phycologia 2022, 61, 299–311. [Google Scholar] [CrossRef]
- Jordan, E.G. Ultrastructural aspects of cell wall synthesis in Spirogyra. Protoplasma 1970, 69, 405–416. [Google Scholar] [CrossRef]
- Doty, K.F.; Betzelberger, A.M.; Kocot, K.M.; Cook, M.E. Immunofluorescence localization of the tubulin cytoskeleton during cell division and cell growth in members of the Coleochaetales (Streptophyta). J. Phycol. 2014, 50, 624–639. [Google Scholar] [CrossRef]
- Blinn, D.W.; Morrison, E. Intercellular cytoplasmic connections in Ctenocladus circinnatus Borzi (Chlorophyceae) with possible ecological significance. Phycologia 1974, 13, 95–97. [Google Scholar] [CrossRef]
- Cook, M.E.; Graham, L.E.; Lavin, C.A. Cytokinesis and nodal anatomy in the charophycean green alga Chara zeylanica. Protoplasma 1998, 203, 65–74. [Google Scholar] [CrossRef]
- Coss, R.A.; Pickett-Heaps, J.D. Gametogenesis in the green alga Oedogonium cardiacum. Protoplasma 1974, 81, 297–311. [Google Scholar] [CrossRef]
- D’Amico, S.-L.; Leonardi, P.-I.; Cáceres, E.-J. Ultrastructure of multicellular dwarf males with external gametangium in Oedogonium macrandrium (Oedogoniales, Chlorophyta). Biocell 2013, 37, 85–90. [Google Scholar] [CrossRef]
- Dawes, C.J. A light and electron microscope survey of algal cell walls. II. Chlorophyceae. Ohio J. Sci. 1966, 66, 317–326. [Google Scholar]
- Deason, T.R.; Darden, W.H.; Ely, S. The development of sperm packets of the M5 strain of Volvox aureus. J. Ultrastruct. Res. 1969, 26, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, C.R.; Blackman, L.M.; Collings, D.A.; Cordwell, S.J.; Overall, R.L. Anti-tropomyosin antibodies co-localise with actin microfilaments and label plasmodesmata. Eur. J. Cell Biol. 2009, 88, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Dainty, J.; Tyree, M. A quantitative investigation of symplasmic transport in Chara corallina. I. Ultrastructure of the nodal complex cell walls. Can. J. Bot. 1974, 52, 1209–1214. [Google Scholar] [CrossRef]
- Floyd, G.L.; Stewart, K.D.; Mattox, K.R. Comparative cytology of ulothrix and stigeoclonium. J. Phycol. 1972, 8, 68–81. [Google Scholar] [CrossRef]
- Fowke, L.C.; Pickett-Heaps, J.D. Cell Division in Spirogyra. II. Cytokinesis. J. Phycol. 1969, 5, 273–281. [Google Scholar] [CrossRef]
- Franceschi, V.R.; Lucas, W.J. The relationship of the charasome to chloride uptake in Chara corallina: Physiological and histochemical investigations. Planta 1982, 154, 525–537. [Google Scholar] [CrossRef]
- Fraser, T.W.; Gunning, B.E.S. Ultrastructure of the Hairs of the Filamentous Green Alga Bulbochaete hiloensis (Nordst.) Tiffany: An Apoplastidic Plant Cell with a Well Developed Golgi Apparatus. Planta 1973, 113, 1–19. [Google Scholar] [CrossRef]
- Hill, G.J.C.; Machlis, L. An ultrastructural study of vegetative cell division in Oedogonium borisianum. J. Phycol. 1968, 4, 261–271. [Google Scholar] [CrossRef]
- Jacob, A.; Wiencke, C.; Lehmann, H.; Kirst, G.O. Physiology and Ultrastructure of Desiccation in the Green Alga Prasiola crispa from Antarctica. Bot. Mar. 1992, 35, 297–304. [Google Scholar] [CrossRef]
