Overexpression of Banana ATG8f Modulates Drought Stress Resistance in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Transformation and Screening of Transgenic Plants
2.2. Plant Materials and Treatments
2.3. RNA Extraction and qRT-PCR
2.4. Determination of ROS and Malondialdehyde (MDA), and Activities of Antioxidant Enzymes
2.5. Measurement of Assimilation Rate
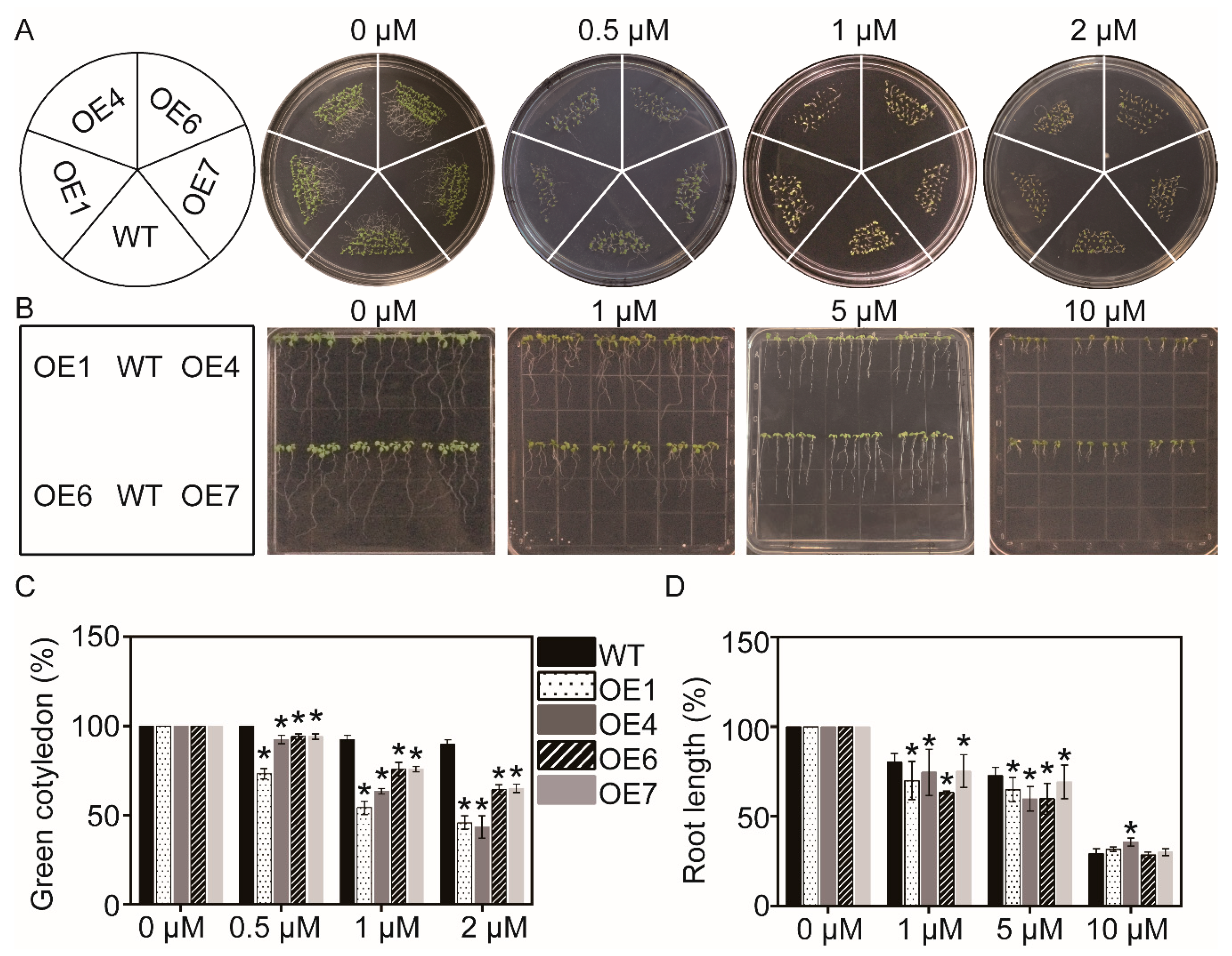
2.6. Analysis of ABA Sensitivity
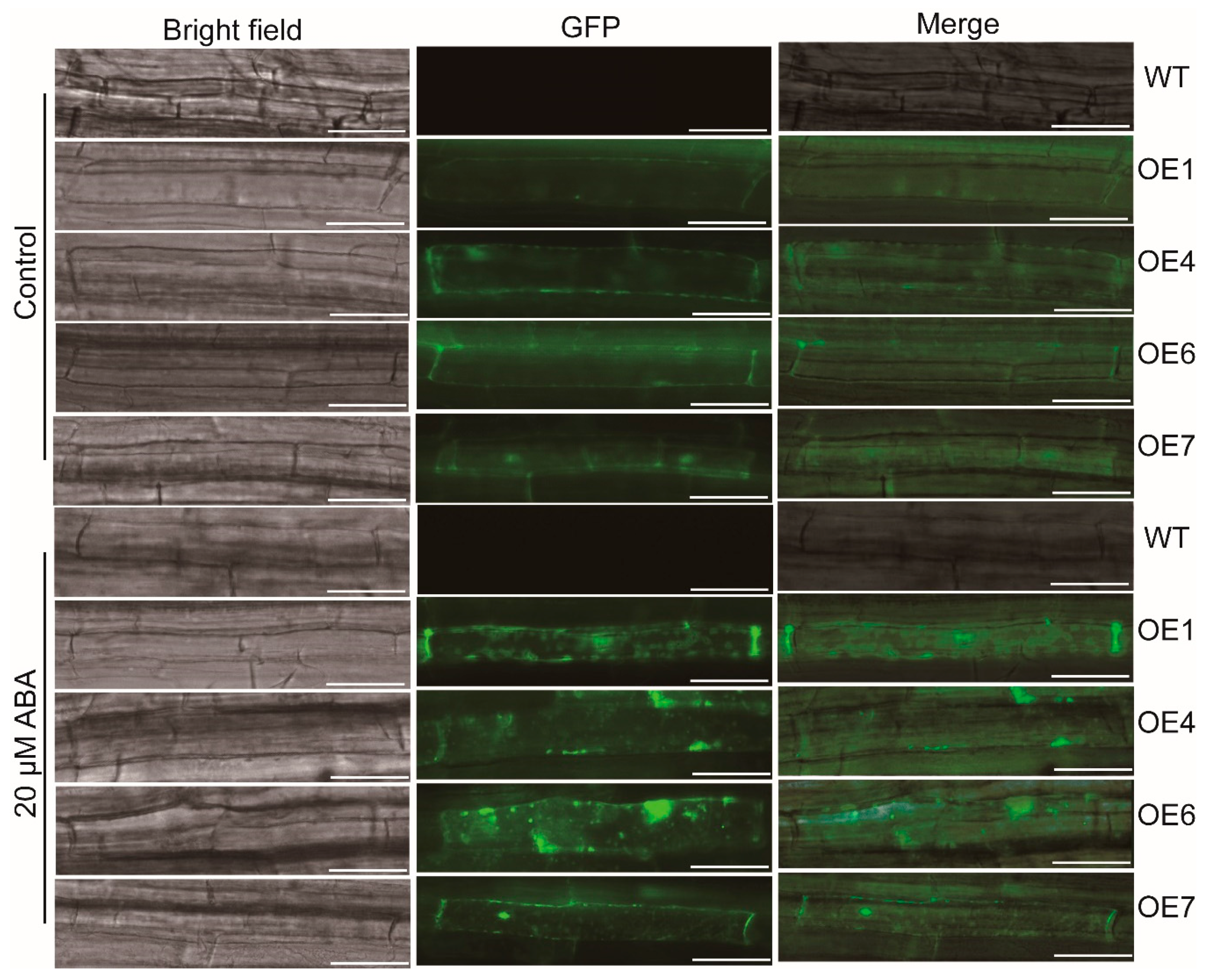
2.7. Observation of Autophagosome
2.8. Statistical Analysis
3. Results
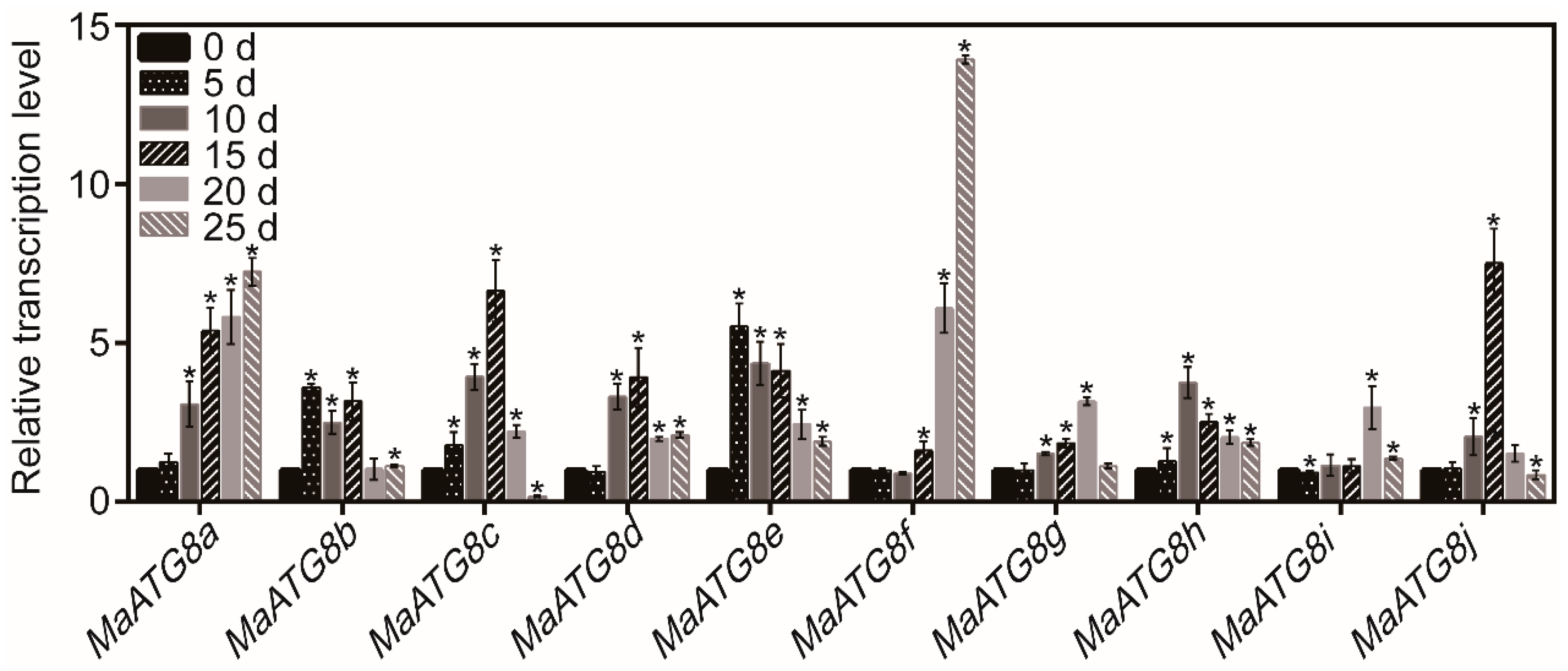
3.1. The Transcript Levels of MaATG8s in Response to Drought Stress
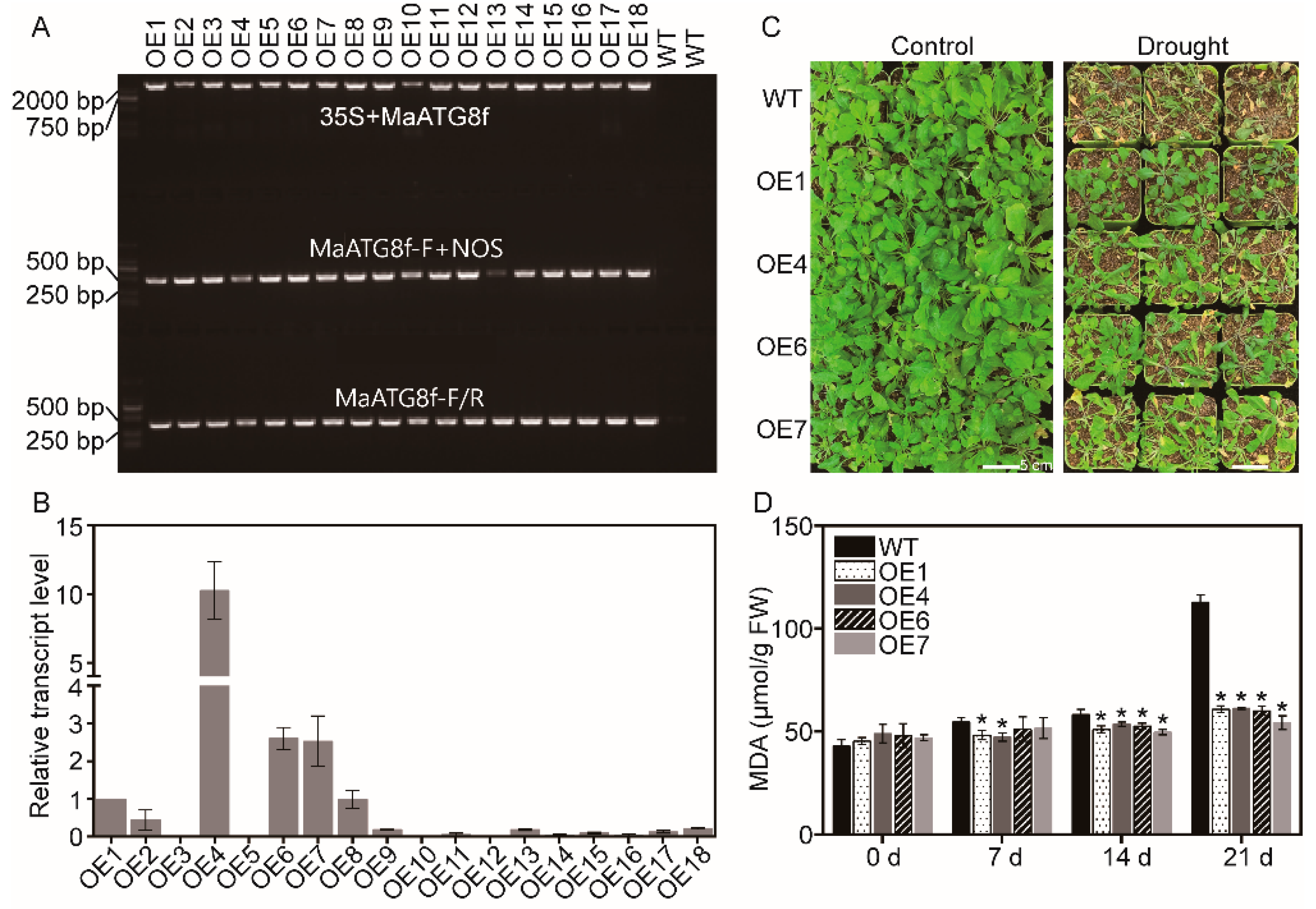
3.2. Overexpression of MaATG8f Increases Drought Stress Resistance in Arabidopsis
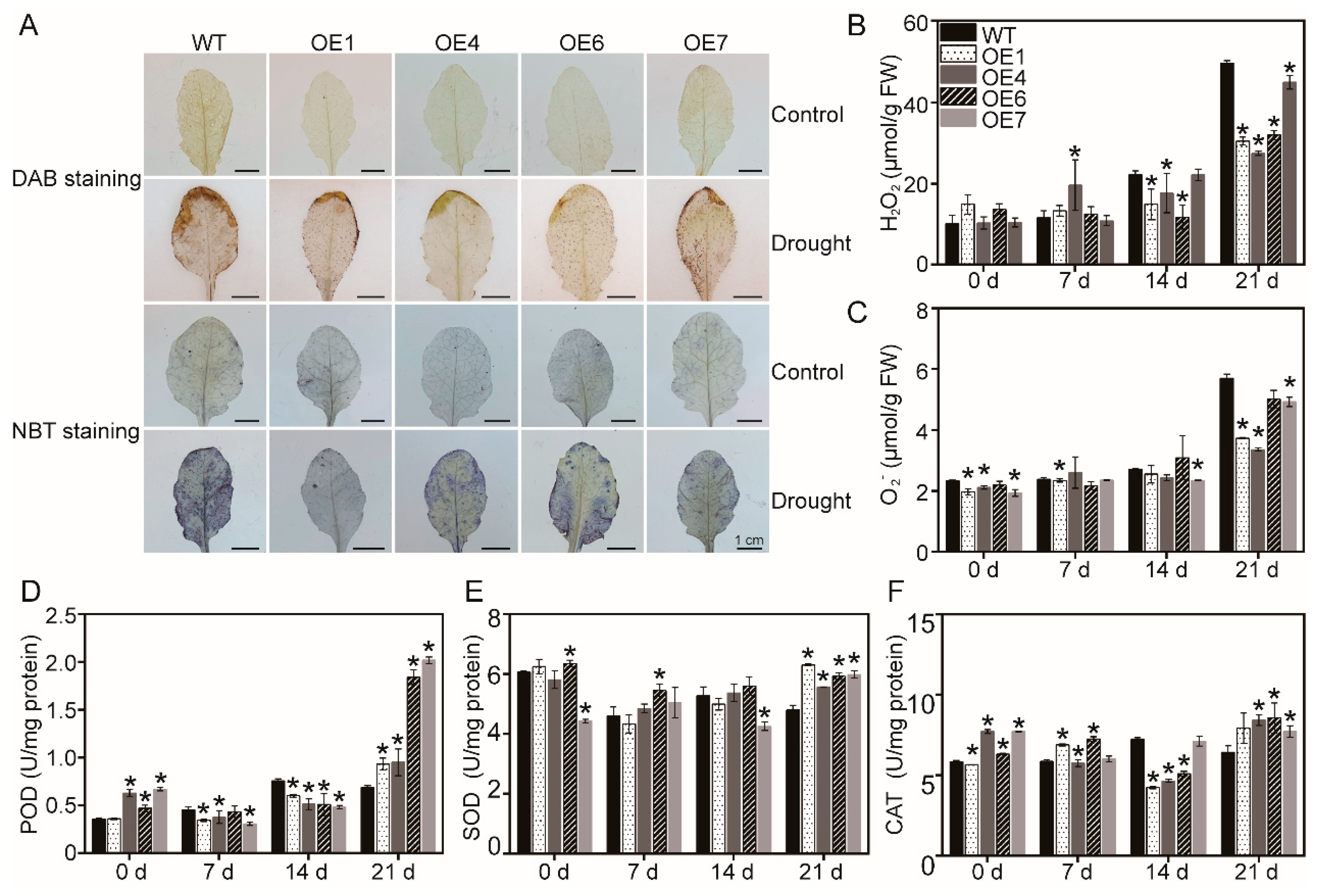
3.3. Determination of ROS and the Activity of Antioxidant Enzymes
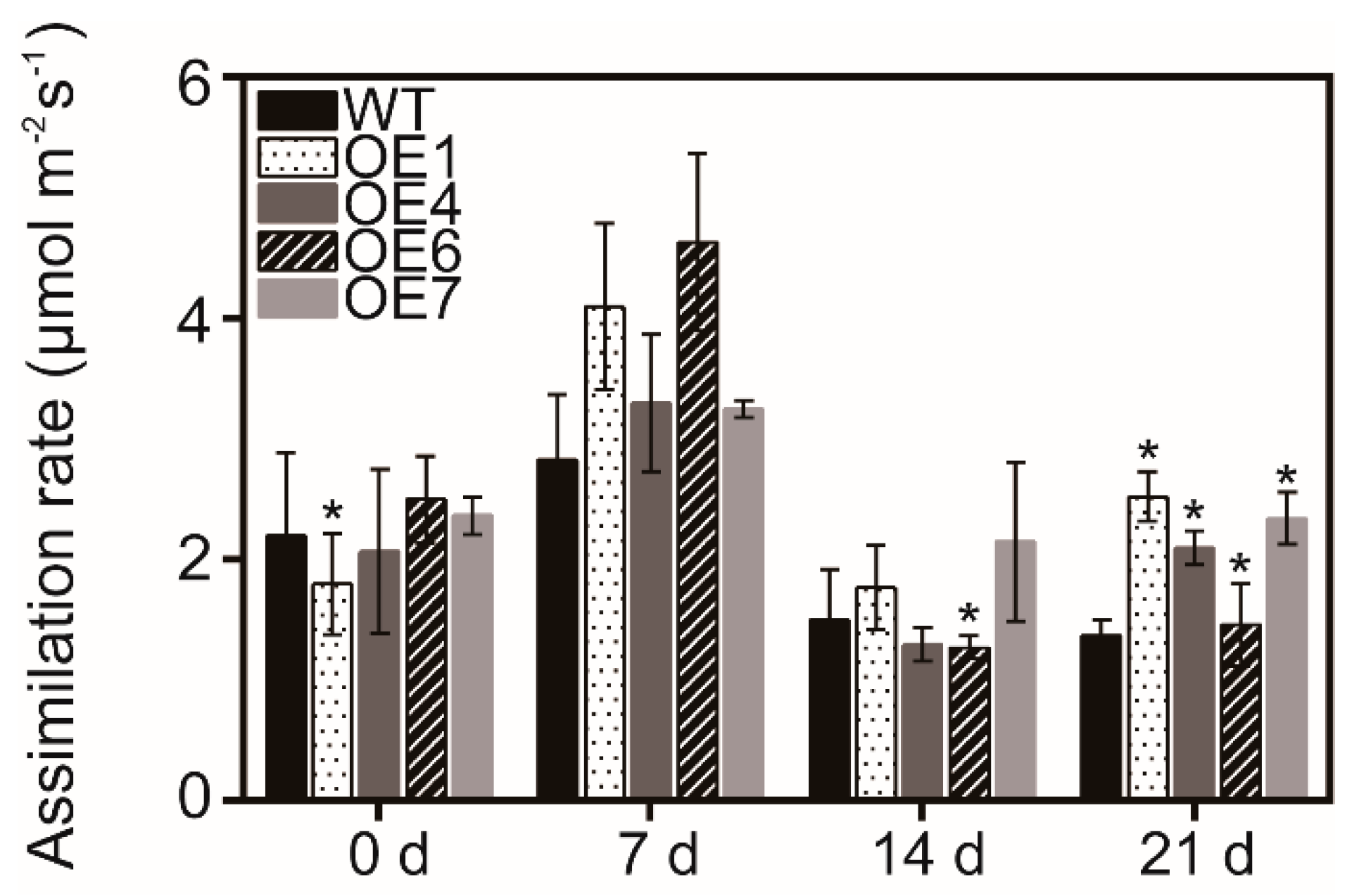
3.4. Overexpression of MaATG8f Promotes Photosynthesis Efficiency
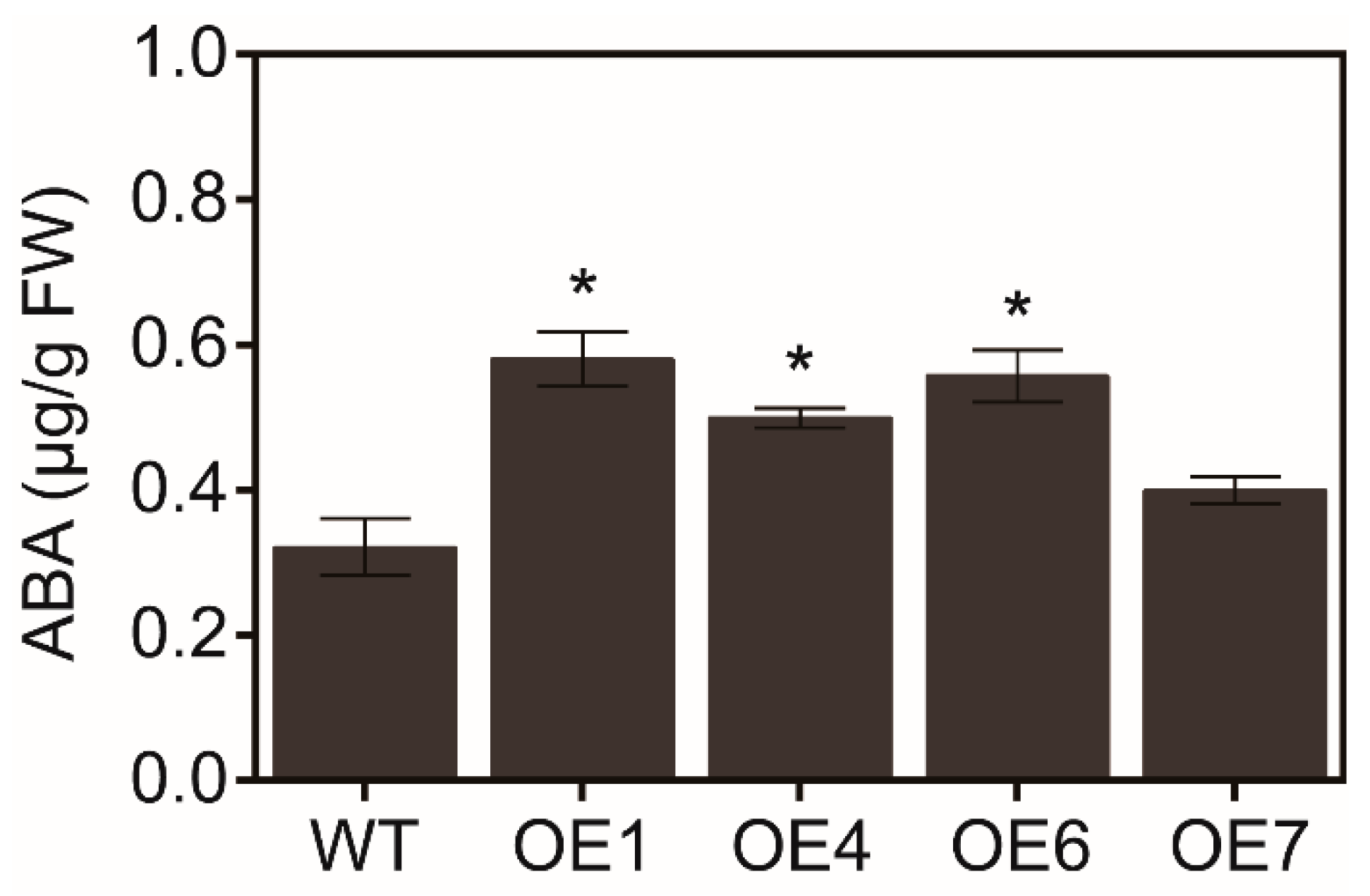
3.5. Overexpression of MaATG8f Increases the Sensitivity to ABA in Arabidopsis
3.6. Overexpression of MaATG8f Promotes the Generation of Autophagosomes byABA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Griffiths, C.A.; Paul, M.J. Targeting carbon for crop yield and drought resilience. J. Sci. Food Agric. 2017, 97, 4663–4671. [Google Scholar] [CrossRef] [PubMed]
- Kaisermann, A.; De Vries, F.T.; Griffiths, R.I.; Bardgett, R.D. Legacy effects of drought on plant-soil feedbacks and plant-plant interactions. New Phytol. 