Transcriptional Regulation of Metabolic and Cellular Processes in Durum Wheat (Triticum turgidum subsp. durum) in the Face of Temperature Increasing
Abstract
1. Introduction
2. Results
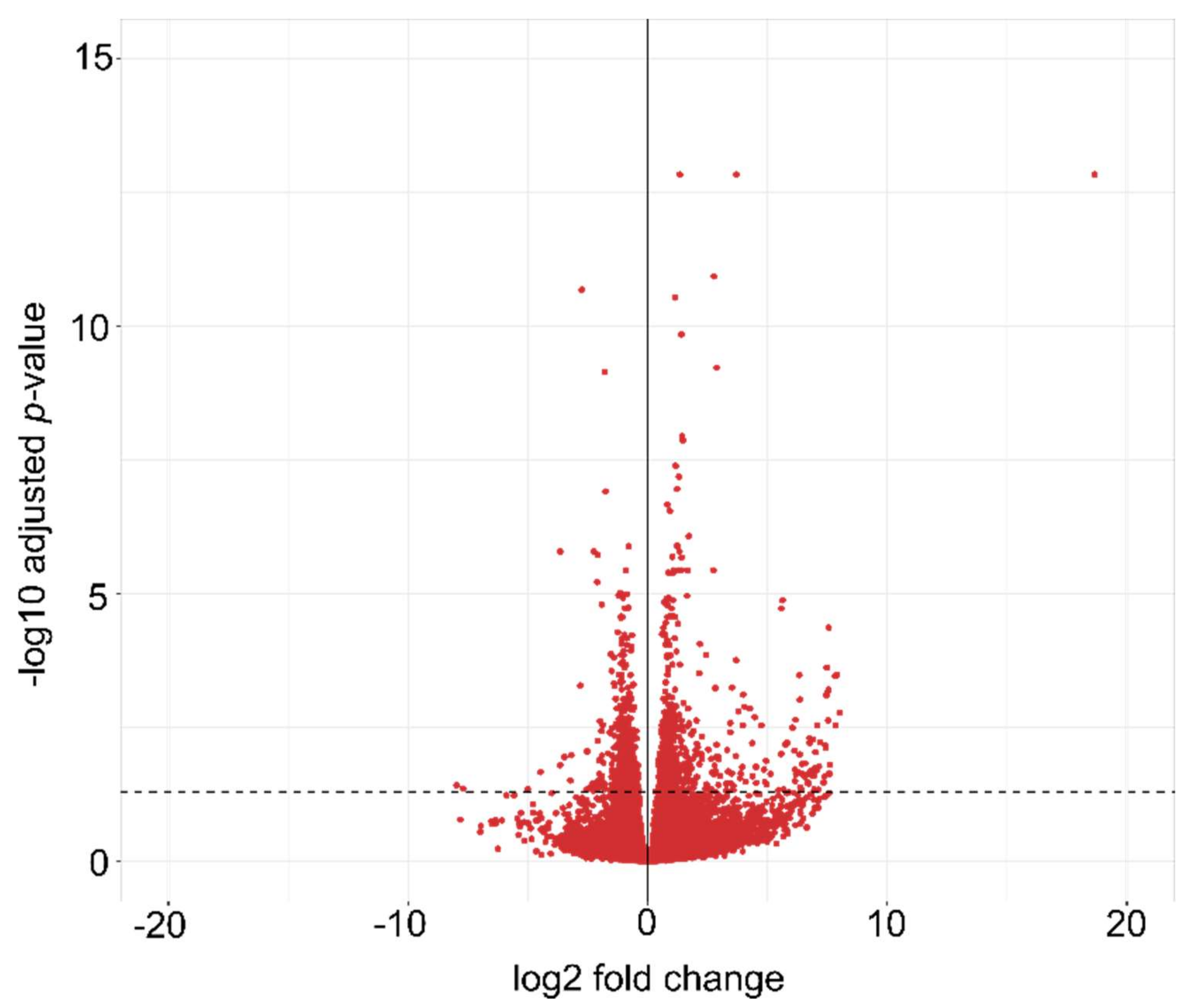
2.1. Validation of RNA Extraction from Durum Wheat Seedlings, Quality, and Sequencing
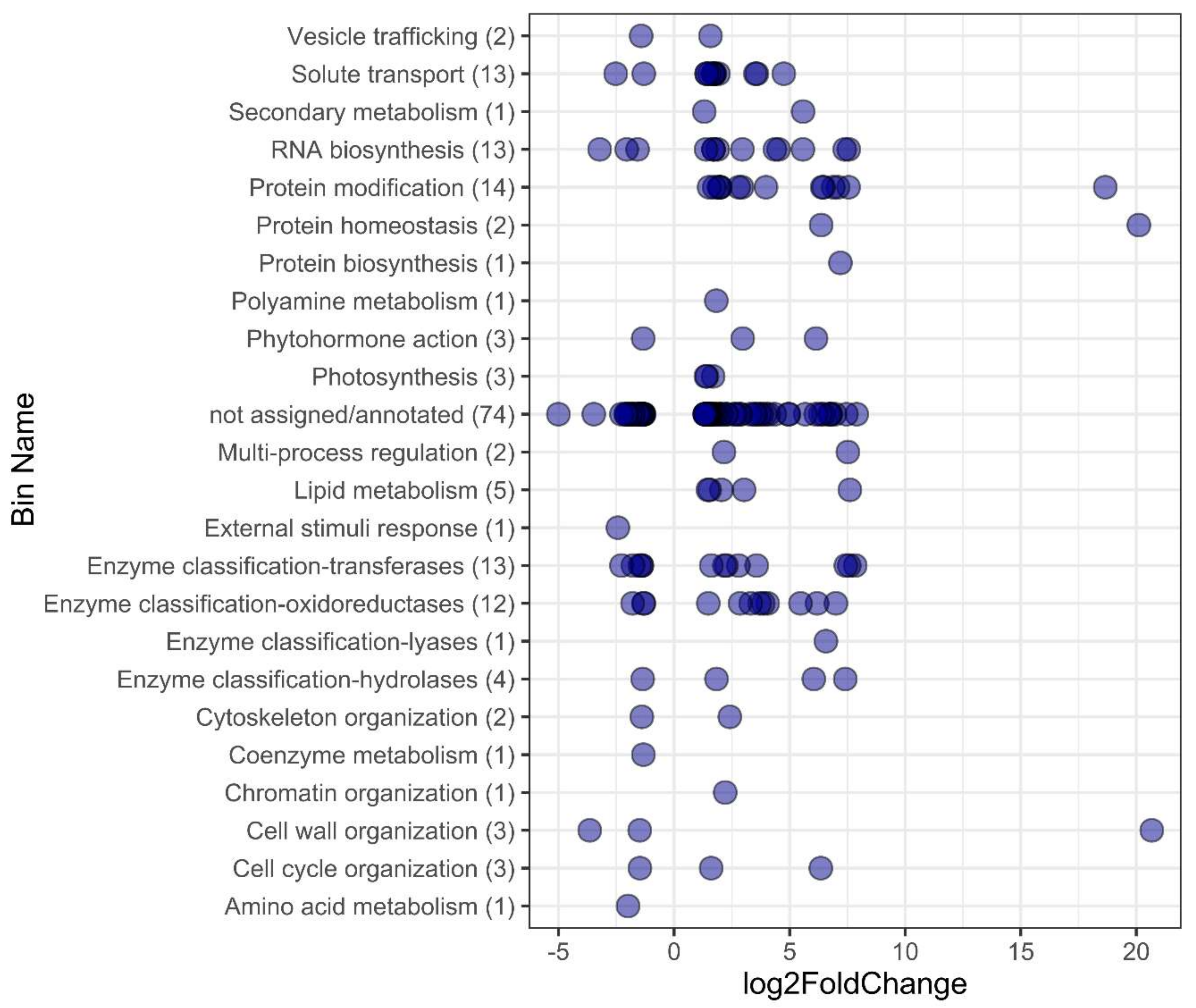
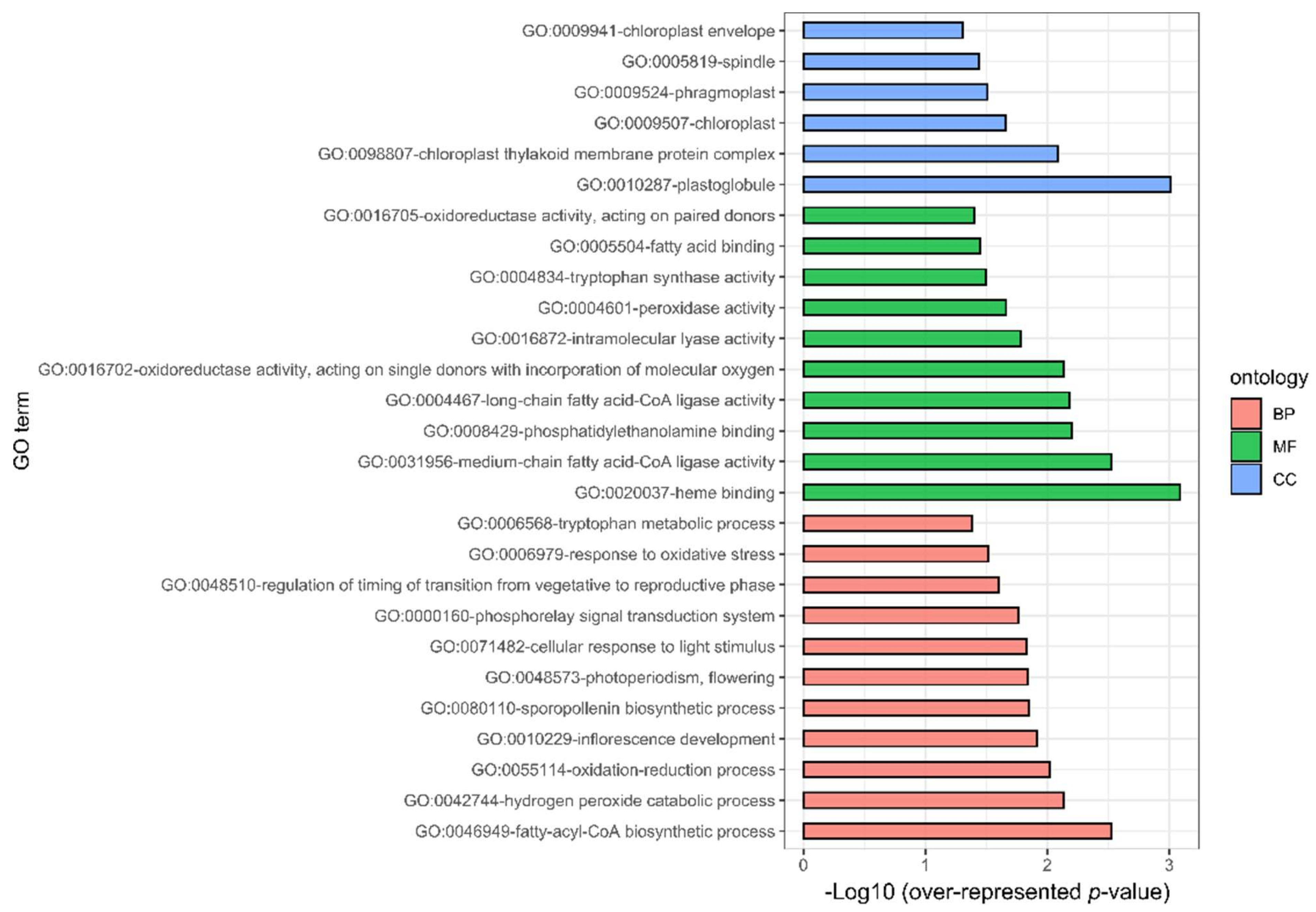
2.2. Enriched Gene Ontology Analysis
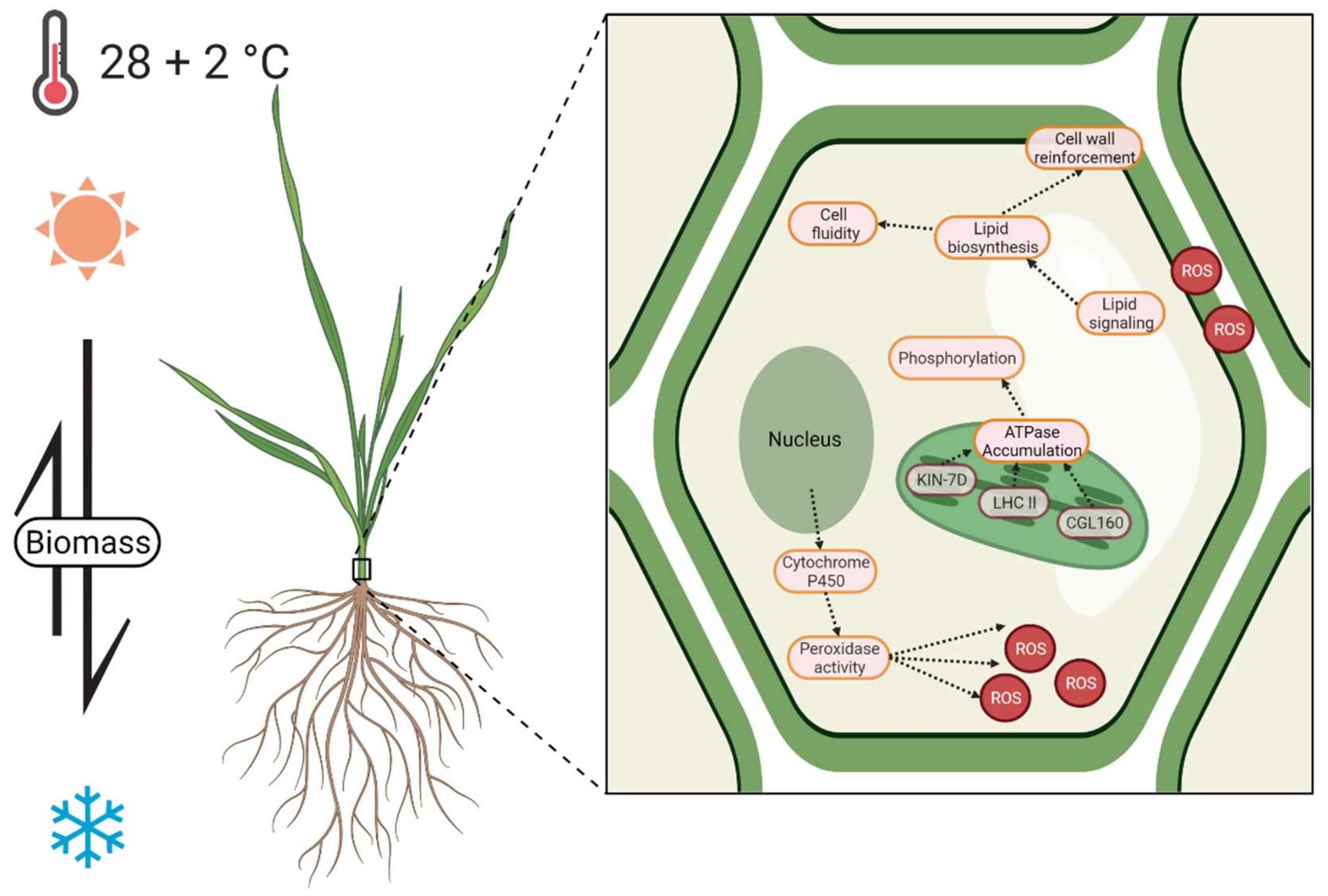
3. Discussion
3.1. Morphometrical Influence of An Elevated Temperature
3.2. Acceleration of Development
3.3. Photosynthesis and ATPase Activity
3.4. Regulation of Lipid Biosynthesis
3.5. ROS Regulation and Detoxification
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. RNA Extraction and RNA-Seq Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- SIAP. Panorama Agroalimentario 2020, 2020th ed.; SIAP, Ed.; Secretaria de Agricultura y Desarrollo Rural: Mexico city, Mexico, 2020. [Google Scholar]
- Matson, P.; Jewett, P. Ecosystems and land-use change in the Yaqui Valley: Does agricultural intensification spare land for nature? In Seeds of Sustainability: Lessons from the Birthplace of the Green Revolution; Island Press/Center for Resource Economics: Washington, DC, USA, 2013; pp. 47–62. ISBN 9781610911771. [Google Scholar]
- Millar, N.; Urrea, A.; Kahmark, K.; Shcherbak, I.; Robertson, G.P.; Ortiz-Monasterio, I. Nitrous oxide (N2O) flux responds exponentially to nitrogen fertilizer in irrigated wheat in the Yaqui Valley, Mexico. Agric. Ecosyst. Environ. 2018, 261, 125–132. [Google Scholar] [CrossRef]
- Tyczewska, A.; Woźniak, E.; Gracz, J.; Kuczyński, J.; Twardowski, T. Towards Food Security: Current State and Future Prospects of Agrobiotechnology. Trends Biotechnol. 2018, 36, 1219–1229. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef]
