Transcriptomic Analysis Reveals the Wound Healing Activity of Mussel Myticin C
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Transcriptomic Experimental Approach
2.3. Bioinformatics: Assembly, RNA-Seq, and Annotation
2.4. Hemocyte Time-Lapse Microscopy and Morphological Analysis
2.5. Histological and Immunofluorescence Assays
2.6. Western Blot of Myticin C
2.7. Wound Healing and Tail Fin Regeneration Assays
3. Results
3.1. Assembly and Annotation of Hemocyte Transcriptome
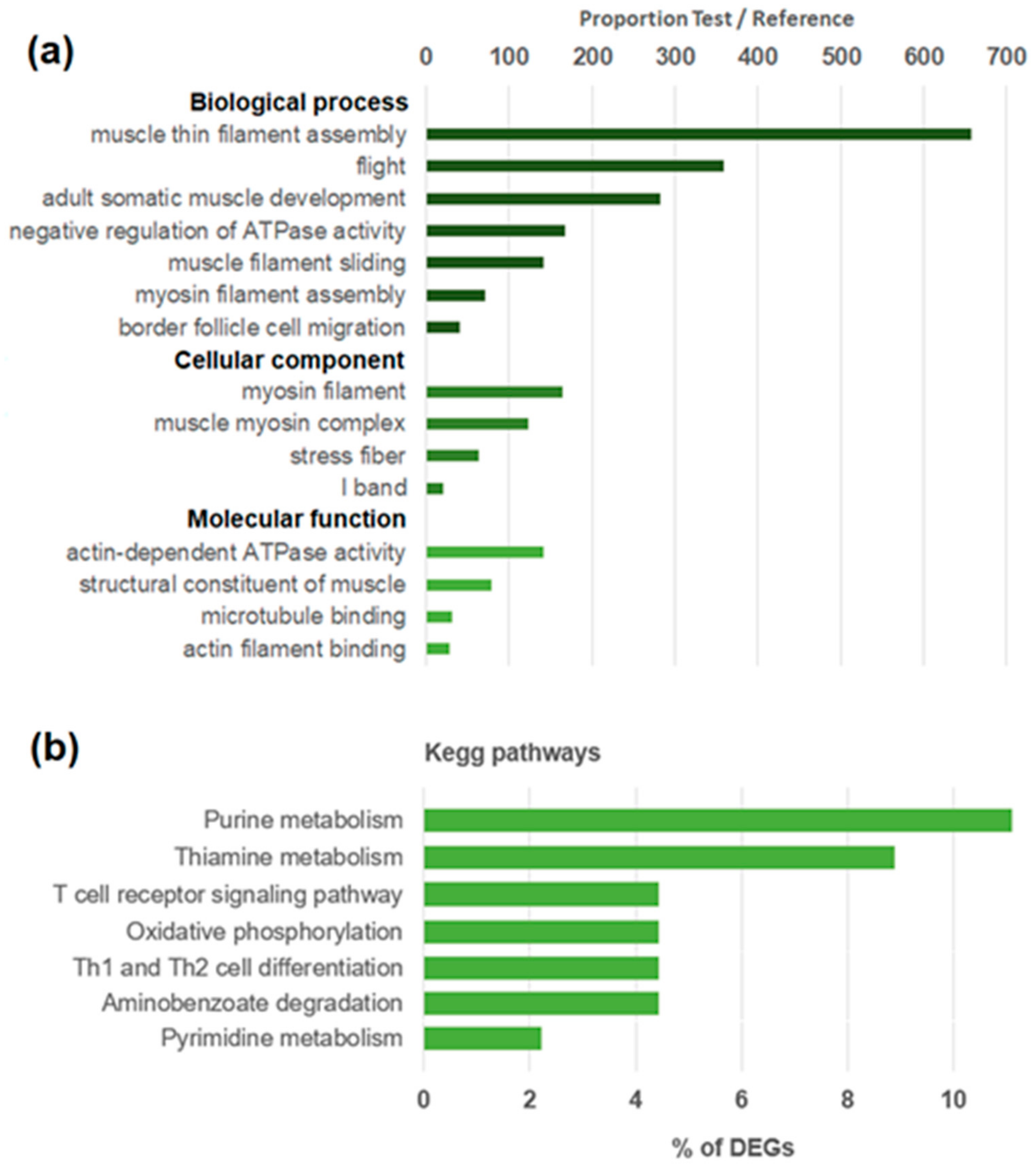
3.2. Hemocyte Transcriptomic Response after a Myticin C Treatment
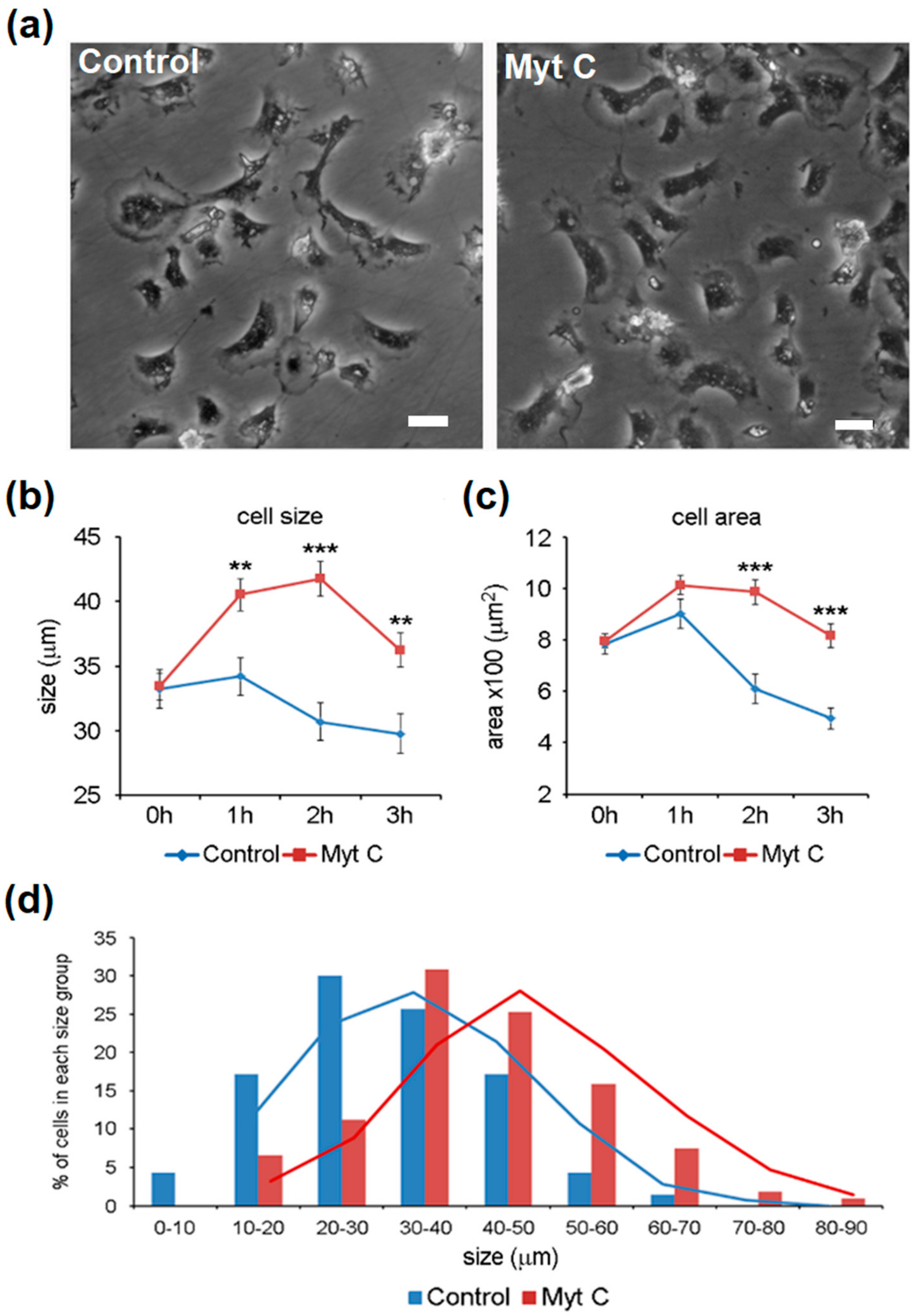
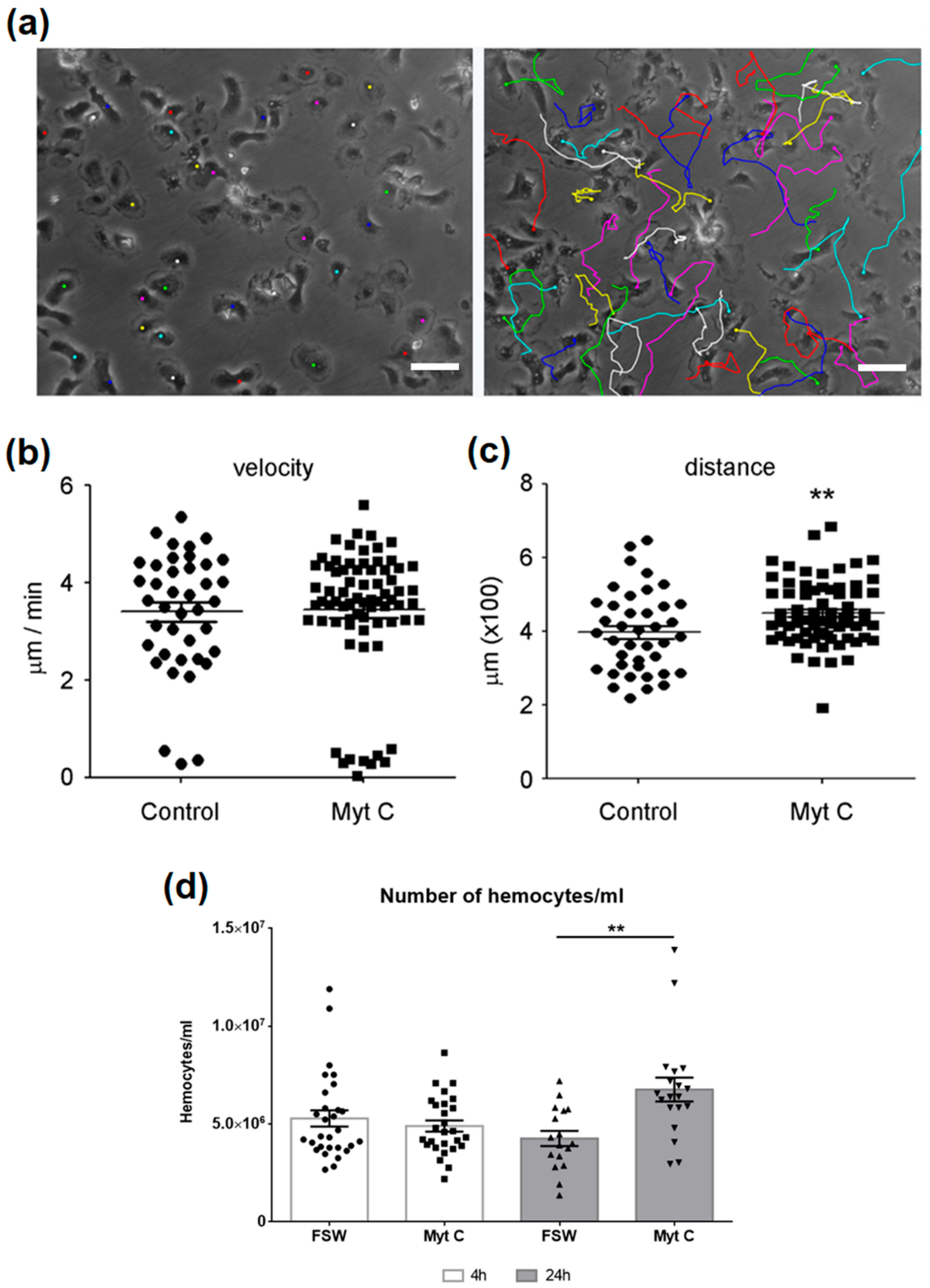