2017, 215, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-Responsive Mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.T. An increase in the “inhibitor-β” content of detached wheat leaves following a period of wilting. Planta 1969, 86, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Most, B.H. Abscisic acid in immature apical tissue of sugar cane and in leaves of plants subjected to drought. Planta 1971, 101, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Beardsell, M.F.; Cohen, D. Relationships between leaf water status, abscisic acid levels, and stomatal resistance in maize and Sorghum. Plant Physiol. 1975, 56, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Henson, I.E. Effects of atmospheric humidity on abscisic acid accumulation and water status in leaves of rice. Ann. Bot. 1984, 54, 569–582. [Google Scholar] [CrossRef]
- Stewart, C.R.; Voetberg, G. Relationship between stress-induced ABA and proline accumulations and ABA-induced proline accumulation in excised barley leaves. Plant Physiol. 1985, 79, 24–27. [Google Scholar] [CrossRef]
- Bensen, R.J.; Boyer, J.S.; Mullet, J.E. Water deficit-induced changes in abscisic acid, growth, polysomes, and translatable RNA in soybean hypocotyls. Plant Physiol. 1988, 88, 289–294. [Google Scholar] [CrossRef]
- Giordani, T.; Natali, L.; D’Ercole, A.; Pugliesi, C.; Fambrini, M.; Vernieri, P.; Vitagliano, C.; Cavallini, A. Expression of a dehydrin gene during embryo development and drought stress in ABA-deficient mutants of sunflower (Helianthus annuus L.). Plant Mol. Biol. 1999, 39, 739–748. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sasaki, Y.; Li, X.; Mori, I.C.; Matsuura, T.; Hirayama, T.; Sato, T.; Yamaguchi, J. ABI1 regulates carbon/nitrogen-nutrient signal transduction independent of ABA biosynthesis and canonical ABA signaling pathways in Arabidopsis. J. Exp. Bot. 2015, 66, 2763–2771. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Park, C.; Kim, J.H.; Joo, H.; Hong, E.; Lee, S.C. Pepper CaREL1, a ubiquitin E3 ligase, regulates drought tolerance via the ABA-signaling pathway. Sci. Rep. 2017, 7, 477. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Vanhee, C.; Zapotoczny, G.; Masquelier, D.; Ghislain, M.; Batoko, H. The Arabidopsis multi-stress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 2011, 23, 785–805. [Google Scholar] [CrossRef]
- Sun, X.; Wang, P.; Jia, X.; Huo, L.Q.; Che, R.M.; Ma, F.W. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 2018, 16, 545–557. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Jia, X.; Ma, F. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017, 256, 53–64. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2019, 25, 1–17. [Google Scholar] [CrossRef]
- Noda, T.; Suzuki, K.; Ohsumi, Y. Yeast autophagosomes: De novo formation of a membrane structure. Trends. Cell Biol. 2002, 12, 231–235. [Google Scholar] [CrossRef]
- Liu, F.; Marshall, R.S.; Li, F.Q. Understanding and exploiting the roles of autophagy in plants through multi-omics approaches. Plant Sci. 2018, 274, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Birgisdottir, Å.B.; Lamark, T.; Johansen, T. The LIR motif-crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [PubMed]
- Kriegenburg, F.; Ungermann, C.; Reggiori, F. Coordination of autophagosome-lysosome fusion by Atg8 family members. Curr. Biol. 2018, 28, R512–R518. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef]
- Kang, C.; Elledge, S.J. How autophagy both activates and inhibits cellular senescence. Autophagy 2016, 12, 898–899. [Google Scholar] [CrossRef]
- Di Berardino, J.; Marmagne, A.; Berger, A.; Yoshimoto, K.; Cueff, G.; Chardon, F.; Masclaux-Daubresse, C.; Reisdorf-Cren, M. Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 2018, 69, 1403–1414. [Google Scholar] [CrossRef]