- Rezaei, E.E.; Siebert, S.; Manderscheid, R.; Müller, J.; Mahrookashani, A.; Ehrenpfordt, B.; Haensch, J.; Weigel, H.J.; Ewert, F. Quantifying the response of wheat yields to heat stress: The role of the experimental setup. Field Crop. Res. 2018, 217, 93–103. [Google Scholar] [CrossRef]
- Liu, B.; Asseng, S.; Müller, C.; Ewert, F.; Elliott, J.; Lobell, D.B.; Martre, P.; Ruane, A.C.; Wallach, D.; Jones, J.W.; et al. Similar estimates of temperature impacts on global wheat yield by three independent methods. Nat. Clim. Chang. 2016, 6, 1130–1136. [Google Scholar] [CrossRef]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; Van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Schachtschabel, J.; Mishkind, M.; Munnik, T.; Arisz, S.A. Hot topic: Thermosensing in plants. Plant Cell Environ. 2021, 44, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Singh, R.; Tiwari, R.; Kumar, R.; Gupta, V.K. Wheat Responses and Tolerance to Terminal Heat Stress: A Review. In Wheat Production in Changing Environments; Springer: Singapore, 2019; pp. 149–173. ISBN 9789811368837. [Google Scholar]
- Balla, K.; Karsai, I.; Bónis, P.; Kiss, T.; Berki, Z.; Horváth, Á.; Mayer, M.; Bencze, S.; Veisz, O. Heat stress responses in a large set of winter wheat cultivars (Triticum aestivum L.) depend on the timing and duration of stress. PLoS ONE 2019, 14, e0222639. [Google Scholar] [CrossRef] [PubMed]
- Eyshi Rezaei, E.; Webber, H.; Gaiser, T.; Naab, J.; Ewert, F. Heat stress in cereals: Mechanisms and modelling. Eur. J. Agron. 2015, 64, 98–113. [Google Scholar] [CrossRef]
- Ni, Z.; Li, H.; Zhao, Y.; Peng, H.; Hu, Z.; Xin, M.; Sun, Q. Genetic improvement of heat tolerance in wheat: Recent progress in understanding the underlying molecular mechanisms. Crop J. 2018, 6, 32–41. [Google Scholar] [CrossRef]