- Kaplan, F.; Lewis, L.A.; Herburger, K.; Holzinger, A. Osmotic stress in Arctic and Antarctic strains of the green alga Zygnema (Zygnematales, Streptophyta): Effects on photosynthesis and ultrastructure. Micron 2013, 44, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, M.; Maszewski, J. Changes in ultrastructure of plasmodesmata during spermatogenesis in Chara vulgaris L. Planta 1985, 166, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, M.; Maszewski, J. Changes in the occurrence and ultrastructure of plasmodesmata in antheridia of Chara vulgaris L. during different stages of spermatogenesis. Protoplasma 1986, 132, 179–188. [Google Scholar] [CrossRef]
- Kwiatkowska, M.; Popłońska, K.; Wojtczak, A. Chara tomentosa Antheridial Plasmodesmata at Various Stages of Spermatogenesis. Biol. Plant. 2003, 46, 233–238. [Google Scholar] [CrossRef]
- Leonardi, P.I.; Correa, J.A.; Cáceres, E.J. Ultrastructure and taxonomy of the genus Endophyton (Ulvales, Ulvophyceae). Eur. J. Phycol. 1997, 32, 175–183. [Google Scholar] [CrossRef]
- Liu, B.; Zhu, H.; Zhao, Z.; Xiong, Q.; Liu, G.; Hu, Z. Oncosaccus: A rare green alga endemic to China belongs to Chaetopeltidales (Chlorophyceae, Chlorophyta). Phycologia 2021, 60, 2–9. [Google Scholar] [CrossRef]
- Lokhorst, G.M. Taxonomic studies on the marine and brackish-water species of Ulothrix (Ulotricales, Chlorophyceae) in western Europe. Blumea Biodivers. Evol. Biogeogr. Plants 1978, 24, 191–299. [Google Scholar]
- Lokhorst, G.M. The ultrastructure of Ulothrix mucosa. I. Mitosis and cytokinesis. Can. J. Bot. 1986, 64, 156–165. [Google Scholar] [CrossRef]
- Lokhorst, G.M.; Bakker, M.E.; Star, W. Ultrastructure of Draparnaldia glomerata (Chaetophorales, Chlorophyceae) II. Mitosis and cytokinesis. Nord. J. Bot. 1984, 4, 553–562. [Google Scholar] [CrossRef]
- Lokhorst, G.M.; Star, W. Pyrenoid ultrastructure in Ulothrix (chlorophyceae). Acta Bot. Neerl. 1980, 29, 1–15. [Google Scholar] [CrossRef]
- Løvlie, A.; Bråten, T. On the division of cytoplasm and chloroplast in the multicellular green alga Ulva mutabilis føyn. Exp. Cell Res. 1968, 51, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Marchant, H.J. Ultrastructure, development and cytoplasmic rotation of seta-bearing cells of Coleochaete scutata (Chlorophyceae). J. Phycol. 1977, 13, 28–36. [Google Scholar] [CrossRef]
- Mattox, K.R.; Stewart, K.D. A Comparative study of cell division in Trichosarcina polymorpha and Pseudendoclonium basiliense (chlorophyceae). J. Phycol. 1974, 10, 447–456. [Google Scholar] [CrossRef]
- Mattox, K.R.; Stewart, K.D.; Floyd, G.L. The cytology and classification of Schizomeris leibleinii (Chlorophyceae). I. The vegetative thallus. Phycologia 1974, 13, 63–69. [Google Scholar] [CrossRef]
- McArthur, D.M.; Moss, B.L. The ultrastructure of cell walls in Enteromorpha intestinalis (L.) link. Br. Phycol. J. 1977, 12, 359–368. [Google Scholar] [CrossRef]
- McArthur, D.M.; Moss, B.L. Ultrastructural studies of vegetative cells, mitosis and cell division in Enteromorpha Intestinalis (L.) Link. Br. Phycol. J. 1978, 13, 255–267. [Google Scholar] [CrossRef]