- Hanamata, S.; Kurusu, T.; Kuchitsu, K. Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 2014, 5, 457. [Google Scholar] [CrossRef]
- Tang, J.; Bassham, D.C. Autophagy in crop plants: What’s new beyond Arabidopsis. Open Biol. 2018, 8, 180162. [Google Scholar] [CrossRef]
- Huang, L.; Yu, L.; Zhang, X.; Fan, B.; Wang, F.; Dai, Y.; Qi, H.; Zhou, Y.; Xie, L. Xiao, S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2019, 15, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, H.; Noda, T.; Shirano, Y.; Kato, T.; Hayash, H.; Shibata, D.; Tabata, S.; Ohsumi, Y. Leaf senescence and starvation induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002, 129, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.L.; Gao, C.; Kang, B.H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E426–E435. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, X.; Jia, X.; Wang, N.; Gong, X.; Ma, F. Characterization of an autophagy-related gene MdATG8i from apple. Front. Plant Sci. 2016, 7, 720. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, Z.; Wang, X.; Li, X.; Zhang, Z.; Fan, B.; Zhu, C.; Chen, Z. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy 2018, 14, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Yoshimoto, K.; Ohsumi, Y.; Jeon, J.S.; An, G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 2009, 27, 67–74. [Google Scholar] [CrossRef]
- Chen, L.; Liao, B.; Qi, H.; Xie, L.J.; Huang, L.; Tan, W.J.; Zhai, N.; Yuan, L.B.; Zhou, Y.; Yu, L.J.; et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 2015, 11, 2233–2246. [Google Scholar] [CrossRef]