- Liu, B.; Asseng, S.; Liu, L.; Tang, L.; Cao, W.; Zhu, Y. Testing the responses of four wheat crop models to heat stress at anthesis and grain filling. Glob. Chang. Biol. 2016, 22, 1890–1903. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Bellstaedt, J.; Trenner, J.; Lippmann, R.; Poeschl, Y.; Zhang, X.; Friml, J.; Quint, M.; Delkera, C. A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 2019, 180, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Lamers, J.; van der Meer, T.; Testerink, C. How Plants Sense and Respond to Stressful Environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Ponce-Soto, G.Y.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.E.; Usadel, B. MapMan4: A Refined Protein Classification and Annotation Framework Applicable to Multi-Omics Data Analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias GOseq GOseq is a method for GO analysis of RNA-seq data that takes into account the length bias inherent in RNA-seq. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Mašková, T.; Herben, T. Root:shoot ratio in developing seedlings: How seedlings change their allocation in response to seed mass and ambient nutrient supply. Ecol. Evol. 2018, 8, 7143–7150. [Google Scholar] [CrossRef]
- Zhang, N.; Belsterling, B.; Raszewski, J.; Tonsor, S.J. Natural populations of Arabidopsis thaliana differ in seedling responses to high temperature stress. AoB Plants 2015, 7, plv101. [Google Scholar] [CrossRef]
- Webb, A.A.R.; Seki, M.; Satake, A.; Caldana, C. Continuous dynamic adjustment of the plant circadian oscillator. Nat. Commun. 2019, 10, 550. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.; Park, C. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef]
