Cytotoxic Psammaplysin Analogues from the Verongid Red Sea Sponge Aplysinella Species
Abstract
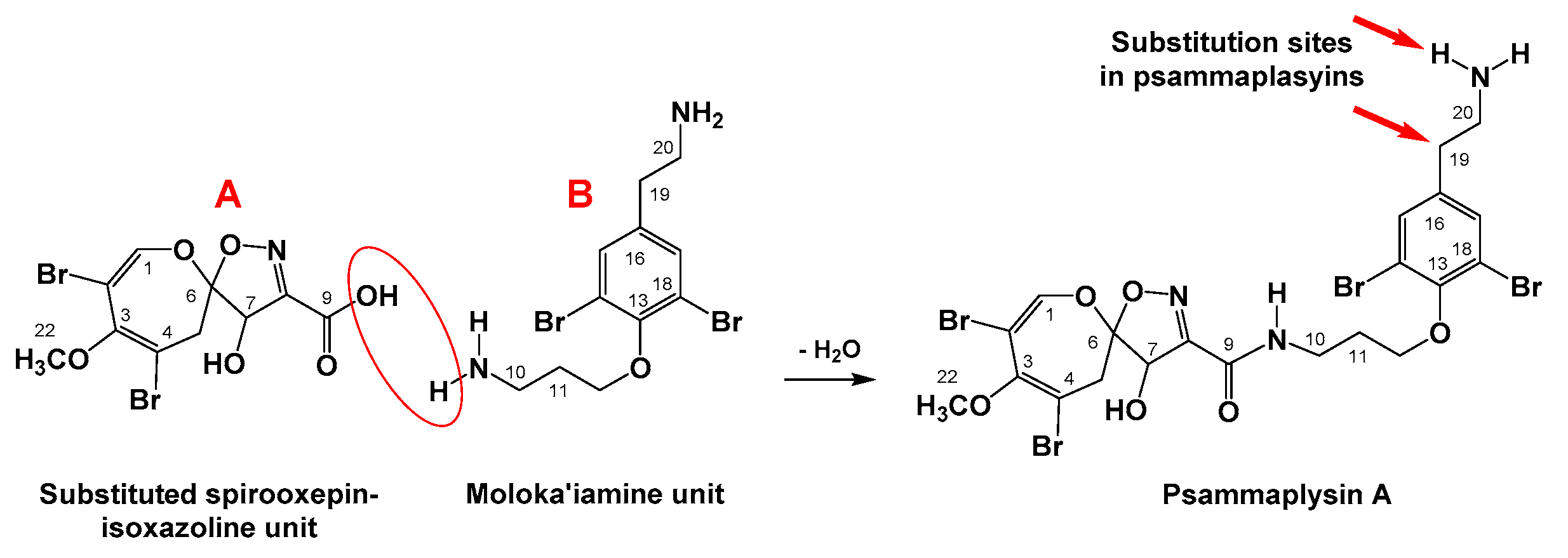
1. Introduction
2. Results and Discussion
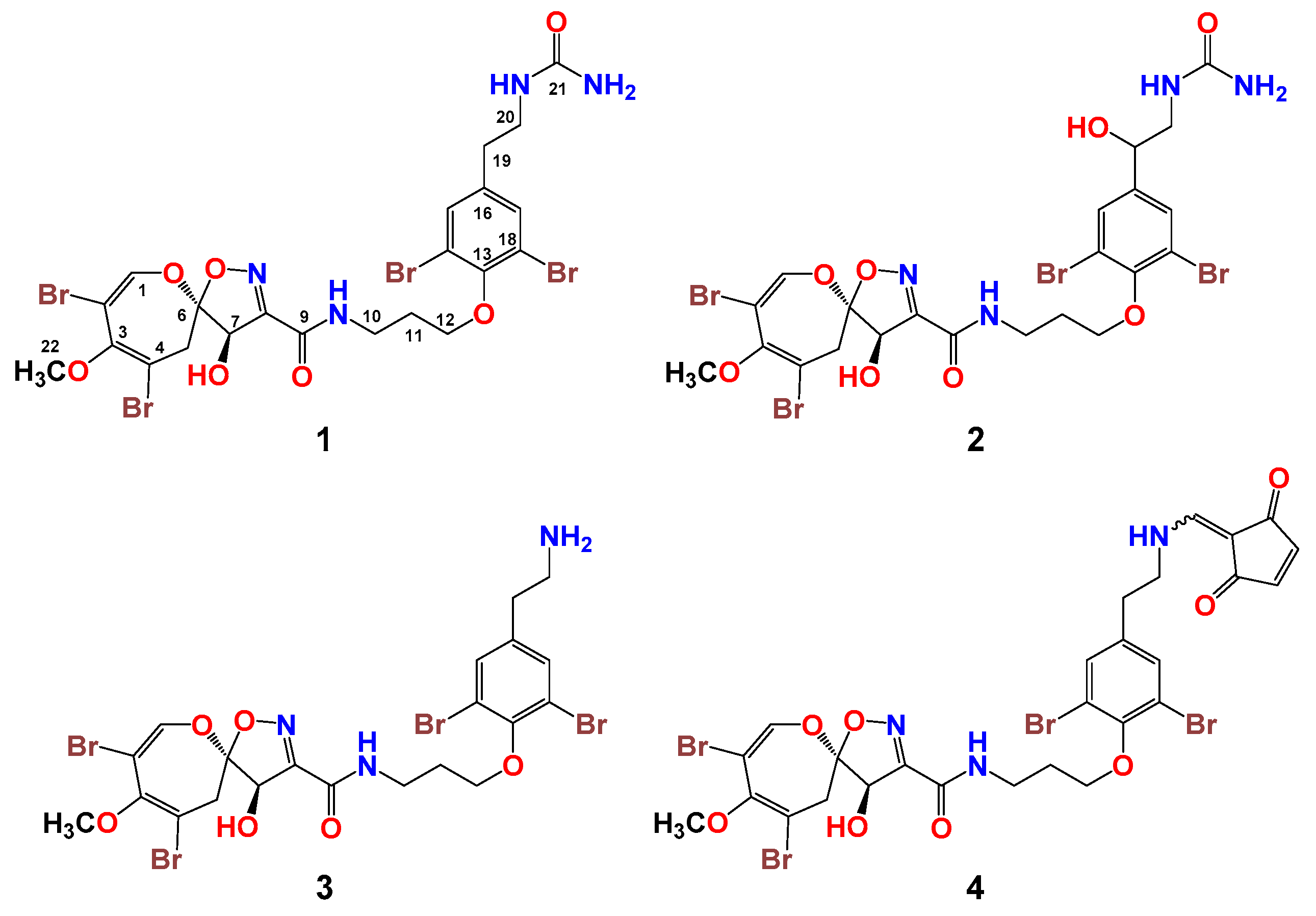
2.1. Isolation of Compounds 1–4
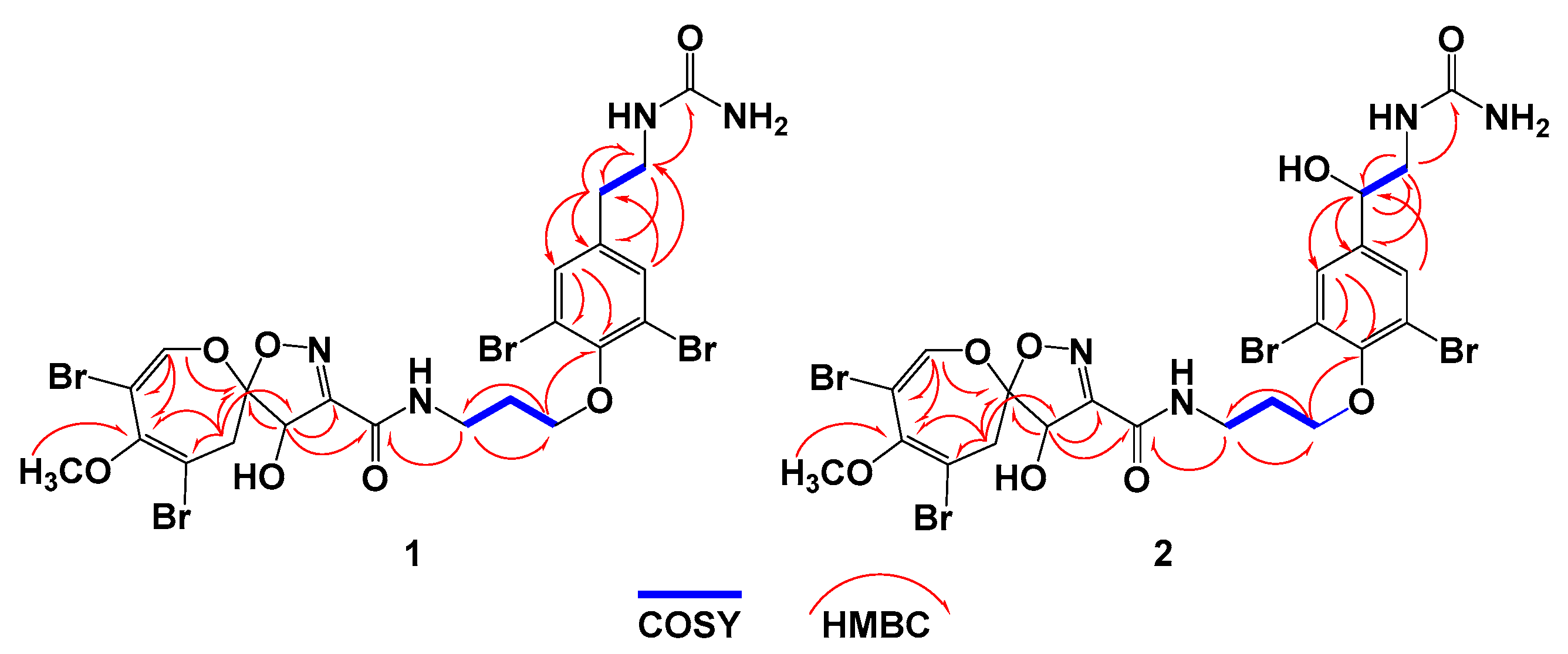
2.2. Structural Determination of Compounds 1–4
2.3. Biological Activities of Compounds 1–4
3. Material and Methods
3.1. General Experimental Procedures
3.2. Biological Materials
3.3. Purification of Compounds 1–4
3.4. Spectral Data of the Compounds
3.5. Cytotoxicity Evaluation of Compounds 1–4
3.5.1. Preparations of Cell Lines and Cell Culture
3.5.2. MTT Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Peng, J.; Li, J.; Hamann, M.T. The marine bromotyrosine derivatives. In The Alkaloids: Chemistry and Biology; Knölker, H.-J., Ed.; Academic Press: San Diego, CA, USA, 2005; Volume 61, pp. 59–262. [Google Scholar]
- Roll, D.M.; Chang, C.W.J.; Scheuer, P.J.; Gray, G.A.; Shoolery, J.N.; Matsumoto, G.K.; Van Duyne, G.D.; Clardy, J. Structure of the psammaplysins. J. Am. Chem. Soc. 1985, 107, 2916–2920. [Google Scholar] [CrossRef]
- Copp, B.R.; Ireland, C.M.; Barrows, L.R. Psammaplysin C: A new cytotoxic dibromotyrosine-derived metabolite from the marine sponge Druinella (= Psammaplysilla) purpurea. J. Nat. Prod. 1992, 55, 822–823. [Google Scholar] [CrossRef] [PubMed]
- Ichiba, T.; Scheuer, P.J.; Kelly-Borges, M. Three bromotyrosine derivatives, one terminating in an unprecedented diketocyclopentenylidene enamine. J. Org. Chem. 1993, 58, 4149–4150. [Google Scholar] [CrossRef]
