ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Proteins and Plasmids
2.2. Biochemical Assays
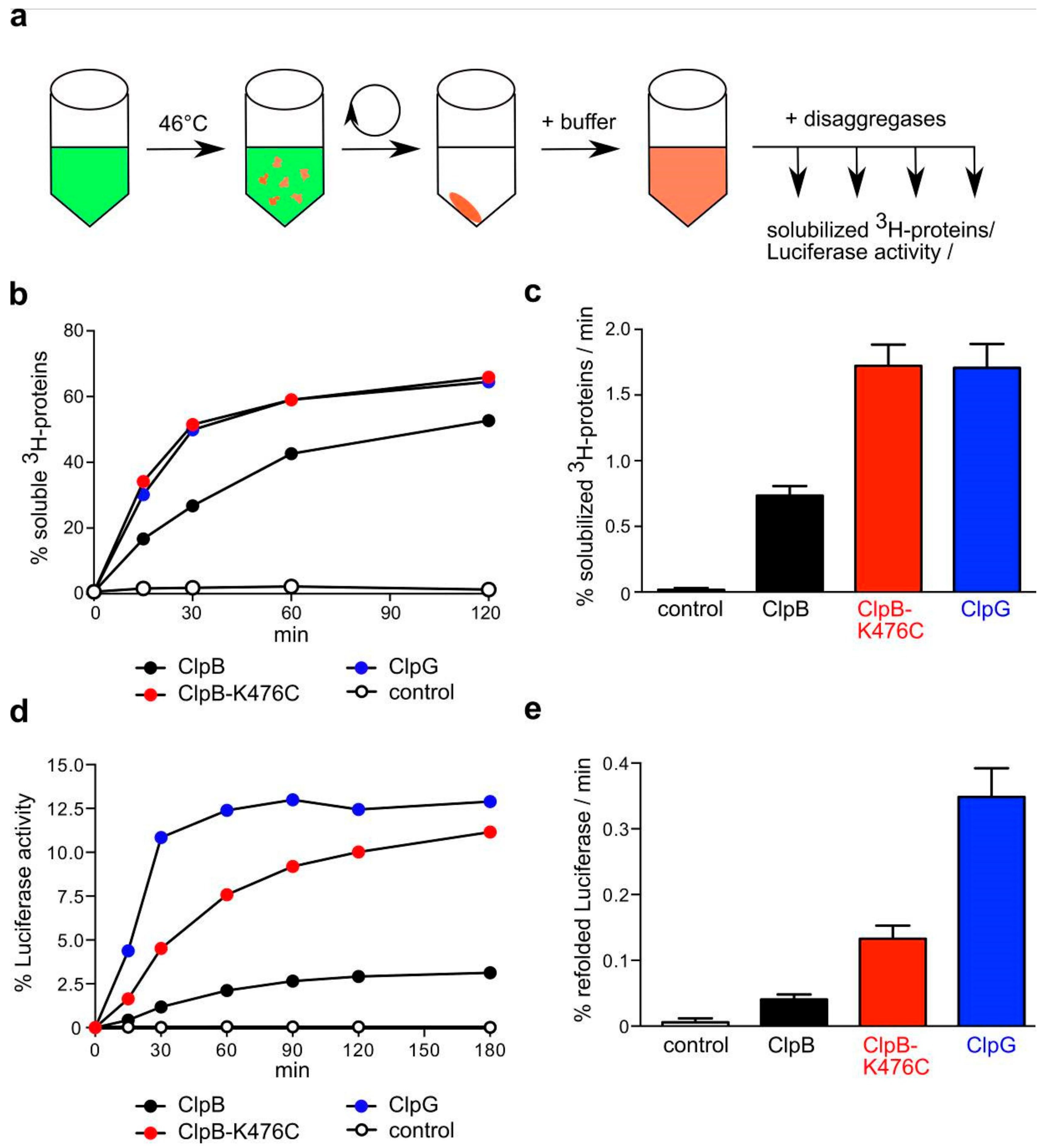
2.2.1. In Vitro Disaggregation Assays
2.2.2. Ex Vivo Disaggregation of Proteins in Heat-Denatured Crude Extracts
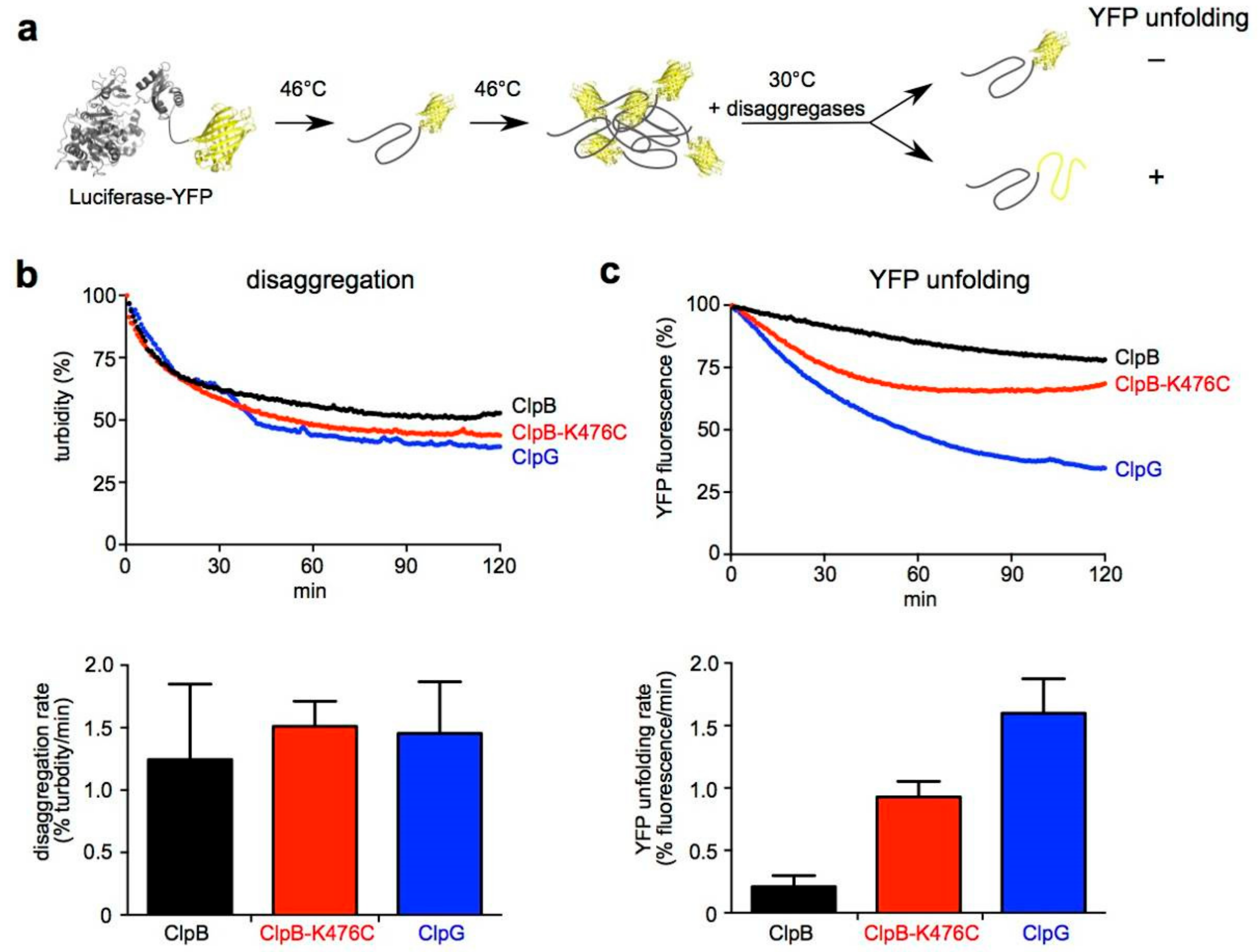
2.2.3. Monitoring YFP Unfolding During Protein Disaggregation
2.3. Characterization of Luciferase Aggregates
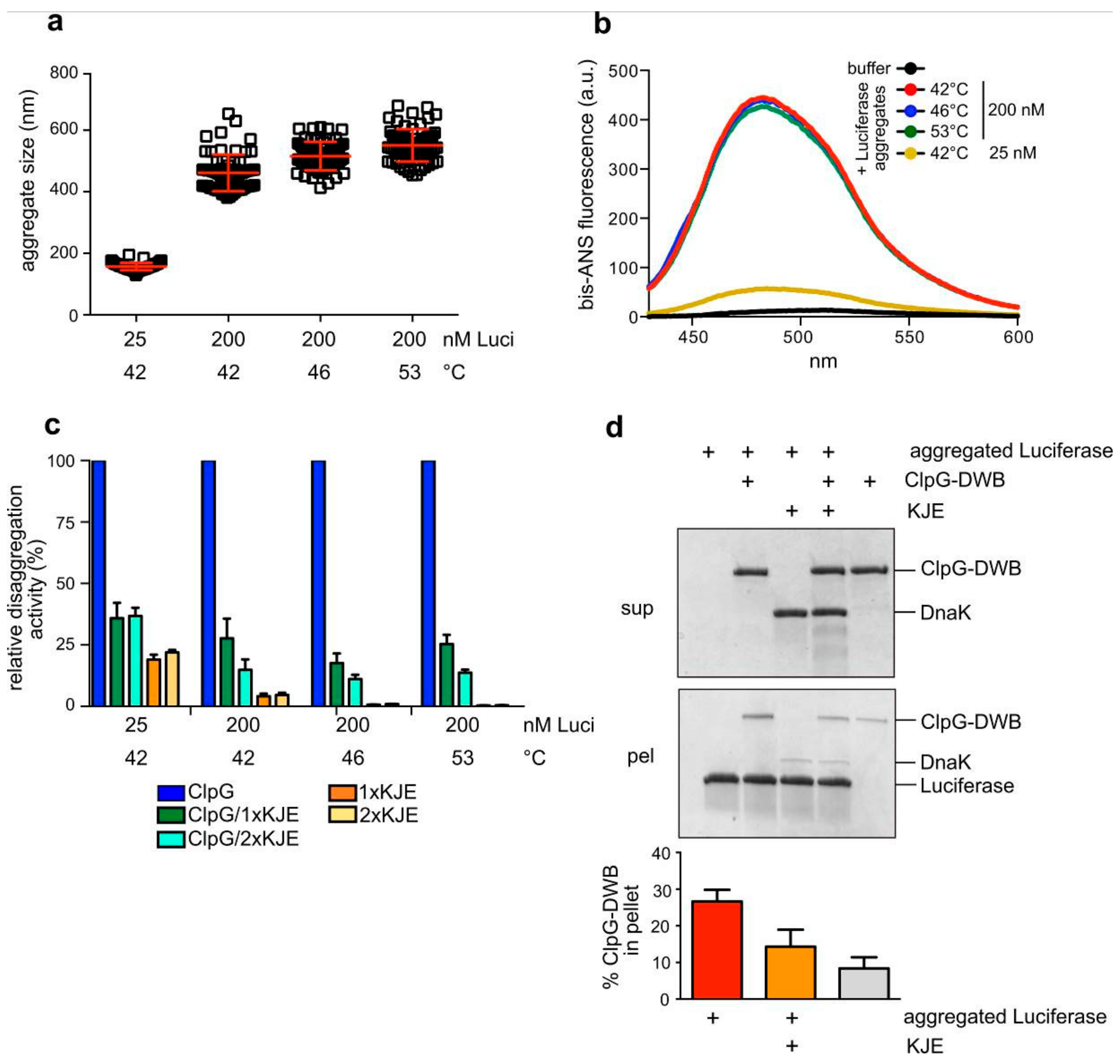
2.3.1. Dynamic Light Scattering Measurements
2.3.2. Bis-ANS Binding
2.4. Aggregate Binding Assay
2.5. In Vivo Disaggregation Assays