3.3. Changes in Hemocytes Induced by Myticin C (Morphology, Mobility, and Number)
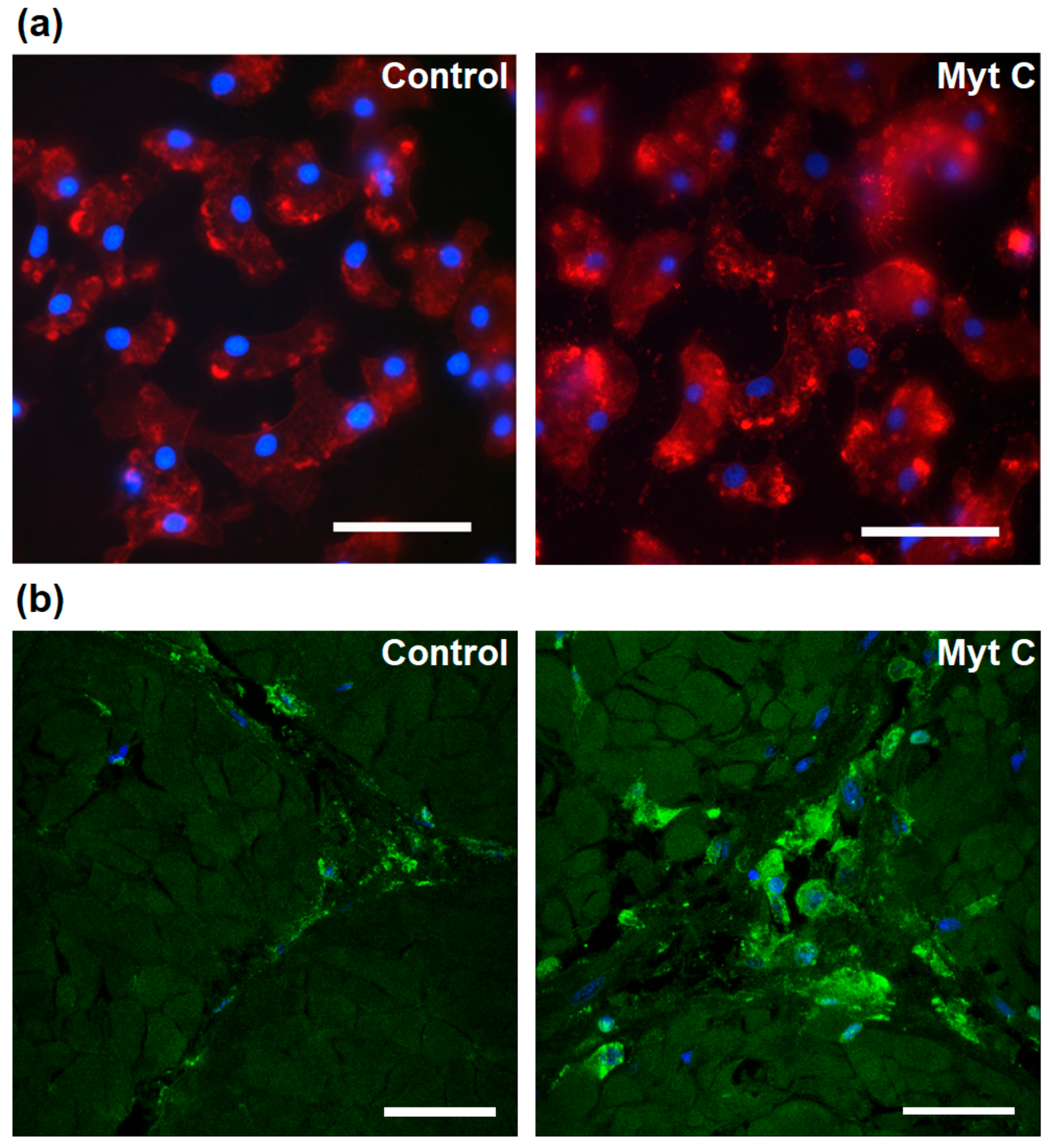
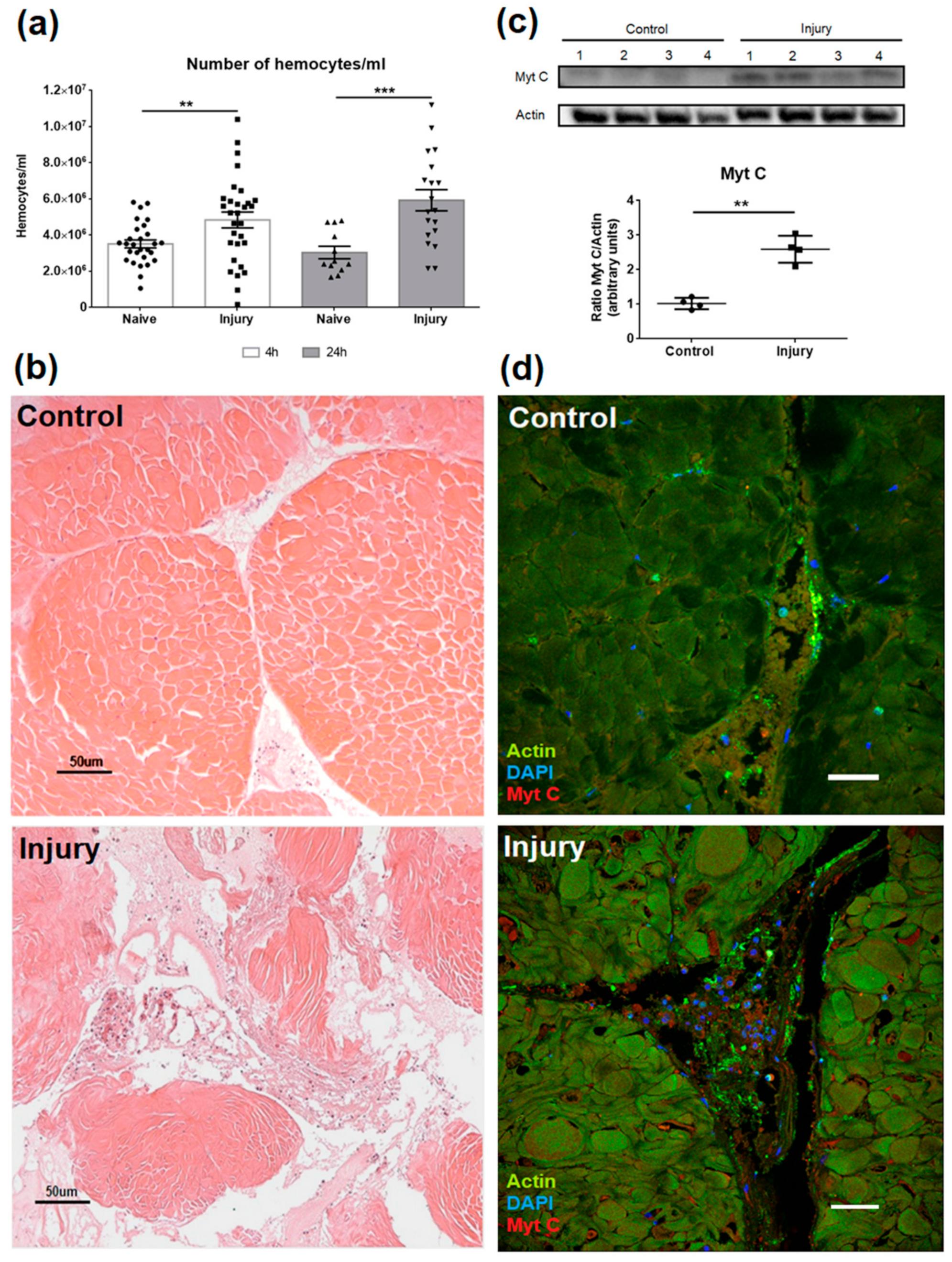
3.4. Effect of Myticin C on Injured Tissues
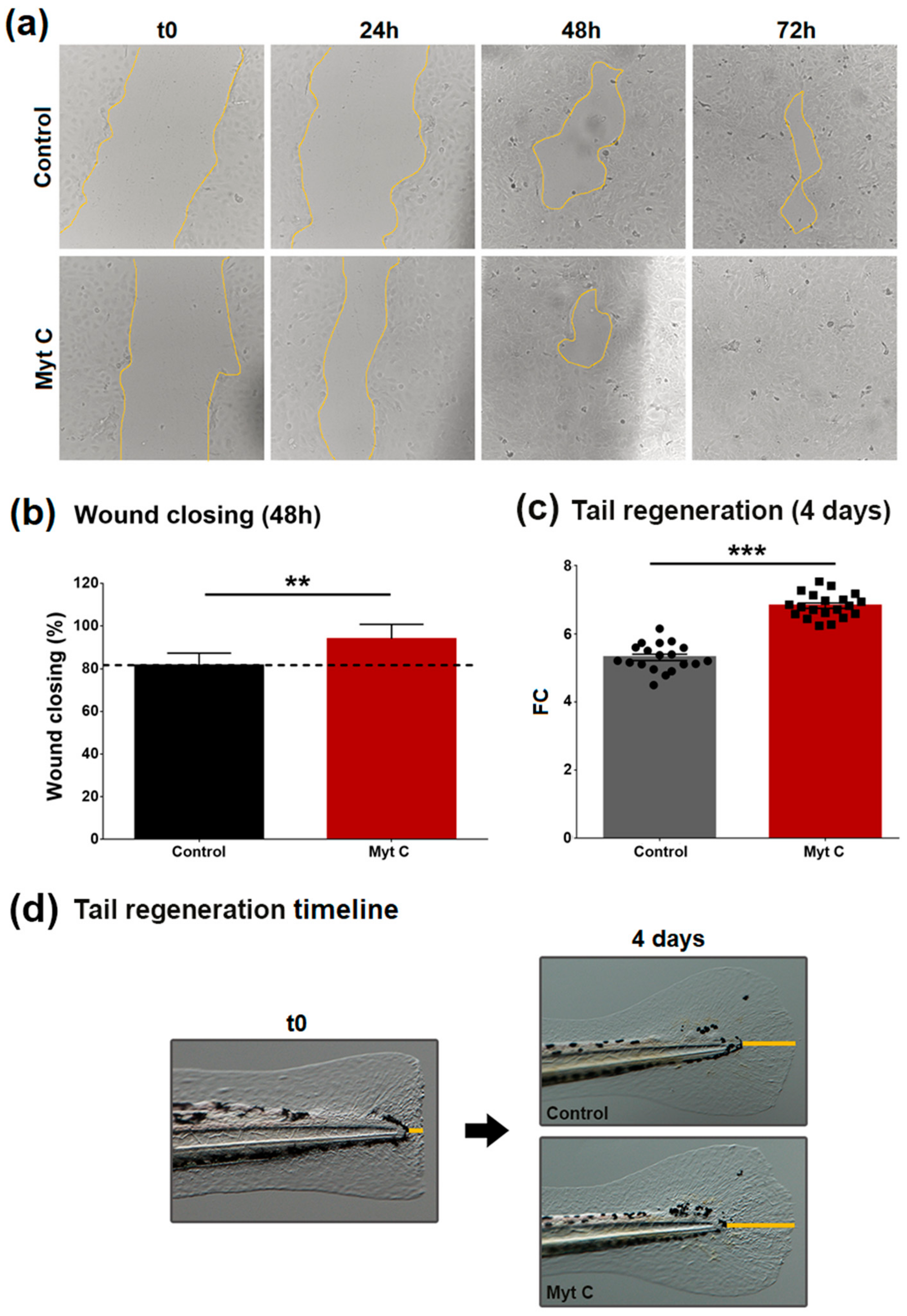
3.5. Effect of Myticin C on Human Keratinocytes, Wound Healing, and Zebrafish Tail Fin Regeneration
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. FAO Home Page. Available online: http://www.fao.org/home/en/ (accessed on 6 December 2019).