- Liu, S.; Fu, X.; Schmitz, F.J.; Kelly-Borges, M. Psammaplysin F, a new bromotyrosine derivative from a sponge, Aplysinella sp. J. Nat. Prod. 1997, 60, 614–615. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Andrews, K.T.; Birrell, G.W.; Tran, T.L.; Camp, D.; Davis, R.A.; Quinn, R.J. Psammaplysin H, a new antimalarial bromotyrosine alkaloid from a marine sponge of the genus Pseudoceratina. Bioorg. Med. Chem. Lett. 2011, 21, 846–848. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.D.; Schupp, P.J.; Schrör, J.-P.; Engemann, A.; Rohde, S.; Kelman, D.; Voogd, N.D.; Carroll, A.; Motti, C.A. Twilight zone sponges from Guam yield theonellin isocyanate and psammaplysins I and J. J. Nat. Prod. 2012, 75, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Mudianta, I.W.; Skinner-Adams, T.; Andrews, K.T.; Davis, R.A.; Hadi, T.A.; Hayes, P.Y.; Garson, M.J. Psammaplysin derivatives from the Balinese marine sponge Aplysinella strongylata. J. Nat. Prod. 2012, 75, 2132–2143. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Han, S.; Lee, H.S.; Kang, J.S.; Yun, J.; Sim, C.J.; Shin, H.J.; Lee, J.S. Cytotoxic psammaplysin analogues from a Suberea sp. marine sponge and the role of the spirooxepinisoxazoline in their activity. J. Nat. Prod. 2013, 76, 1731–1736. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. Ceratinamides A and B: New antifouling dibromotyrosine derivatives from the marine sponge Pseudoceratina Purpurea. Tetrahedron 1996, 52, 8181–8186. [Google Scholar] [CrossRef]
- Yang, X.; Davis, R.A.; Buchanan, M.S.; Duffy, S.; Avery, V.M.; Camp, D.; Quinn, R.J. Antimalarial bromotyrosine derivatives from the Australian marine sponge Hyattella sp. J. Nat. Prod. 2010, 73, 985–987. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, S.I.; Ohno, T.; Hokari, R.; Ishiyama, A.; Iwatsuki, M.; Ōmura, S.; Kobayashi, J.; Kubota, T. Ceratinadins E and F, new bromotyrosine alkaloids from an Okinawan marine sponge Pseudoceratina sp. Mar. Drugs. 