Are Protein Cavities and Pockets Commonly Used by Redox Active Signalling Molecules?
Abstract
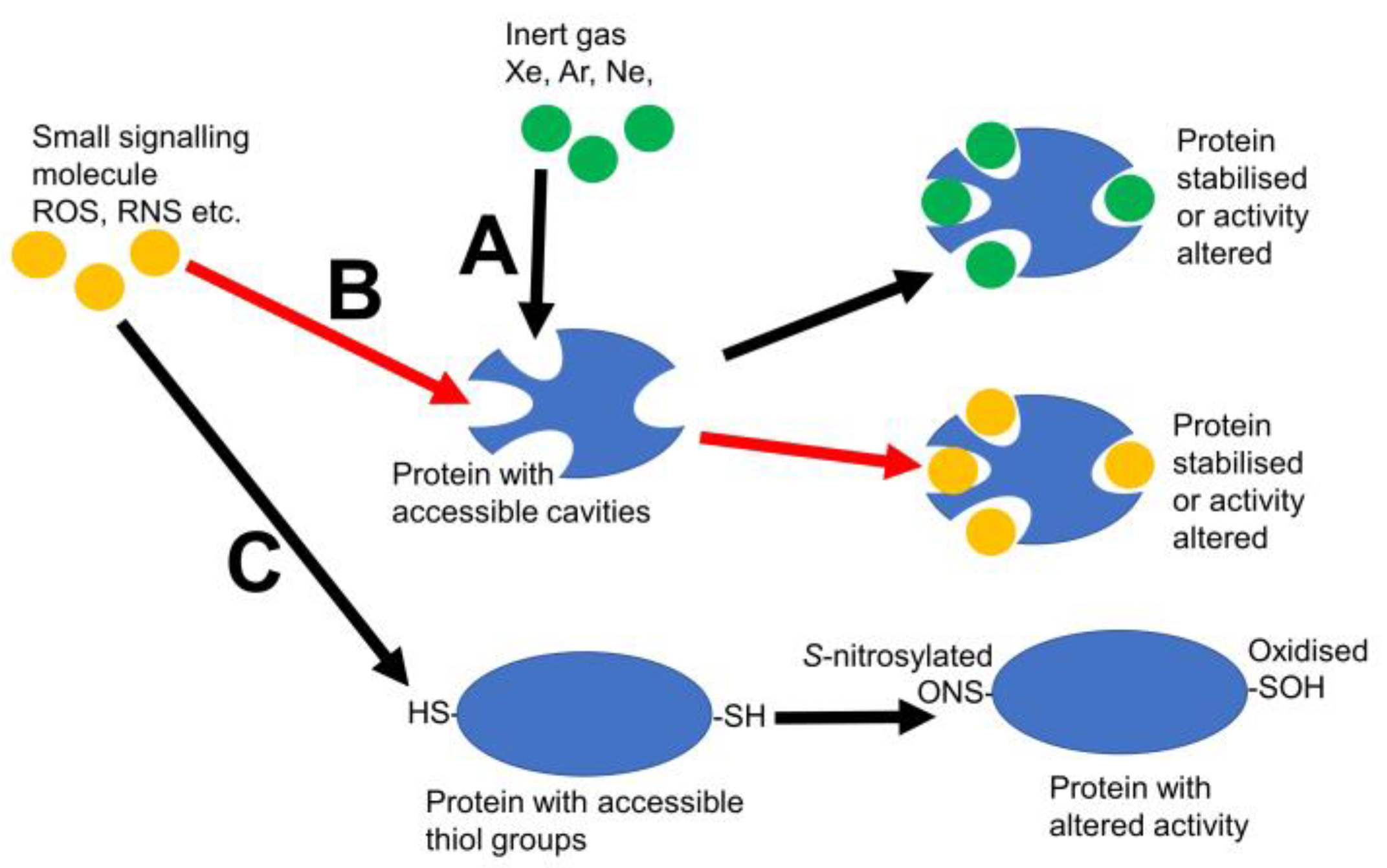
1. Introduction
2. Other Inert Gases
3. Other Small Signalling Molecules That May Need to Be Considered When Discussing Protein Cavities
4. Do Other Small Signalling Molecules Use Xe Pockets?
5. Conclusions and Future
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Chaki, M.; Valderrama, R.; Sánchez-Calvo, B.; Mata-Pérez, C.; Padilla, M.N.; Corpas, F.J.; Barroso, J.B. Nitric oxide buffering and conditional nitric oxide release in stress response. J. Exp. Bot. 2018, 69, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Bayr, H. Reactive oxygen species. Crit. Care Med. 2005, 33, S498–S501. [Google Scholar] [CrossRef]
- Bruckdorfer, R. The basics about nitric oxide. Mol. Asp. Med. 2005, 26, 3–31. [Google Scholar] [CrossRef]
- Kleniewska, P.; Piechota, A.; Skibska, B.; Gorąca, A. The NADPH oxidase family and its inhibitors. Arch. Immunol. Ther. Exp. 2012, 60, 277–294. [Google Scholar] [CrossRef] [PubMed]
- Millar, T.M.; Stevens, C.R.; Benjamin, N.; Eisenthal, R.; Harrison, R.; Blake, D.R. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett. 1998, 427, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Geller, D.A.; Billiar, T.R. Molecular biology of nitric oxide synthases. Cancer Metastasis Rev. 1998, 17, 7–23. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H. Gasotransmitter hydrogen sulfide signaling in neuronal health and disease. Biochem. Pharmacol. 2018, 149, 101–109. [Google Scholar] [CrossRef]
- Jiang, J.; Chan, A.; Ali, S.; Saha, A.; Haushalter, K.J.; Lam, W.L.M.; Glasheen, M.; Parker, J.; Brenner, M.; Mahon, S.B.; et al. Hydrogen sulfide—Mechanisms of toxicity and development of an antidote. Sci. Rep. 2016, 6, 20831. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Desikan, R.; Clarke, A.; Hurst, R.D.; Neill, S.J. Cell signalling following plant/pathogen interactions involves the generation of reactive oxygen and reactive nitrogen species. Plant Physiol. Biochem. 2002, 40, 611–617. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef]
- Berni, R.; Luyckx, M.; Xu, X.; Legay, S.; Sergeant, K.; Hausman, J.F.; Lutts, S.; Cai, G.; Guerriero, G. Reactive oxygen species and heavy metal stress in plants: Impact on the cell wall and secondary metabolism. Environ. Exp. Bot. 2019, 161, 98–106. [Google Scholar] [CrossRef]
- Suzuki, N.; Mittler, R. Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiol. Plant. 2006, 126, 45–51. [Google Scholar] [CrossRef]
- Schilter, D. Thiol oxidation: A slippery slope. Nat. Rev. Chem. 2017, 1, 0013. [Google Scholar] [CrossRef]
- Astier, J.; Kulik, A.; Koen, E.; Besson-Bard, A.; Bourque, S.; Jeandroz, S.; Lamotte, O.; Wendehenne, D. Protein S-nitrosylation: What’s going on in plants? Free Radic. Biol. Med. 2012, 53, 1101–1110. [Google Scholar] [CrossRef]