- Messyasz, B.; Czerwik-Marcinkowska, J.; Massalski, A.; Uher, B.; Rybak, A.; Szendzina, L.; Pikosz, M. Morphological and ultrastructural studies on Ulva flexuosa subsp. pilifera (Chlorophyta) from Poland. Acta Soc. Bot. Pol. 2013, 82, 157–163. [Google Scholar] [CrossRef]
- Messyasz, B.; Czerwik-Marcinkowska, J.; Uher, B.; Rybak, A.; Szendzina, L.; Pikosz, M. Ulva flexuosa subsp. pilifera (Chlorophyta, Ulvophyceae) from the Wielkopolska region (West Poland): A new observation on the ultrastructure of vegetative cells. Oceanol. Hydrobiol. Stud. 2013, 42, 209–215. [Google Scholar] [CrossRef]
- Michetti, K. A light and electron microscopic study on the formation, structure and germination of akinetes of Stigeoclonium tenue (Chaetophorales, Chlorophyceae). Algol. Stud. 2002, 105, 111–124. [Google Scholar]
- Michetti, K.M.; Leonardi, P.I.; Cáceres, E.J. Precisiones sobre la morfología Y ultraestructura del talo de Chaetophora elegans (chaetophorales, chlorophyta). Darwiniana 2003, 41, 37–41. [Google Scholar]
- Molnar, K.E.; Stewart, K.D.; Mattox, K.R. Cell division in the filamentous pleurastrum and its comparison with the unicellular platymonas (chlorophyceae). J. Phycol. 1975, 11, 287–296. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D. Cell division and wall structure in microspora. New Phytol. 1973, 72, 347–355. [Google Scholar] [CrossRef]
- Pickett-Heaps, J. Ultrastructure and differentiation in Chara (fibrosa) IV. Spermatogenesis. Aust. J. Biol. Sci. 1968, 21, 655–690. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D. Reproduction by zoospores in Oedogonium. Protoplasma 1971, 72, 275–314. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D. Some Ultrastructural Features of Volvox, with Particular Reference to the Phenomenon of Inversion. Planta 1970, 90, 174–190. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D. Cell division in Stichococcus. Br. Phycol. J. 1974, 9, 63–73. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.D.; McDonald, K.L. Cylindrocapsa: Cell Division and Phylogenetic Affinities. New Phytol. 1975, 74, 235–241. [Google Scholar] [CrossRef]
- Retallack, B.; Butler, R.D. The development and structure of the zoospore vesicle in Bulbochaete hiloensis. Arch. Für Mikrobiol. 1970, 72, 223–237. [Google Scholar] [CrossRef]
- Retallack, B.; Butler, R.D. Reproduction in Bulbochaete hiloensis (Nordst.) Tiffany. Arch. Für Mikrobiol. 1973, 90, 343–364. [Google Scholar] [CrossRef]
- Rindi, F.; Guiry, M.D.; Lopez-Bautista, J.M. New records of Trentepohliales (Ulvophyceae, Chlorophyta) from Africa. Nova Hedwig. 2006, 83, 431–449. [Google Scholar] [CrossRef]
- Rindi, F.; Lam, D.W.; López-Bautista, J.M. Trentepohliales (Ulvophyceae, Chlorophyta) from Panama. Nova Hedwig. 2008, 87, 421–444. [Google Scholar] [CrossRef]
- Rindi, F.; López-Bautista, J.M. New and interesting records of Trentepohlia (Trentepohliales, Chlorophyta) from French Guiana, including the description of two new species. Phycologia 2007, 46, 698–708. [Google Scholar] [CrossRef]