- Munch, D.; Rodriguez, E.; Bressendorff, S.; Park, O.K.; Hofius, D.; Petersen, M. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 2014, 10, 1579–1587. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef]
- Yue, W.; Nie, X.; Cui, L.; Zhi, Y.; Zhang, T.; Du, X.; Song, W. Genome-wide sequence and expressional analysis of autophagy gene family in bread wheat (Triticum aestivum L.). J. Plant Physiol. 2018, 229, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Avila-Ospina, L.; Marmagne, A.; Soulay, F.; Masclaux-Daubresse, C. Identification of barley (Hordeum vulgare L.) autophagy genes and their expression levels during leaf senescence, chronic nitrogen limitation and in response to dark exposure. Agronomy 2016, 6, 15. [Google Scholar] [CrossRef]
- Zhai, Y.; Guo, M.; Wang, H.; Lu, J.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic Stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Liu, W.; Hu, W.; Liu, G.; Wu, C.; Liu, W.; Zeng, H.; He, C.; Shi, H. Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 2017, 36, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought stress responses in crops. Funct. Integr. Genomics 2014, 14, 11–22. [Google Scholar] [CrossRef]
- Liu, G.Y.; Zeng, H.Q.; Li, X.; Wei, Y.X.; Shi, H.T. Functional analysis of MaWRKY24 in transcriptional activation of autophagy-related genes and plant disease resistance against soil-borne Fusarium oxysporum f. sp. cubense. Pathogens 2019, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, N.; Pelletier, G. In Planta agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar]
- Chen, L.; Zhong, H.Y.; Kuang, J.F.; Li, J.G.; Lu, W.J.; Chen, J.Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef]