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical biology of H2S signaling through persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef]
- Miles, J.A.; Egan, J.L.; Fowler, J.A.; Machattou, P.; Millard, A.D.; Perry, C.J.; Scanlan, D.J.; Taylor, P.C. The evolutionary origins of peroxynitrite signalling. Biochem. Biophys. Res. Commun. 2021, 580, 107–112. [Google Scholar] [CrossRef]
- Whiteman, M.; Li, L.; Kostetski, I.; Chu, S.H.; Siau, J.L.; Bhatia, M.; Moore, P.K. Evidence for the formation of a novel nitrosothiol from the gaseous mediators nitric oxide and hydrogen sulphide. Biochem. Biophys. Res. Commun. 2006, 343, 303–310. [Google Scholar] [CrossRef]
- Ming, Y.; Ma, Q.H.; Han, X.L.; Li, H.Y. Molecular hydrogen improves type 2 diabetes through inhibiting oxidative stress. Exp. Ther. Med. 2020, 20, 359–366. [Google Scholar] [CrossRef]
- Ohno, K.; Ito, M.; Ichihara, M.; Ito, M. Molecular hydrogen as an emerging therapeutic medical gas for neurodegenerative and other diseases. Oxidative Med. Cell. Longev. 2012, 2012, 353152. [Google Scholar] [CrossRef]
- Alwazeer, D.; Liu, F.F.C.; Wu, X.Y.; LeBaron, T.W. Combating oxidative stress and inflammation in COVID-19 by molecular hydrogen therapy: Mechanisms and perspectives. Oxidative Med. Cell. Longev. 2021, 2021, 5513868. [Google Scholar] [CrossRef]
- Ohsawa, I.; Ishikawa, M.; Takahashi, K.; Watanabe, M.; Nishimaki, K.; Yamagata, K.; Katsura, K.; Katayama, Y.; Asoh, S.; Ohta, S. Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat. Med. 2007, 13, 688. [Google Scholar] [CrossRef]
- Hancock, J.T.; LeBaron, T.W.; Russell, G. Molecular hydrogen: Redox reactions and possible biological interactions. React. Oxyg. Species 2021, 11, 17–25. [Google Scholar] [CrossRef]
- Lawrence, J.H.; Loomis, W.F.; Tobias, C.A.; Turpin, F.H. Preliminary observations on the narcotic effect of xenon with a review of values for solubilities of gases in water and oils. J. Physiol. 1946, 105, 197. [Google Scholar] [CrossRef] [PubMed]
- Turan, H.T.; Boittier, E.; Meuwly, M. Interaction at a distance: Xenon migration in Mb. J. Chem. Phys. 2023, 158, 125103. [Google Scholar] [CrossRef] [PubMed]
- Duff, A.P.; Trambaiolo, D.M.; Cohen, A.E.; Ellis, P.J.; Juda, G.A.; Shepard, E.M.; Langley, D.B.; Dooley, D.M.; Freeman, H.C.; Guss, J.M. Using xenon as a probe for dioxygen-binding sites in copper amine oxidases. J. Mol. Biol. 2004, 344, 599–607. [Google Scholar] [CrossRef]
- Murray, J.W.; Maghlaoui, K.; Kargul, J.; Sugiura, M.; Barber, J. Analysis of xenon binding to photosystem II by X-ray crystallography. Photosynth. Res. 2008, 98, 523–527. [Google Scholar] [CrossRef]
- Hancock, J.T.; Russell, G.; Craig, T.J.; May, J.; Morse, H.R.; Stamler, J.S. Understanding hydrogen: Lessons to be learned from physical interactions between the inert gases and the globin superfamily. Oxygen 2022, 2, 578–590. [Google Scholar] [CrossRef]