- Farrington, J.W.; Tripp, B.W.; Tanabe, S.; Subramanian, A.; Sericano, J.L.; Wade, T.L.; Knap, A.H.; Edward, D. Goldberg’s proposal of “the Mussel Watch”: Reflections after 40 years. Mar. Pollut. Bull. 2016, 110, 501–510. [Google Scholar] [CrossRef]
- Gardner, J.P.; Zbawicka, M.; Westfall, K.M.; Wenne, R. Invasive blue mussels threaten regional scale genetic diversity in mainland and remote offshore locations: The need for baseline data and enhanced protection in the Southern Ocean. Glob. Chang. Biol. 2016, 22, 3182–3195. [Google Scholar] [CrossRef] [PubMed]
- Figueras, A.; Moreira, R.; Sendra, M.; Novoa, B. Genomics and immunity of the Mediterranean mussel Mytilus galloprovincialis in a changing environment. Fish Shellfish Immunol. 2019, 90, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Costa, M.; Forn-Cuni, G.; Balseiro, P.; Chamorro, R.; Dios, S.; Figueras, A.; Novoa, B. Occurrence, seasonality and infectivity of Vibrio strains in natural populations of mussels Mytilus galloprovincialis. Dis. Aquat. Organ. 2014, 108, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Thébault, A.; Dégremont, L.; Arzul, I.; Miossec, L.; Robert, M.; Chollet, B.; François, C.; Joly, J.P.; Ferrand, S.; et al. Ostreid herpesvirus 1 detection and relationship with Crassostrea gigas spat mortality in France between 1998 and 2006. Vet. Res. 2011, 42, 73. [Google Scholar] [CrossRef]
- Barbosa-Solomieu, V.; Renault, T.; Travers, M.A. Mass mortality in bivalves and the intricate case of the Pacific oyster, Crassostrea gigas. J. Invertebr. Pathol. 2015, 131, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Prado-Alvarez, M.; Gestal, C.; Li, H.; Roch, P.; Novoa, B.; Figueras, A. Functional and molecular immune response of Mediterranean mussel (Mytilus galloprovincialis) haemocytes against pathogen-associated molecular patterns and bacteria. Fish Shellfish Immunol. 2009, 26, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Mitta, G.; Vandenbulckeb, F.; Rocha, P. Original involvement of antimicrobial peptides in mussel innate immunity. FEBS Lett. 2000, 486, 185–190. [Google Scholar] [CrossRef]
- Novoa, B.; Romero, A.; Álvarez, Á.L.; Moreira, R.; Pereiro, P.; Costa, M.M.; Dios, S.; Estepa, A.; Parra, F.; Figueras, A. Antiviral activity of myticin C peptide from mussel: An ancient defense against herpesviruses. J. Virol. 2016, 90, 7692–7702. [Google Scholar] [CrossRef]
- Hubert, F.; Noel, T.; Roch, P. A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis). Eur. J. Biochem. 1996, 240, 302–306. [Google Scholar] [CrossRef]
- Mitta, G.; Hubert, F.; Noël, T.; Roch, P. Myticin, a novel cysteine-rich antimicrobial peptide isolated from haemocytes and plasma of the mussel Mytilus galloprovincialis. Eur. J. Biochem. 1999, 265, 71–78. [Google Scholar] [CrossRef]
- Mitta, G.; Vandenbulcke, F.; Hubert, F.; Salzet, M.; Roch, P. Involvement of mytilins in mussel antimicrobial defense. J. Biol. Chem. 2000, 275, 12954–12962. [Google Scholar] [CrossRef] [PubMed]
- Sonthi, M.; Toubiana, M.; Pallavicini, A.; Venier, P.; Roch, P. Diversity of coding sequences and gene structures of the antifungal peptide mytimycin (MytM) from the Mediterranean mussel, Mytilus galloprovincialis. Mar. Biotechnol. 2011, 13, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; De Moro, G.; Manfrin, C.; Venier, P.; Pallavicini, A. Big defensins and mytimacins, new AMP families of the Mediterranean mussel Mytilus galloprovincialis. Dev. Comp. Immunol. 2012, 36, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Wang, X.C.; Liu, H.H.; Fan, M.H.; Sun, J.J.; Shen, W. Molecular characterization of a novel antimicrobial peptide from Mytilus coruscus. Fish Shellfish Immunol. 2013, 34, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.L.; Huang, W.; Zhou, S.Q.; Wang, X.C.; Liu, H.H.; Fan, M.H.; Wang, R.X.; Gao, P.; Liao, Z. Characterization of a novel antimicrobial peptide with chitin-biding domain from Mytilus coruscus. Fish Shellfish Immunol. 2014, 41, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Leoni, G.; De Poli, A.; Mardirossian, M.; Gambato, S.; Florian, F.; Venier, P.; Wilson, D.N.; Tossi, A.; Pallavicini, A.; Gerdol, M. Myticalins: A novel multigenic family of linear, cationic antimicrobial peptides from marine mussels (Mytilus spp.). Mar. Drugs 2017, 15, 261. [Google Scholar] [CrossRef]
- Pallavicini, A.; Costa, M.M.; Gestal, C.; Dreos, R.; Figueras, A.; Venier, P.; Novoa, B. High sequence variability of myticin transcripts in hemocytes of immune-stimulated mussels suggests ancient host-pathogen interactions. Dev. Comp. Immunol. 2008, 32, 213–226. [Google Scholar] [CrossRef]
- Costa, M.M.; Dios, S.; Alonso-Gutierrez, J.; Romero, A.; Novoa, B.; Figueras, A. Evidence of high individual diversity on myticin C in mussel (Mytilus galloprovincialis). Dev. Comp. Immunol. 2009, 33, 162–170. [Google Scholar] [CrossRef]
- Balseiro, P.; Falcó, A.; Romero, A.; Dios, S.; Martínez-López, A.; Figueras, A.; Estepa, A.; Novoa, B. Mytilus galloprovincialis myticin C: A chemotactic molecule with antiviral activity and immunoregulatory properties. PLoS ONE 2011, 6, e23140. [Google Scholar] [CrossRef]
- Rey-Campos, M.; Moreira, R.; Valenzuela-Muñoz, V.; Gallardo-Escárate, C.; Novoa, B.; Figueras, A. High individual variability in the transcriptomic response of Mediterranean mussels to Vibrio reveals the involvement of myticins in tissue injury. Sci. Rep. 2019, 9, 3569. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, A.; Encinar, J.A.; Medina-Gali, R.M.; Balseiro, P.; Garcia-Valtanen, P.; Figueras, A.; Novoa, B.; Estepa, A. pH-dependent solution structure and activity of a reduced form of the host-defense peptide myticin C (Myt C) from the mussel Mytilus galloprovincialis. Mar. Drugs 2013, 11, 2328–2346. [Google Scholar] [CrossRef] [PubMed]
- Qiagen. Quiagen Digital Insights Home Page. Available online: http://www.clcbio.com (accessed on 6 December 2019).