2.5.1. Luciferase Reactivation
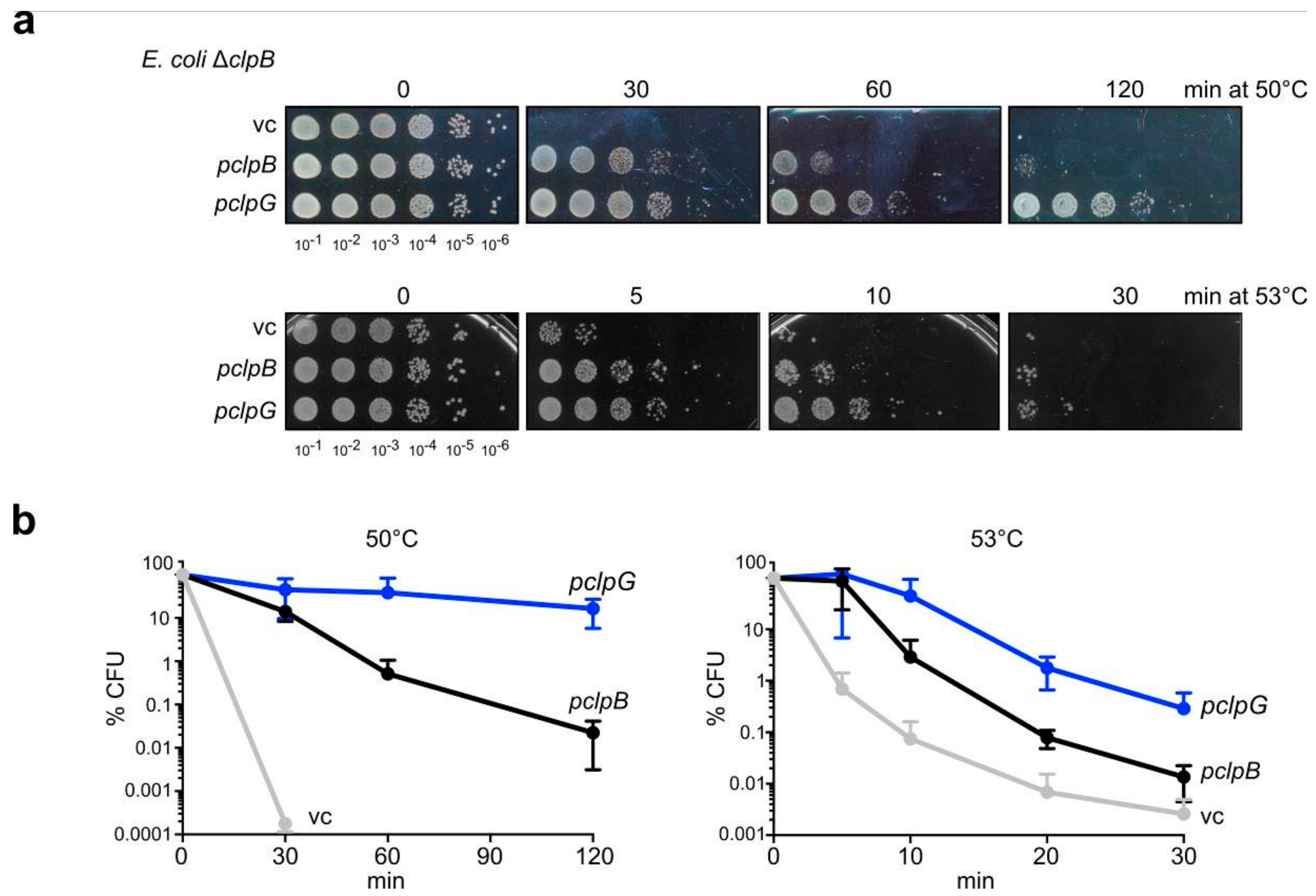
2.5.2. Heat Resistance Assay
2.6. Cellular toxicity assay
3. Results
3.1. ClpG Provides Efficient Protection Towards Severe Heat Stress
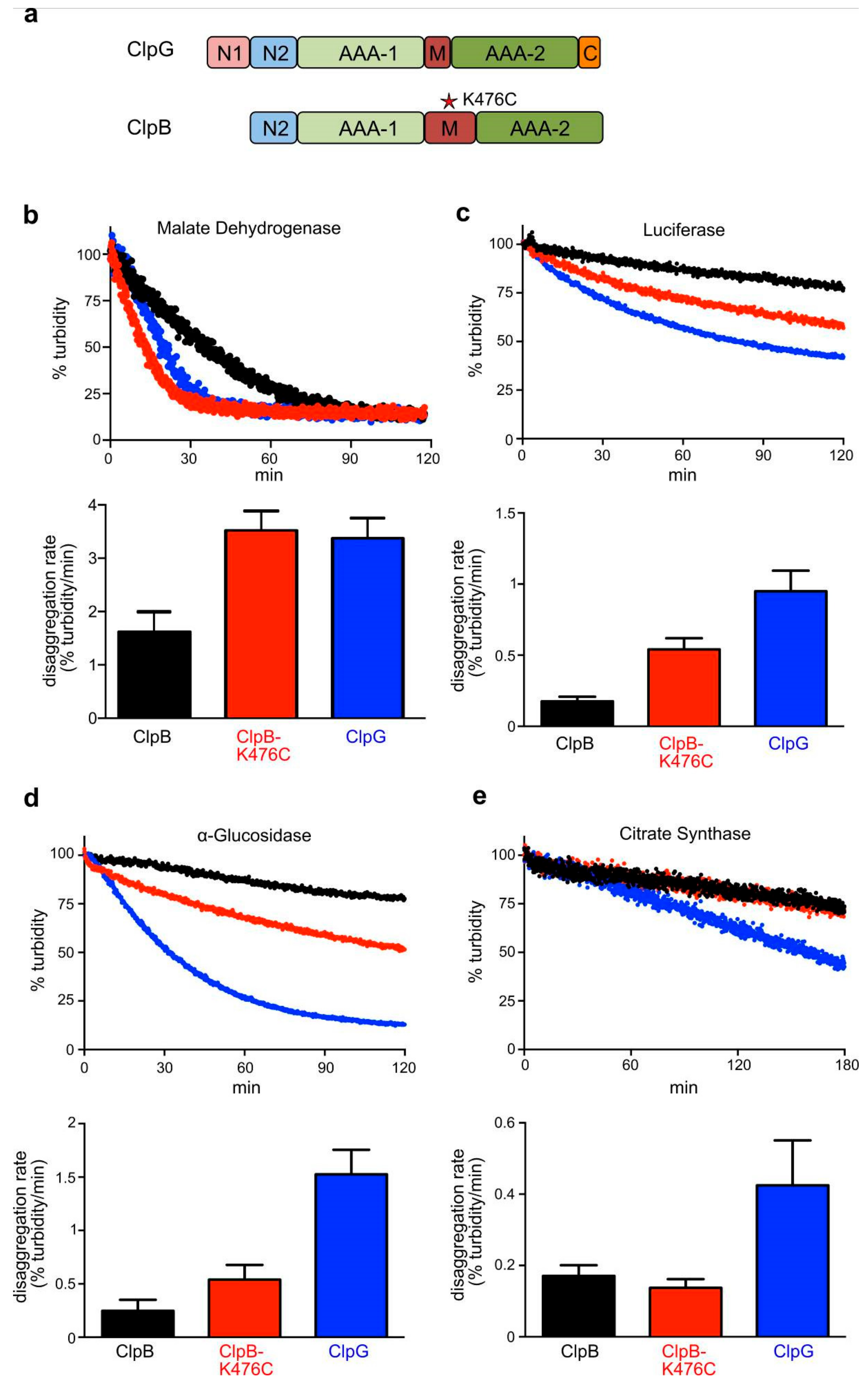
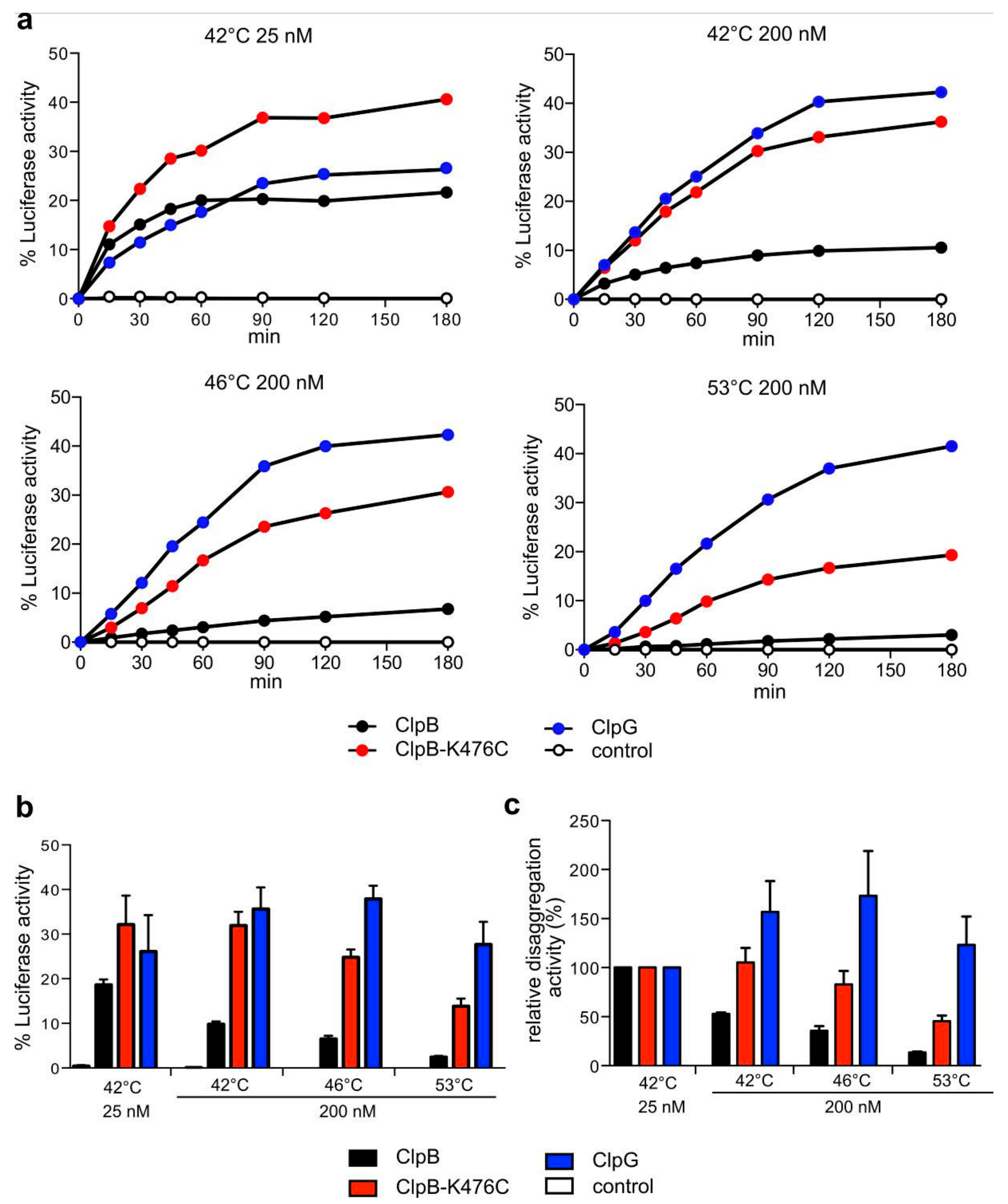
3.2. In Vitro ClpG is More Efficient in the Disaggregation of Diverse Aggregated Model Substrates
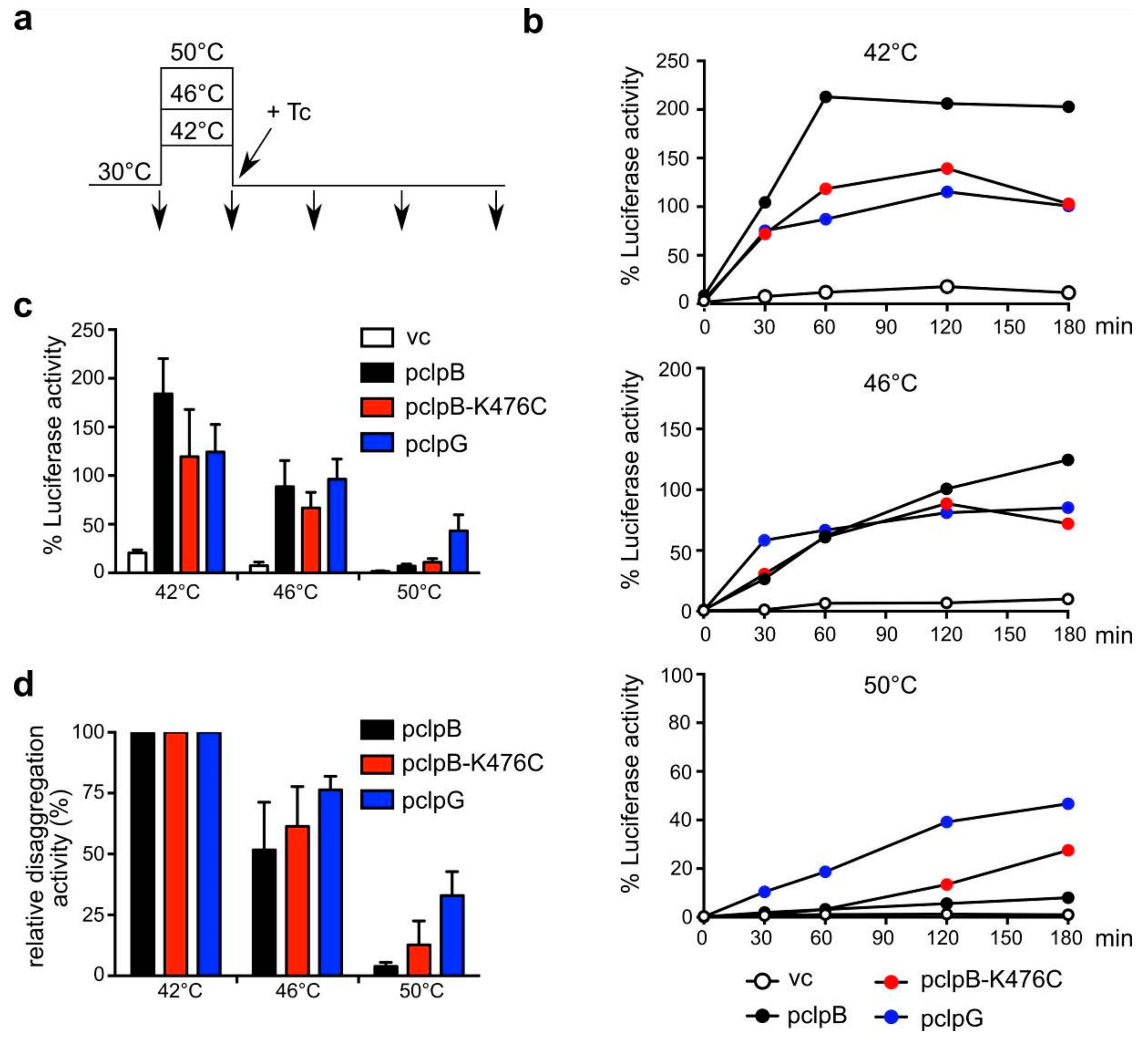
3.3. ClpB and ClpG Reveal Temperature-Dependent Differences in Disaggregation Potential In Vivo