- Masuda, K.; Yamada, T.; Kagawa, Y.; Fukuda, H. Time Lag Between Light and Heat Diurnal Cycles Modulates CIRCADIAN CLOCK ASSOCIATION 1 Rhythm and Growth in Arabidopsis thaliana. Front. Plant Sci. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fristedt, R.; Martins, N.F.; Strenkert, D.; Clarke, C.A.; Suchoszek, M.; Thiele, W.; Schöttler, M.A.; Merchant, S.S. The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS ONE 2015, 10, e0121658. [Google Scholar] [CrossRef] [PubMed]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.; Jansson, S. The Light-Harvesting Chlorophyll a/b Binding Proteins Lhcb1 and Lhcb2 Play Complementary Roles during State Transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Y.; Chong, K. The novel functions of kinesin motor proteins in plants. Protoplasma 2012, 249, 95–100. [Google Scholar] [CrossRef]
- Grahl, S.; Reiter, B.; Gügel, I.L.L.; Vamvaka, E.; Gandini, C.; Jahns, P.; Soll, J.; Leister, D.; Rühle, T. The Arabidopsis Protein CGLD11 Is Required for Chloroplast ATP Synthase Accumulation. Mol. Plant 2016, 9, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Qian, D.; Liu, B.; Ma, J.; Wan, D.; Wang, X.; He, W.; Xiang, Y. ALA6, a P4-type ATPase, is involved in heat stress responses in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Okumura, M.; Inoue, S.I.; Kuwata, K.; Kinoshita, T. Photosynthesis activates plasma membrane H+-ATPase via sugar accumulation. Plant Physiol. 2016, 171, 580–589. [Google Scholar] [CrossRef]
- Estravis-Barcala, M.; Mattera, M.G.; Soliani, C.; Bellora, N.; Opgenoorth, L.; Heer, K.; Arana, M.V. Molecular bases of responses to abiotic stress in trees. J. Exp. Bot. 2020, 71, 3765–3779. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef]
- Mueller, S.P.; Unger, M.; Guender, L.; Fekete, A.; Mueller, M.J. Phospholipid: Diacylglycerol acyltransferase-mediated triacylglyerol synthesis augments basal thermotolerance. Plant Physiol. 2017, 175, 486–497. [Google Scholar] [CrossRef]