- Jin, Z.; Piazza, O.; Ma, D.; Scarpati, G.; De Robertis, E. Xenon anesthesia and beyond: Pros and cons. Minerva Anestesiol. 2018, 85, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, B.P.; Watson, H.C.; Kendrew, J.C. Binding of xenon to sperm whale myoglobin. Nature 1965, 207, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Hermans, J.; Shankar, S. The free energy of xenon binding to myoglobin from molecular dynamics simulation. Isr. J. Chem. 1986, 27, 225–227. [Google Scholar] [CrossRef]
- Tilton, R.F., Jr.; Singh, U.C.; Weiner, S.J.; Connolly, M.L.; Kuntz, I.D., Jr.; Kollman, P.A.; Max, N.; Case, D.A. Computational studies of the interaction of myoglobin and xenon. J. Mol. Biol. 1986, 192, 443–456. [Google Scholar] [CrossRef]
- Conn, H.L., Jr. Equilibrium distribution of radioxenon in tissue: Xenon-hemoglobin association curve. J. Appl. Physiol. 1961, 16, 1065–1070. [Google Scholar] [CrossRef]
- Tilton, R.F., Jr.; Kuntz, I.D., Jr. Nuclear magnetic resonance studies of xenon-129 with myoglobin and hemoglobin. Biochemistry 1982, 21, 6850–6857. [Google Scholar] [CrossRef] [PubMed]
- Lepeshkevich, S.V.; Gilevich, S.N.; Parkhats, M.V.; Dzhagarov, B.M. Molecular oxygen migration through the xenon docking sites of human hemoglobin in the R-state. Biochim. Biophys. Acta BBA-Proteins Proteom. 2016, 1864, 1110–1121. [Google Scholar] [CrossRef]
- Prangé, T.; Schiltz, M.; Pernot, L.; Colloc’h, N.; Longhi, S.; Bourguet, W.; Fourme, R. Exploring hydrophobic sites in proteins with xenon or krypton. Proteins Struct. Funct. Bioinform. 1998, 30, 61–73. [Google Scholar] [CrossRef]
- Rubin, S.M.; Lee, S.Y.; Ruiz, E.J.; Pines, A.; Wemmer, D.E. Detection and characterization of xenon-binding sites in proteins by 129Xe NMR spectroscopy. J. Mol. Biol. 2002, 322, 425–440. [Google Scholar] [CrossRef]
- Pirrat, P.; Smith, M.A.; Pearson, A.R.; McPherson, M.J.; Phillips, S.E. Structure of a xenon derivative of Escherichia coli copper amine oxidase: Confirmation of the proposed oxygen-entry pathway. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Marassio, G.; Prangé, T.; David, H.N.; Santos, J.S.D.O.; Gabison, L.; Delcroix, N.; Abraini, J.H.; Colloc’h, N. Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: A crystallographic study. FASEB J. 2011, 25, 2266–2275. [Google Scholar] [CrossRef] [PubMed]
- Stoppe, C.; Ney, J.; Brenke, M.; Goetzenich, A.; Emontzpohl, C.; Schälte, G.; Grottke, O.; Moeller, M.; Rossaint, R.; Coburn, M. Sub-anesthetic xenon increases erythropoietin levels in humans: A randomized controlled trial. Sports Med. 2016, 46, 1753–1766. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.A.; Katz, I.; Warden, A.; Thornton, A.W.; Farjot, G. Identifying medically relevant xenon protein targets by in silico screening of the structural proteome. Med. Gas Res. 2023, 13, 33–38. [Google Scholar] [CrossRef]