- Robinson, M.D.; Smyth, G.K. Small-sample estimation of negative binomial dispersion, with applications to sage data. Biostatistics 2008, 9, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ozato, K.; Shin, D.M.; Chang, T.H.; Morse, H.C., 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008, 8, 849–860. [Google Scholar] [CrossRef]
- Mende, K.; Petoukhova, O.; Koulitchkova, V.; Schaub, G.A.; Lange, U.; Kaufmann, R.; Nowak, G. Dipetalogastin, a potent thrombin inhibitor from the blood-sucking insect Dipetalogaster maximus cDNA cloning, expression and characterization. Eur. J. Biochem. 1999, 266, 583–590. [Google Scholar] [CrossRef]
- Mende, K.; Lange, U.; Nowak, G. Three recombinant serine proteinase inhibitors expressed from the coding region of the thrombin inhibitor dipetalogastin. Insect Biochem. Mol. Biol. 2004, 34, 971–979. [Google Scholar] [CrossRef]
- Bazzi, Z.A.; Lanoue, D.; El-Youssef, M.; Romagnuolo, R.; Tubman, J.; Cavallo-Medved, D.; Porter, L.A.; Boffa, M.B. Activated thrombin-activatable fibrinolysis inhibitor (TAFIα) attenuates breast cancer cell metastatic behaviors through inhibition of plasminogen activation and extracellular proteolysis. BMC Cancer 2016, 16, 328. [Google Scholar] [CrossRef]
- Velle, K.B.; Fritz-Laylin, L.K. Diversity and evolution of actin-dependent phenotypes. Curr. Opin. Genet. Dev. 2019, 58–59, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Gimona, M.; Kaverina, I.; Resch, G.P.; Vignal, E.; Burgstaller, G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell 2003, 14, 2482–2491. [Google Scholar] [CrossRef] [PubMed]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Myosin: The actin motor protein. In Molecular Cell Biology, 4th ed.; Freeman, W.H., Ed.; EEUU: New York, NY, USA, 2000; Volume 29, pp. 126–133. [Google Scholar]
- Hankeln, T.; Wystub, S.; Laufs, T.; Schmidt, M.; Gerlach, F.; Saaler-Reinhardt, S.; Reuss, S.; Burmester, T. The cellular and subcellular localization of neuroglobin and cytoglobin—A clue to their function? IUBMB Life 2004, 56, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Timón-Gómez, A.; Nývltová, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef]
- Shellard, A.; Mayor, R. Chemotaxis during neural crest migration. Semin. Cell Dev. Biol. 2016, 55, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, H.Y. Lung inflammation: Role of endogenous chemotactic factors in attracting polymorphonuclear granulocytes. Am. Rev. Respir. Dis. 1983, 127, 16–25. [Google Scholar]
- Rovin, B.H.; Phan, L.T. Chemotactic factors and renal inflammation. Am. J. Kidney Dis. 1998, 31, 1065–1084. [Google Scholar] [CrossRef]
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar] [CrossRef]
- Collins, C.; Nelson, W.J. Running with neighbors: Coordinating cell migration and cell-cell adhesion. Curr. Opin. Cell Biol. 2015, 36, 62–70. [Google Scholar] [CrossRef]
- Barriga, E.H.; Mayor, R. Embryonic cell-cell adhesion: A key player in collective neural crest migration. Curr. Top. Dev. Biol. 2015, 112, 301–323. [Google Scholar]
- Huse, M. Mechanical forces in the immune system. Nat. Rev. Immunol. 2017, 17, 679–690. [Google Scholar] [CrossRef]
- Bellavia, G.; Fasanaro, P.; Melchionna, R.; Capogrossi, M.C.; Napolitano, M. Transcriptional control of skin reepithelialization. J. Dermatol. Sci. 2014, 73, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Kasuya, A.; Tokura, Y. Attempts to accelerate wound healing. J. Dermatol. Sci. 2014, 76, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Gosain, A.; DiPietro, L.A. Aging and wound healing. World J. Surg. 2004, 28, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Ishida, Y. Molecular pathology of wound healing. Forensic. Sci. Int. 2010, 203, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.G.; Wu, X.; Guan, J.L. Wound-healing assay. Methods Mol. Biol. 2005, 294, 23–29. [Google Scholar]
- Jonkman, J.E.; Cathcart, J.A.; Xu, F.; Bartolini, M.E.; Amon, J.E.; Stevens, K.M.; Colarusso, P. An introduction to the wound healing assay using live-cell microscopy. Cell Adh. Migr. 2014, 8, 440–451. [Google Scholar] [CrossRef]