- Demski, K.; Łosiewska, A.; Jasieniecka-Gazarkiewicz, K.; Klińska, S.; Banaś, A. Phospholipid:Diacylglycerol Acyltransferase1 Overexpression Delays Senescence and Enhances Post-heat and Cold Exposure Fitness. Front. Plant Sci. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Guo, W. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef]
- Ravichandran, S.; Ragupathy, R.; Edwards, T.; Domaratzki, M.; Cloutier, S. Microrna-guided regulation of heat stress response in wheat. BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Sihag, P.; Sagwal, V.; Kumar, A.; Balyan, P.; Mir, R.R.; Dhankher, O.P.; Kumar, U. Discovery of miRNAs and Development of Heat-Responsive miRNA-SSR Markers for Characterization of Wheat Germplasm for Terminal Heat Tolerance Breeding. Front. Genet. 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant responses to heat stress: Physiology, transcription, noncoding rnas, and epigenetics. Int. J. Mol. Sci. 2021, 22, 117. [Google Scholar] [CrossRef]
- Figueroa López, P.; Félix Fuentes, J.; Fuentes Dávila, G.; Valenzuela Herrera, V.; Chávez Villalba, G.; Mendoza Lugo, J. Cirno c2008, nueva variedad de trigo cristalino con alto rendimiento potencial para el estado de Sonora. Rev. Mex. Ciencias Agrícolas 2010, 1, 745–749. [Google Scholar]
- Robles Montoya, R.I.; Chaparro Encinas, L.A.; Parra Cota, F.I.; De los Santos Villalobos, S. Mejorando rasgos biométricos de plántulas de trigo con la inoculación de un consorcio nativo de Bacillus. Rev. Mex. Ciencias Agrícolas 2020, 11, 229–235. [Google Scholar] [CrossRef][Green Version]
- Lees, K.; Fitzsimons, M.; Snape, J.; Tappin, A.; Comber, S. Soil sterilisation methods for use in OECD 106: How effective are they? Chemosphere 2018, 209, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, N.; Deckers, J.; Govaerts, B. Classification of the Soil at CIMMYT ’s Experimental Station in the Yaqui Valley Near Ciudad Obregón, Sonora, México; CIMMYT Report; CIMMYT: Ciudad de Mexico, Mexico, 2009. [Google Scholar]