- Yeh, S.Y.; Peterson, R.E. Solubility of krypton and xenon in blood, protein solutions, and tissue homogenates. J. Appl. Physiol. 1965, 20, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yuan, K.; Wang, X.; Zhang, L.; Hu, J. Influence of krypton gas nanobubbles on the activity of pepsin. Langmuir 2020, 36, 14070–14075. [Google Scholar] [CrossRef]
- Sanders, R.D.; Ma, D.; Maze, M. Argon neuroprotection. Crit. Care 2010, 14, 117. [Google Scholar] [CrossRef]
- Höllig, A.; Schug, A.; Fahlenkamp, A.V.; Rossaint, R.; Coburn, M.; the Argon Organo-Protective Network (AON). Argon: Systematic review on neuro-and organoprotective properties of an “inert” gas. Int. J. Mol. Sci. 2014, 15, 18175–18196. [Google Scholar] [CrossRef]
- Grottke, O.; Coburn, M. Argon: Neuroprotection in in vitro models of cerebral ischemia and traumatic brain injury. Crit. Care 2009, 13, R206. [Google Scholar]
- Ulbrich, F.; Kaufmann, K.; Roesslein, M.; Wellner, F.; Auwärter, V.; Kempf, J.; Loop, T.; Buerkle, H.; Goebel, U. Argon mediates anti-apoptotic signaling and neuroprotection via inhibition of toll-like receptor 2 and 4. PLoS ONE 2015, 10, e0143887. [Google Scholar] [CrossRef]
- Fahlenkamp, A.V.; Rossaint, R.; Haase, H.; Al Kassam, H.; Ryang, Y.M.; Beyer, C.; Coburn, M. The noble gas argon modifies extracellular signal-regulated kinase 1/2 signaling in neurons and glial cells. Eur. J. Pharmacol. 2012, 674, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhang, R.; Sun, X. Bustling argon: Biological effect. Med. Gas Res. 2013, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Martusevich, A.; Surovegina, A.; Popovicheva, A.; Didenko, N.; Artamonov, M.; Nazarov, V. Some beneficial effects of inert gases on blood oxidative metabolism: In vivo study. BioMed Res. Int. 2022, 2022, 5857979. [Google Scholar] [CrossRef]
- Schreiner, H.R.; Gregoire, R.C.; Lawrie, J.A. New biological effect of the gases of the helium group. Science 1962, 136, 653–654. [Google Scholar] [CrossRef] [PubMed]
- Jawad, N.; Rizvi, M.; Gu, J.; Adeyi, O.; Tao, G.; Maze, M.; Ma, D. Neuroprotection (and lack of neuroprotection) afforded by a series of noble gases in an in vitro model of neuronal injury. Neurosci. Lett. 2009, 460, 232–236. [Google Scholar] [CrossRef]
- Rizvi, M.; Jawad, N.; Li, Y.; Vizcaychipi, M.P.; Maze, M.; Ma, D. Effect of noble gases on oxygen and glucose deprived injury in human tubular kidney cells. Exp. Biol. Med. 2010, 235, 886–891. [Google Scholar] [CrossRef]
- Krumova, K.; Cosa, G. Overview of reactive oxygen species. In Singlet Oxygen: Applications in Biosciences and Nanosciences; Nonell, S., Flors, C., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2016; pp. 1–21. [Google Scholar]
- Antunes, F.; Brito, P.M. Quantitative biology of hydrogen peroxide signaling. Redox Biol. 2017, 13, 1–7. [Google Scholar] [CrossRef]