- Farmer, E.E.; Mousavi, S.A.; Lenglet, A. Leaf numbering for experiments on long distance signalling in Arabidopsis. Protoc. Exch. 2013. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Chan, Z. Comparative proteomic and metabolomic analyses reveal mechanisms of improved cold stress tolerance in bermudagrass (Cynodondactylon (L.) Pers.) by exogenous calcium. J. Integr. Plant Biol. 2014, 56, 1064–1079. [Google Scholar] [CrossRef]
- Fan, S.; Chang, Y.; Liu, G.; Shang, S.; Tian, L.; Shi, H. Molecular functional analysis of auxin/indole-3-acetic acid proteins (Aux/IAAs) in plant disease resistance in cassava. Physiol. Plant 2019. [Google Scholar] [CrossRef] [PubMed]
- Afridi, M.S.; Amna1, S.; Mahmood, T.; Salam, A.; Mukhtar, T.; Mehmood, S.; Ali, J.; Khatoon, Z.; Bibi, M.; Javed, M.T.; et al. Induction of tolerance to salinity in wheat genotypes by plant growth promoting endophytes: Involvement of ACC deaminase and antioxidant enzymes. Plant Physiol. Biochem. 2019, 139, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Bozhkov, P.V. Plant autophagy: Mechanisms and functions. J. Exp. Bot. 2018, 69, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Rahal, A.; Kumar, A.; Singh, V.; Yadav, B.; Tiwari, R.; Chakraborty, S.; Dhama, K. Oxidative stress, prooxidants, and antioxidants: The interplay. Biomed. Res. Int. 2014, 761264. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. ROS, mitochondria and the regulation of autophagy Trends in cell biology. Trends Cell Biol. 2007, 17, 422–427. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-derived bioactive peptides on inflammation and oxidative stress. Biomed. Res. Int. 2014, 608979. [Google Scholar] [CrossRef]