2018, 16, 463. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.H.; Li, J.; Zhang, M.M.; Cui, J.; Gui, Y.H.; Zhang, Y.; Li, J.Y.; Liu, K.C.; Lin, H.W. Frondoplysins A and B, unprecedented terpene-alkaloid bioconjugates from Dysidea frondosa. Org. Lett. 2019, 21, 6190–6193. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.T.; Scheuer, P.J.; Kelly-Borges, M. Biogenetically diverse, bioactive constituents of a sponge, order Verongida: Bromotyramines and sesquiterpene-shikimate derived metabolites. J. Org. Chem. 1993, 58, 6565–6569. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Sulaiman, M.; Khedr, A.; El Sayed, K.A. Bioactive alkaloids from the Red Sea marine Verongid sponge Pseudoceratina arabica. Tetrahedron 2015, 71, 7837–7841. [Google Scholar] [CrossRef]
- Mándi, A.; Mudianta, I.W.; Kurtán, T.; Garson, M.J. Absolute configuration and conformational study of psammaplysins A and B from the Balinese marine sponge Aplysinella strongylata. J. Nat. Prod. 2015, 78, 2051–2056. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, D.M.; Amirul, I.M.; Turnbull, L.; Davis, R.A.; Whitchurch, C.B.; McAlpine, S.R. Psammaplysin F: A unique inhibitor of bacterial chromosomal partitioning. Bioorg. Med. Chem. Lett. 2013, 23, 4862–4866. [Google Scholar] [CrossRef] [PubMed]
- Christen, K.E.; Davis, R.A.; Kennedy, D. Psammaplysin F increases the efficacy of bortezomib and sorafenib through regulation of stress granule formation. Int. J. Biochem. Cell. Biol. 2019, 112, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Youssef, D.T.A.; Mooberry, S.L. Hurghadolide A and swinholide I, potent actin-microfilament disrupters from the Red Sea sponge Theonella