- Dogra, V.; Kim, C. Singlet oxygen metabolism: From genesis to signaling. Front. Plant Sci. 2020, 10, 1640. [Google Scholar] [CrossRef]
- Molassiotis, A.; Fotopoulos, V. Oxidative and nitrosative signaling in plants: Two branches in the same tree? Plant Signal. Behav. 2011, 6, 210–214. [Google Scholar] [CrossRef]
- Hancock, J.T. Nitric oxide signaling in plants. Plants 2020, 9, 1550. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P. L Signaling by sulfur-containing molecules. Quantitative aspects. Arch. Biochem. Biophys. 2017, 617, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Reactive sulfur species (RSS): Possible new players in the oxidative metabolism of plant peroxisomes. Front. Plant Sci. 2015, 6, 116. [Google Scholar] [CrossRef]
- Aroca, A.; Gotor, C.; Romero, L.C. Hydrogen sulfide signaling in plants: Emerging roles of protein persulfidation. Front. Plant Sci. 2018, 9, 1369. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Lara, L.O.; Medrano-Macías, J.; Pérez-Labrada, F.; Rivas-Martínez, E.N.; García-Enciso, E.L.; González-Morales, S.; Juárez-Maldonado, A.; Rincón-Sánchez, F.; Benavides-Mendoza, A. From elemental sulfur to hydrogen sulfide in agricultural soils and plants. Molecules 2019, 24, 2282. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.E.; Gibson, Q.H. Ligand migration in sperm whale myoglobin. Biochemistry 1997, 36, 11909–11917. [Google Scholar] [CrossRef]
- Tetreau, C.; Blouquit, Y.; Novikov, E.; Quiniou, E.; Lavalette, D. Competition with xenon elicits ligand migration and escape pathways in myoglobin. Biophys. J. 2004, 86, 435–447. [Google Scholar] [CrossRef]
- Abraini, J.H.; Marassio, G.; David, H.N.; Vallone, B.; Prangé, T.; Colloc’h, N. Crystallographic studies with xenon and nitrous oxide provide evidence for protein-dependent processes in the mechanisms of general anesthesia. Anesthesiology 2014, 121, 1018–1027. [Google Scholar] [CrossRef]
- LaBella, F.S.; Stein, D.; Queen, G. The site of general anesthesia and cytochrome P450 monooxygenases: Occupation of the enzyme heme pocket by xenon and nitrous oxide. Eur. J. Pharmacol. 1999, 381, R1–R3. [Google Scholar] [CrossRef]
- Winter, M.B.; Herzik, M.A., Jr.; Kuriyan, J.; Marletta, M.A. Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain. Proc. Natl. Acad. Sci. USA 2011, 108, E881–E889. [Google Scholar] [CrossRef]
- Brunori, M. Nitric oxide moves myoglobin centre stage. Trends Biochem. Sci. 2001, 26, 209–210. [Google Scholar] [CrossRef]
- Brunori, M. Structural dynamics of myoglobin. Biophys. Chem. 2000, 86, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Tilton, R.F., Jr.; Kuntz, I.D., Jr.; Petsko, G.A. Cavities in proteins: Structure of a metmyoglobin xenon complex solved to 1.9. ANG. Biochemistry 1984, 23, 2849–2857. [Google Scholar] [CrossRef]