- Valenzuela-Aragon, B.; Parra-Cota, F.I.; Santoyo, G.; Arellano-Wattenbarger, G.L.; de los Santos-Villalobos, S. Plant-assisted selection: A promising alternative for in vivo identification of wheat (Triticum turgidum L. subsp. Durum) growth promoting bacteria. Plant Soil 2019, 435, 367–384. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A Decimal Code for the Growth Stages of Cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Rojas Padilla, J.; Chaparro Encinas, L.A.; Robles Montoya, R.I.; De los Santos Villalobos, S. Promoción de crecimiento en trigo (Triticum turgidum L. subsp. durum) por la co-inoculación de cepas nativas de Bacillus aisladas del Valle del Yaqui, México. Nov. Sci. 2020, 12. [Google Scholar] [CrossRef]
- Tüzmen, S.; Baskin, Y.; Nursal, A.F.; Eraslan, S.; Esemen, Y.; Çalibas, G.; Demir, A.B.; Abbasoglu, D.; Hizel, C. Techniques for nucleic acid engineering: The foundation of gene manipulation. In Omics Technologies and Bio-Engineering: Towards Improving Quality of Life; Elsevier: Belo Horizonte, Brazil, 2018; Volume 1, pp. 247–315. ISBN 9780128047491. [Google Scholar]
- Chaparro-Encinas, L.A.; Arellano-Wattenbarger, G.L.; Parra-Cota, F.I.; de los Santos-Villalobos, S. A modified CTAB and Trizol® protocol for high-quality RNA extraction from whole wheat seedlings, including rhizosphere. Cereal Res. Commun. 2020, 48, 275–282. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Smedley, D.; Haider, S.; Durinck, S.; Pandini, L.; Provero, P.; Allen, J.; Arnaiz, O.; Awedh, M.H.; Baldock, R.; Barbiera, G.; et al. The BioMart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015, 43, W589–W598. [Google Scholar] [CrossRef] [PubMed]
| Biometrical Traits | Optimal Temperature (28 °C) | Increased Temperature (30 °C) | Difference (%) |
|---|---|---|---|