swinhoei. J. Nat. Prod. 2006, 69, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Youssef, D.T.A.; Shaala, L.A.; Mohamed, G.A.; Badr, J.M.; Bamanie, F.H.; Ibrahim, S.R. Theonellamide G, a potent antifungal and cytotoxic bicyclic glycopeptide from the Red Sea marine sponge Theonella swinhoei. Mar. Drugs. 2014, 12, 1911–1923. [Google Scholar] [CrossRef] [PubMed]
| Position | δC (mult.) * | δH (mult., J (Hz)) | HMBC |
|---|---|---|---|
| 1 | 146.9, CH | 7.16 (1H, s) | C-2, C-3, C-6 |
| 2 | 104.5, qC | ||
| 3 | 150.0, qC | ||
| 4 | 104.6, qC | ||
| 5a 5b | 38.4, CH2 | 3.38 (1H, d, 16.0) 3.08 (1H, d, 16.0) | C-3, C-4, C-6 C-3, C-4, C-6 |
| 6 | 121.0, qC | ||
| 7 | 80.5, CH | 5.00 (1H, s) | C-6, C-8, C-9 |
| 8 | 159.0, qC | ||
| 9 | 160.8, qC | ||
| 10 | 38.1, CH2 | 3.64 (2H, t, 6.5) | C-9, C-11, C-12 |
| 11 | 30.7, CH2 | 2.15 (2H, quin., 6.5) | C-10, C-12 |
| 12 | 72.2, CH2 | 4.08 (2H, t, 6.5) | C-10, C-11, C-13 |
| 13 | 153.0, qC | ||
| 14 | 119.1, qC | ||
| 15 | 134.4, CH | 7.49 (1H, s) | C-13, C-14, C-18, C-19 |
| 16 | 139.8, qC | ||
| 17 | 134.4, CH | 7.49 (1H, s) | C-13, C-14, C-18, C-19 |
| 18 | 119.1, qC | ||
| 19 | 35.1, CH2 | 2.78 (2H, t, 6.6) | C-16, C-17, C-20 |
| 20 | 40.1, CH2 | 3.46 (2H, t, 6.6) | C-16, C-19, C-21 |
| 21 | 161.6, qC | ||
| 22 | 59.4, CH3 | 3.67 (3H, s) | C-3 |
| Position | δC (mult.) * | δH (mult., J (Hz)) | HMBC |
|---|---|---|---|
| 1 | 146.8, CH | 7.14 (1H, s) | C-2, C-3, C-6 |
| 2 | 104.2, qC | ||
| 3 | 149.8, qC | ||
| 4 | 104.5, qC | ||
| 5a 5b | 38.2, CH2 | 3.38 (1H, d, 16.0) 3.05 (1H, d, 16.0) | C-3, C-4, C-6 C-3, C-4, C-6 |
| 6 | 120.8, qC | ||
| 7 | 80.4, CH | 4.97 (1H, s) | C-6, C-8, C-9 |
| 8 | 158.8, qC | ||
| 9 | 160.7, qC | ||
| 10 | 37.9, CH2 | 3.61 (2H, t, 6.5) | C-9, C-11, C-12 |