- Daigle, R.; Rousseau, J.A.; Guertin, M.; Lagüe, P. Theoretical investigations of nitric oxide channeling in Mycobacterium tuberculosis truncated hemoglobin N. Biophys. J. 2009, 97, 2967–2977. [Google Scholar] [CrossRef]
- Ye, X.; Yu, A.; Champion, P.M. Dynamics of nitric oxide rebinding and escape in horseradish peroxidase. J. Am. Chem. Soc. 2006, 128, 1444–1445. [Google Scholar] [CrossRef] [PubMed]
- Terasaka, E.; Yamada, K.; Wang, P.H.; Hosokawa, K.; Yamagiwa, R.; Matsumoto, K.; Ishii, S.; Mori, T.; Yagi, K.; Sawai, H.; et al. Dynamics of nitric oxide controlled by protein complex in bacterial system. Proc. Natl. Acad. Sci. USA 2017, 114, 9888–9893. [Google Scholar] [CrossRef]
- Chu, K.; Vojtchovský, J.; McMahon, B.H.; Sweet, R.M.; Berendzen, J.; Schlichting, I. Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature 2000, 403, 921–923. [Google Scholar] [CrossRef]
- Elber, R.; Karplus, M. Enhanced sampling in molecular dynamics: Use of the time-dependent Hartree approximation for a simulation of carbon monoxide diffusion through myoglobin. J. Am. Chem. Soc. 1990, 112, 9161–9175. [Google Scholar] [CrossRef]
- Nishihara, Y.; Sakakura, M.; Kimura, Y.; Terazima, M. The escape process of carbon monoxide from myoglobin to solution at physiological temperature. J. Am. Chem. Soc. 2004, 126, 11877–11888. [Google Scholar] [CrossRef]
- Bossa, C.; Anselmi, M.; Roccatano, D.; Amadei, A.; Vallone, B.; Brunori, M.; Di Nola, A. Extended molecular dynamics simulation of the carbon monoxide migration in sperm whale myoglobin. Biophys. J. 2004, 86, 3855–3862. [Google Scholar] [CrossRef]
- Anselmi, M.; Di Nola, A.; Amadei, A. The kinetics of ligand migration in crystallized myoglobin as revealed by molecular dynamics simulations. Biophys. J. 2008, 94, 4277–4281. [Google Scholar] [CrossRef]
- Gee, L.B.; Leontyev, I.; Stuchebrukhov, A.; Scott, A.D.; Pelmenschikov, V.; Cramer, S.P. Docking and migration of carbon monoxide in nitrogenase: The case for gated pockets from infrared spectroscopy and molecular dynamics. Biochemistry 2015, 54, 3314–3319. [Google Scholar] [CrossRef] [PubMed]
- Goldet, G.; Brandmayr, C.; Stripp, S.T.; Happe, T.; Cavazza, C.; Fontecilla-Camps, J.C.; Armstrong, F.A. Electrochemical kinetic investigations of the reactions of [FeFe]-hydrogenases with carbon monoxide and oxygen: Comparing the importance of gas tunnels and active-site electronic/redox effects. J. Am. Chem. Soc. 2009, 131, 14979–14989. [Google Scholar] [CrossRef] [PubMed]
- Cundari, T.R.; Wilson, A.K.; Drummond, M.L.; Gonzalez, H.E.; Jorgensen, K.R.; Payne, S.; Braunfeld, J.; De Jesus, M.; Johnson, V.M. CO2-formatics: How do proteins bind carbon dioxide? J. Chem. Inf. Model. 2009, 49, 2111–2115. [Google Scholar] [CrossRef] [PubMed]
- Hiromoto, T.; Fujiwara, S.; Hosokawa, K.; Yamaguchi, H. Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site. J. Mol. Biol. 2006, 364, 878–896. [Google Scholar] [CrossRef]