- Chen, Y.; Azad, M.B.; Gibson, S.B. Superoxide is the major reactive oxygen species regulating autophagy Cell death and differentiation. Cell Death Differ. 2009, 16, 1040–1052. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Crosstalk and redox signaling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- Ehonen, S.; Yarmolinsky, D.; Kollist, H.; Kangasjärvi, J. Reactive oxygen species, photosynthesis, and environment in the regulation of stomata. Antioxid Redox Signal. 2019, 30, 1220–1237. [Google Scholar] [CrossRef]
- Ishida, H.; Izumi, M.; Wada, S.; Makino, A. Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta. 2014, 1837, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant. Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Zhuang, L.; Gao, Y.; Huang, B. Abscisic acid mediation of drought priming-enhanced heat tolerance in tall fescue (Festuca arundinacea) and Arabidopsis. Physiol. Plant. 2019. [Google Scholar] [CrossRef] [PubMed]
- Loveys, B.R.; Kriedemann, P.E. Internal control of stomatal physiology and photosynthesis. I. Stomatal regulation and associated changes in endogenous levels of abscisic and phaseic acids. Funct. Plant Biol. 1974, 1, 407–415. [Google Scholar] [CrossRef]
- Zhang, J.; Schurr, U.; Davies, W.J. Control of stomatal behaviour by abscisic acid which apparently originates in the roots. J. Exp. Bot. 1987, 38, 1174–1181. [Google Scholar] [CrossRef]
- Zhang, J.; Davies, W.J. Changes in the concentration of ABA in xylem sap as a function of changing soil water status can account for changes in leaf conductance and growth. Plant Cell Environ. 1990, 13, 277–285. [Google Scholar] [CrossRef]
- Munns, R.; Sharp, R.E. Involvement of abscisic acid in controlling plant growth in soil of low water potential. Funct. Plant Biol. 1993, 20, 425–437. [Google Scholar] [CrossRef]