- Bilgi, A.; Zumrut Biber Muftuler, F.; Akman, L.; Ilker Medine, E.; Tonbaklar Bilgi, P.; Kozgus Guldu, O.; Goksun Gokulu, S.; Tekin, V.; Cosan Terek, M. In vitro determination of wound healing potential of axonge. Wounds 2017, 29, 209–214. [Google Scholar]
- Chiu, Y.H.; Ritchlin, C.T. DC-STAMP: A key regulator in osteoclast differentiation. J. Cell Physiol. 2016, 231, 2402–2407. [Google Scholar] [CrossRef]
- Martinez, J.R.; Dhawan, A.; Farach-Carson, M.C. Modular proteoglycan perlecan/HSPG2: Mutations, phenotypes, and functions. Genes 2018, 9, e556. [Google Scholar] [CrossRef]
| Reads Origin | Raw | Trimmed |
| Control 1 | 90,967,138 | 99.53% |
| Control 2 | 84,688,626 | 99.49% |
| Control 3 | 53,800,576 | 94.49% |
| MytC 1 | 73,793,348 | 99.42% |
| MytC 2 | 80,981,306 | 98.79% |
| MytC 3 | 51,346,136 | 98.88% |
| Assembly | ||
| Contigs | 154,093 | |
| Range contig length | 200–16,293 | |
| Average contig length | 509 | |
| N50 | 568 | |
| Blast | ||
| Contigs identified by Uniprot/SwissProt | 30,596 | |
| Contigs identified by molluscs database | 48,876 | |
| GO analysis | ||
| Annotated contigs | 30,416 | |
| KEGG analysis | ||
| Pathway assigned contigs | 9118 | |
| Name | FC | Description |
|---|---|---|
| MytC_contig_121683 | 59.81 | Cytochrome c oxidase subunit 1 |
| MytC_contig_151173 | 14.74 | Cytoglobin-1 |
| MytC_contig_40784 | 7.55 | LKD-rich protein-1 |
| MytC_contig_135551 | 5.87 | Vitelline envelope zona pellucida domain 9 |
| MytC_contig_53155 | 4.97 | Basement membrane-specific heparan sulfate proteoglycan core protein (HSPG2) |
| MytC_contig_137132 | 4.85 | Vitelline envelope zona pellucida domain 9 |
| MytC_contig_73904 | 4.56 | DnaJ homolog subfamily B member 5 (DNAJB5/HSP40) |
| MytC_contig_135670 | 4.31 | Serine protease inhibitor dipetalogastin |
| MytC_contig_99492 | 4.22 | Tripartite motif-containing protein 56 (TRIM56) |
| MytC_contig_136869 | 4.21 | Serine protease inhibitor dipetalogastin |
| MytC_contig_74563 | 4.02 | Sarcoplasmic calcium-binding protein |
| MytC_contig_34823 | 3.62 | RS-rich protein-1 |
| MytC_contig_34824 | 3.50 | RS-rich protein-2 |
| MytC_contig_35853 | 3.26 | Transgelin-like protein-6 |
| MytC_contig_22493 | 3.23 | DC-STAMP domain-containing protein 2 (DCST2) |
| MytC_contig_12073 | 3.07 | Myosin heavy chain |
| MytC_contig_65743 | 3.04 | RS-rich protein-1 |
| MytC_contig_30352 | 2.95 | Calponin-like protein |
| MytC_contig_90861 | 2.93 | LKD-rich protein-1 |
| MytC_contig_60714 | 2.88 | Myosin heavy chain |
| MytC_contig_40783 | 2.83 | LKD-rich protein-1 |
| MytC_contig_25094 | 2.76 | Protein SOGA3 |
| MytC_contig_19629 | 2.70 | Calponin-like protein |
| MytC_contig_268 | 2.69 | Myosin regulatory light chain |
| MytC_contig_18832 | 2.57 | Small heat shock protein 22 |
| MytC_contig_17930 | 2.57 | Nicotinamidase |
| MytC_contig_31755 | 2.49 | RS-rich protein-2 |
| MytC_contig_49773 | 2.46 | Myosin |
| MytC_contig_34500 | 2.29 | DBH-like monooxygenase protein 1 homolog |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey-Campos, M.; Moreira, R.; Romero, A.; Medina-Gali, R.M.; Novoa, B.; Gasset, M.; Figueras, A. Transcriptomic Analysis Reveals the Wound Healing Activity of Mussel Myticin C. Biomolecules 2020, 10, 133. https://doi.org/10.3390/biom10010133
Rey-Campos M, Moreira R, Romero A, Medina-Gali RM, Novoa B, Gasset M, Figueras A. Transcriptomic Analysis Reveals the Wound Healing Activity of Mussel Myticin C. Biomolecules. 2020; 10(1):133. https://doi.org/10.3390/biom10010133
Chicago/Turabian StyleRey-Campos, Magalí, Rebeca Moreira, Alejandro Romero, Regla M. Medina-Gali, Beatriz Novoa, María Gasset, and Antonio Figueras. 2020. "Transcriptomic Analysis Reveals the Wound Healing Activity of Mussel Myticin C" Biomolecules 10, no. 1: 133. https://doi.org/10.3390/biom10010133
APA StyleRey-Campos, M., Moreira, R., Romero, A., Medina-Gali, R. M., Novoa, B., Gasset, M., & Figueras, A. (2020). Transcriptomic Analysis Reveals the Wound Healing Activity of Mussel Myticin C. Biomolecules, 10(1), 133. https://doi.org/10.3390/biom10010133