- Chen, L.; Lyubimov, A.Y.; Brammer, L.; Vrielink, A.; Sampson, N.S. The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase. Biochemistry 2008, 47, 5368–5377. [Google Scholar] [CrossRef]
- Zhao, P.; Kong, F.; Jiang, Y.; Qin, X.; Tian, X.; Cong, Z. Enabling peroxygenase activity in by engineering hydrogen peroxide tunnels. J. Am. Chem. Soc. 2023, 145, 5506–5511. [Google Scholar] [CrossRef]
- Moreno, D.M.; Martí, M.A.; De Biase, P.M.; Estrin, D.A.; Demicheli, V.; Radi, R.; Boechi, L. Exploring the molecular basis of human manganese superoxide dismutase inactivation mediated by tyrosine 34 nitration. Arch. Biochem. Biophys. 2011, 507, 304–309. [Google Scholar] [CrossRef]
- Eckenhoff, R.G. Promiscuous ligands and attractive cavities. Mol. Interv. 2001, 1, 258. [Google Scholar]
- Otting, G.; Liepinsh, E.; Halle, B.; Frey, U. NMR identification of hydrophobic cavities with low water occupancies in protein structures using small gas molecules. Nat. Struct. Biol. 1997, 4, 396–404. [Google Scholar] [CrossRef]
- Carugo, O.; Argos, P. Accessibility to internal cavities and ligand binding sites monitored by protein crystallographic thermal factors. Proteins 1998, 31, 201–213. [Google Scholar] [CrossRef]
- Colloc’h, N.; Carpentier, P.; Montemiglio, L.C.; Vallone, B.; Prangé, T. Mapping hydrophobic tunnels and cavities in neuroglobin with noble gas under pressure. Biophys. J. 2017, 113, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Zhao, P.; Gong, W.; Ding, W.; He, Q. Fe-porphyrin: A redox-related biosensor of hydrogen molecule. Nano Res. 2023, 16, 2020–2025. [Google Scholar] [CrossRef]
- Kim, S.A.; Jong, Y.C.; Kang, M.S.; Yu, C.J. Antioxidation activity of molecular hydrogen via protoheme catalysis in vivo; an insight from ab initio calculations. Res. Sq. 2022. pre-print. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S. Molecular hydrogen may activate the transcription factor Nrf2 to alleviate oxidative stress through the hydrogen-targeted porphyrin. Aging Pathobiol. Ther. 2023, 13, 25–32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hancock, J.T. Are Protein Cavities and Pockets Commonly Used by Redox Active Signalling Molecules? Plants 2023, 12, 2594. https://doi.org/10.3390/plants12142594
Hancock JT. Are Protein Cavities and Pockets Commonly Used by Redox Active Signalling Molecules? Plants. 2023; 12(14):2594. https://doi.org/10.3390/plants12142594
Chicago/Turabian StyleHancock, John T. 2023. "Are Protein Cavities and Pockets Commonly Used by Redox Active Signalling Molecules?" Plants 12, no. 14: 2594. https://doi.org/10.3390/plants12142594
APA StyleHancock, J. T. (2023). Are Protein Cavities and Pockets Commonly Used by Redox Active Signalling Molecules? Plants, 12(14), 2594. https://doi.org/10.3390/plants12142594