- Saradadevi, R.; Palta, J.A.; Siddique, K.H.M. ABA-Mediated stomatal response in regulating water use during the development of terminal drought in wheat. Front. Plant Sci. 2017, 8, 1251. [Google Scholar]
- Hong, E.; Lim, C.W.; Han, S.W.; Lee, S.C. Functional Analysis of the Pepper Ethylene-Responsive Transcription Factor, CaAIEF1, in Enhanced ABA Sensitivity and Drought Tolerance. Front Plant Sci. 2017, 8, 1407. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, G.; Li, Y.; Kong, X.; Zhang, L.; Wang, J.; Li, X.; Yang, Y. ABA Receptor Subfamily III Enhances Abscisic Acid Sensitivity and Improves the Drought Tolerance of Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1938. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.W.; Feng, H.; Deng, J.; Zhang, R.X.; Wen, J.Q.; Dong, J.L.; Wang, T. dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef]
- Vanhee, C.; Batoko, H. Autophagy involvement in responses to abscisic acid by plant cells. Autophagy 2011, 7, 655–656. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Liu, G.; Wang, Y.; Wei, Y.; Shi, H. Overexpression of Banana ATG8f Modulates Drought Stress Resistance in Arabidopsis. Biomolecules 2019, 9, 814. https://doi.org/10.3390/biom9120814
Li B, Liu G, Wang Y, Wei Y, Shi H. Overexpression of Banana ATG8f Modulates Drought Stress Resistance in Arabidopsis. Biomolecules. 2019; 9(12):814. https://doi.org/10.3390/biom9120814
Chicago/Turabian StyleLi, Bing, Guoyin Liu, Yuqi Wang, Yunxie Wei, and Haitao Shi. 2019. "Overexpression of Banana ATG8f Modulates Drought Stress Resistance in Arabidopsis" Biomolecules 9, no. 12: 814. https://doi.org/10.3390/biom9120814
APA StyleLi, B., Liu, G., Wang, Y., Wei, Y., & Shi, H. (2019). Overexpression of Banana ATG8f Modulates Drought Stress Resistance in Arabidopsis. Biomolecules, 9(12), 814. https://doi.org/10.3390/biom9120814