| Shoot length (cm) | 22.6 ± 2.9 | 21.1 ± 4.9 | −6.73% |
| Root length (cm) | 16.6 ± 3.6 * | 13.2 ± 5.0 | −20.00% |
| Dry shoot weight (g) | 38.0 ± 7.1 * | 28.7 ± 10.3 | −24.50% |
| Dry root weight (g) | 52.9 ± 12.3 | 58.6 ± 21.6 | 10.80% |
| Root:shoot ratio (dry mass, g) | 1.5 ± 0.5 | 2.4 ± 1.4 * | 57.92% |
| Biovolume index | 130.8 ± 9.2 * | 93.9 ± 29.2 | −28.20% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaparro-Encinas, L.A.; Santoyo, G.; Peña-Cabriales, J.J.; Castro-Espinoza, L.; Parra-Cota, F.I.; Santos-Villalobos, S.d.l. Transcriptional Regulation of Metabolic and Cellular Processes in Durum Wheat (Triticum turgidum subsp. durum) in the Face of Temperature Increasing. Plants 2021, 10, 2792. https://doi.org/10.3390/plants10122792
Chaparro-Encinas LA, Santoyo G, Peña-Cabriales JJ, Castro-Espinoza L, Parra-Cota FI, Santos-Villalobos Sdl. Transcriptional Regulation of Metabolic and Cellular Processes in Durum Wheat (Triticum turgidum subsp. durum) in the Face of Temperature Increasing. Plants. 2021; 10(12):2792. https://doi.org/10.3390/plants10122792
Chicago/Turabian StyleChaparro-Encinas, Luis Abraham, Gustavo Santoyo, Juan José Peña-Cabriales, Luciano Castro-Espinoza, Fannie Isela Parra-Cota, and Sergio de los Santos-Villalobos. 2021. "Transcriptional Regulation of Metabolic and Cellular Processes in Durum Wheat (Triticum turgidum subsp. durum) in the Face of Temperature Increasing" Plants 10, no. 12: 2792. https://doi.org/10.3390/plants10122792
APA StyleChaparro-Encinas, L. A., Santoyo, G., Peña-Cabriales, J. J., Castro-Espinoza, L., Parra-Cota, F. I., & Santos-Villalobos, S. d. l. (2021). Transcriptional Regulation of Metabolic and Cellular Processes in Durum Wheat (Triticum turgidum subsp. durum) in the Face of Temperature Increasing. Plants, 10(12), 2792. https://doi.org/10.3390/plants10122792