| 11 | 30.5, CH2 | 2.12 (2H, quin., 6.5) | C-10, C-12 |
| 12 | 72.0, CH2 | 4.06 (2H, t, 6.5) | C-10, C-11, C-13 |
| 13 | 153.5, qC | ||
| 14 | 119.2, qC | ||
| 15 | 131.7, CH | 7.60 (1H, s) | C-13, C-14, C-18, C-19 |
| 16 | 143.2, qC | ||
| 17 | 131.7, CH | 7.60 (1H, s) | C-13, C-14, C-18, C-19 |
| 18 | 119.2, qC | ||
| 19 | 71.5, CH | 4.69 (1H, dd, 9.5, 3.0) | C-15, C-16, C-17, C-20 |
| 20a 20b | 46.2, CH2 | 3.47 (1H, dd, 13.0, 3.0) 3.31 (1H, m) | C-16, C-19, C-21 C-16, C-19, C-21 |
| 21 | 161.2, qC | ||
| 22 | 59.3, CH3 | 3.63 (3H, s) | C-3 |
| Compound | IC50 (μM) | ||
|---|---|---|---|
| MDA-MB-231 | HeLa | HCT 116 | |
| 1 | 19.4 ± 1.80 | 22.2 ± 2.0 | 8.2 ± 0.72 |
| 2 | 13.2 ± 0.45 | 17.6 ± 1.90 | 7.0 ± 0.65 |
| 3 | 3.90 ± 0.20 | 8.50 ± 0.81 | 5.1 ± 0.41 |
| 4 | 0.29 ± 0.05 | 2.10 ± 0.12 | 3.7 ± 0.31 |
| 5-fluorouracil (5-FU) * | 13.0 ± 0.30 | 12.3 ± 0.25 | 4.6 ± 0.23 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaala, L.A.; Youssef, D.T.A. Cytotoxic Psammaplysin Analogues from the Verongid Red Sea Sponge Aplysinella Species. Biomolecules 2019, 9, 841. https://doi.org/10.3390/biom9120841
Shaala LA, Youssef DTA. Cytotoxic Psammaplysin Analogues from the Verongid Red Sea Sponge Aplysinella Species. Biomolecules. 2019; 9(12):841. https://doi.org/10.3390/biom9120841
Chicago/Turabian StyleShaala, Lamiaa A., and Diaa T. A. Youssef. 2019. "Cytotoxic Psammaplysin Analogues from the Verongid Red Sea Sponge Aplysinella Species" Biomolecules 9, no. 12: 841. https://doi.org/10.3390/biom9120841
APA StyleShaala, L. A., & Youssef, D. T. A. (2019). Cytotoxic Psammaplysin Analogues from the Verongid Red Sea Sponge Aplysinella Species. Biomolecules, 9(12), 841. https://doi.org/10.3390/biom9120841