- Rindi, F.; Lopez-Bautista, J.M. Diversity and ecology of Trentepohliales (Ulvophyceae, Chlorophyta) in French Guiana. Cryptogam. Algol. 2008, 29, 13–43. [Google Scholar]
- Rindi, F.; Sherwood, A.R.; Guiry, M.D. Taxonomy and distribution of Trentepohlia and Printzina (Trentepohliales, Chlorophyta) in the Hawaiian Islands. Phycologia 2005, 44, 270–284. [Google Scholar] [CrossRef]
- Sanchez Puerta, M.V.; Leonardi, P.I.; Cceres, E.J.; O’Kelly, C.J. 141 New Entities in the Order Chaetopeltidales. J. Phycol. 2003, 39, 49. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Leonardi, P.I.; O’Kelly, C.J.; Cáceres, E.J. Pseudulvella americana belongs to the order chaetopeltidales (class chlorophyceae), evidence from ultrastructure and Ssu rdna sequence data. J. Phycol. 2006, 42, 943–950. [Google Scholar] [CrossRef]
- Segaar, P.J.; Lokhorst, G.M. Cell division in the green alga Ulothrix palusalsa (Ulvophyceae, Chlorophyta): A combined immunofluorescence and transmission electron microscopy study. Phycologia 1987, 26, 100–110. [Google Scholar] [CrossRef]
- Segaar, P.J.; Lokhorst, G.M. Dynamics of the microtubular cytoskeleton in the green alga Aphanochaete magna (Chlorophyta). I. Late mitotic stages and the origin and development of the phycoplast. Protoplasma 1988, 142, 176–187. [Google Scholar] [CrossRef]
- Sluiman, H.J. A pathway of plasma membrane biogenesis bypassing the Golgi apparatus during cell division in the green alga Cylindrocapsa geminella. J. Cell Sci. 1984, 72, 89–100. [Google Scholar] [CrossRef]
- Watanabe, S.; Fučíková, K.; Lewis, L.A.; Lewis, P.O. Hiding in plain sight: Koshicola spirodelophila gen. et sp. nov. (Chaetopeltidales, Chlorophyceae), a novel green alga associated with the aquatic angiosperm Spirodela polyrhiza. Am. J. Bot. 2016, 103, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, E.; Santiañez, W.J.E.; Kogame, K. Asterocladon ednae sp. nov. (Asterocladales, Phaeophyceae) from the Philippines. Phycol. Res. 2022, 70, 185–191. [Google Scholar] [CrossRef]
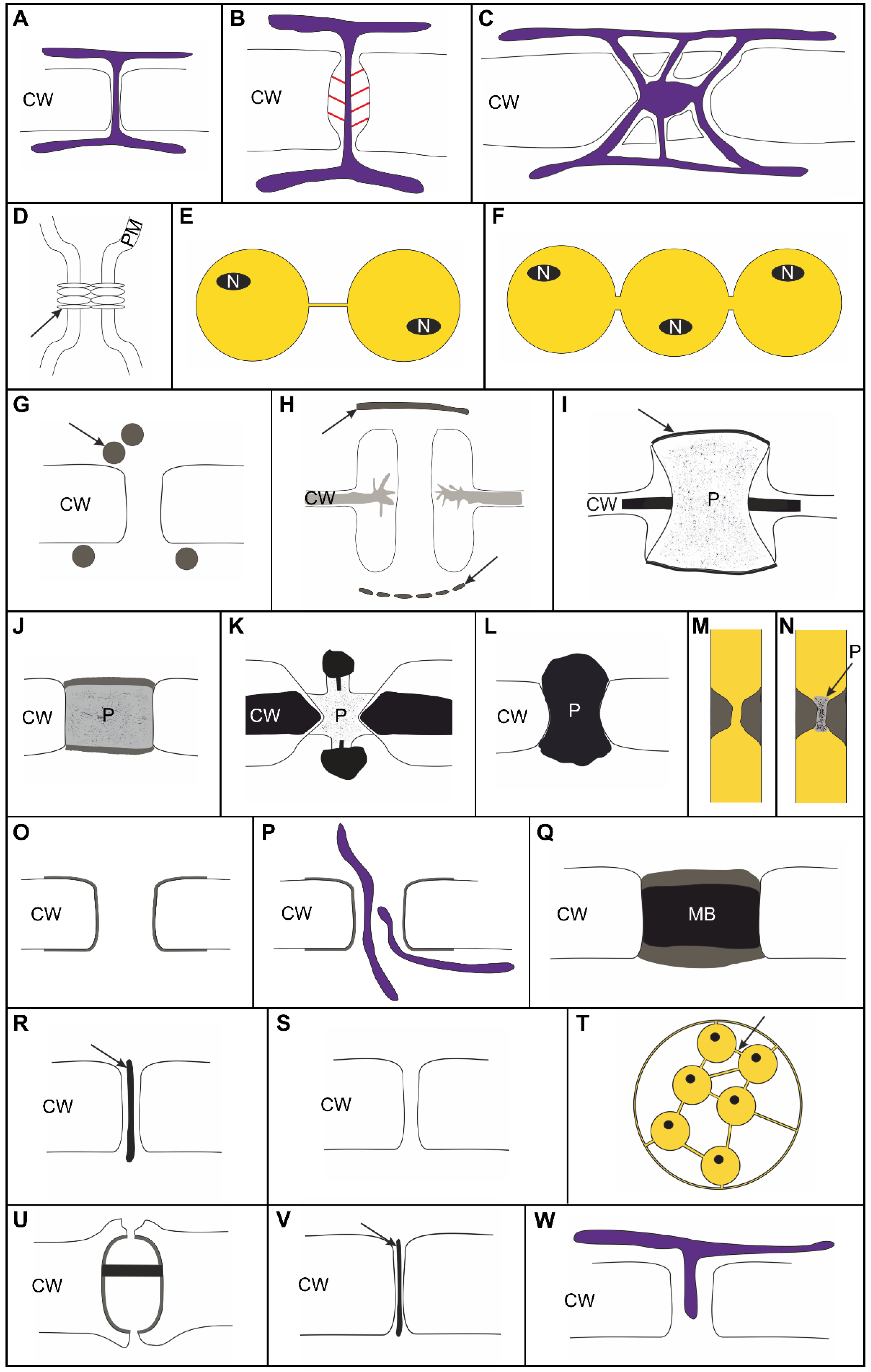
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wegner, L.; Porth, M.L.; Ehlers, K. Multicellularity and the Need for Communication—A Systematic Overview on (Algal) Plasmodesmata and Other Types of Symplasmic Cell Connections. Plants 2023, 12, 3342. https://doi.org/10.3390/plants12183342
Wegner L, Porth ML, Ehlers K. Multicellularity and the Need for Communication—A Systematic Overview on (Algal) Plasmodesmata and Other Types of Symplasmic Cell Connections. Plants. 2023; 12(18):3342. https://doi.org/10.3390/plants12183342
Chicago/Turabian StyleWegner, Linus, Merlin Leon Porth, and Katrin Ehlers. 2023. "Multicellularity and the Need for Communication—A Systematic Overview on (Algal) Plasmodesmata and Other Types of Symplasmic Cell Connections" Plants 12, no. 18: 3342. https://doi.org/10.3390/plants12183342
APA StyleWegner, L., Porth, M. L., & Ehlers, K. (2023). Multicellularity and the Need for Communication—A Systematic Overview on (Algal) Plasmodesmata and Other Types of Symplasmic Cell Connections. Plants, 12(18), 3342. https://doi.org/10.3390/plants12183342