3.4. ClpG but not ClpB Shows Robust Disaggregation Activity towards Luciferase Aggregates Formed at Increasing Temperatures
3.5. Surface Properties of Luciferase Aggregates Do Not Explain Differing Disaggregation Activities
3.6. ClpG Exhibits High Unfolding Power in Contrast to ClpB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mogk, A.; Tomoyasu, T.; Goloubinoff, P.; Rüdiger, S.; Röder, D.; Langen, H.; Bukau, B. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999, 18, 6934–6949. [Google Scholar] [CrossRef]
- Pu, Y.; Li, Y.; Jin, X.; Tian, T.; Ma, Q.; Zhao, Z.; Lin, S.Y.; Chen, Z.; Li, B.; Yao, G.; et al. ATP-Dependent Dynamic Protein Aggregation Regulates Bacterial Dormancy Depth Critical for Antibiotic Tolerance. Mol. Cell 2019, 73, 143–156. [Google Scholar] [CrossRef]
- Li, H.; Gänzle, M. Some Like It Hot: Heat Resistance of Escherichia coli in Food. Front. Microbiol. 2016, 7, 1763. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Bukau, B.; Kampinga, H.H. Cellular Handling of Protein Aggregates by Disaggregation Machines. Mol. Cell 2018, 69, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Puchades, C.; Rampello, A.J.; Shin, M.; Giuliano, C.J.; Wiseman, R.L.; Glynn, S.E.; Lander, G.C. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 2017, 358, eaao0464. [Google Scholar] [CrossRef] [PubMed]
- De la Pena, A.H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362, eaav0725. [Google Scholar] [CrossRef]
- Squires, C.L.; Pedersen, S.; Ross, B.M.; Squires, C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991, 173, 4254–4262. [Google Scholar] [CrossRef]
- Sanchez, Y.; Lindquist, S.L. HSP104 required for induced thermotolerance. Science 1990, 248, 1112–1115. [Google Scholar] [CrossRef]
- Queitsch, C.; Hong, S.W.; Vierling, E.; Lindquist, S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef]
- Schlothauer, T.; Mogk, A.; Dougan, D.A.; Bukau, B.; Turgay, K. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. USA 2003, 100, 2306–2311. [Google Scholar] [CrossRef]
- Zolkiewski, M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 1999, 274, 28083–28086. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Goloubinoff, P.; Mogk, A.; Peres Ben Zvi, A.; Tomoyasu, T.; Bukau, B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 1999, 96, 13732–13737. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012, 198, 387–404. [Google Scholar] [CrossRef]
- Acebron, S.P.; Martin, I.; del Castillo, U.; Moro, F.; Muga, A. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009, 583, 2991–2996. [Google Scholar] [CrossRef]
- Oguchi, Y.; Kummer, E.; Seyffer, F.; Berynskyy, M.; Anstett, B.; Zahn, R.; Wade, R.C.; Mogk, A.; Bukau, B. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 2012, 19, 1338–1346. [Google Scholar] [CrossRef]
- Lee, J.; Kim, J.H.; Biter, A.B.; Sielaff, B.; Lee, S.; Tsai, F.T. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. USA 2013, 110, 8513–8518. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Moradi, S.; Zarrine-Afsar, A.; Glover, J.R.; Kay, L.E. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 2013, 339, 1080–1083. [Google Scholar] [CrossRef]
- Carroni, M.; Kummer, E.; Oguchi, Y.; Wendler, P.; Clare, D.K.; Sinning, I.; Kopp, J.; Mogk, A.; Bukau, B.; Saibil, H.R. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife 2014, 3, e02481. [Google Scholar] [CrossRef]
- Lipinska, N.; Zietkiewicz, S.; Sobczak, A.; Jurczyk, A.; Potocki, W.; Morawiec, E.; Wawrzycka, A.; Gumowski, K.; Slusarz, M.; Rodziewicz-Motowidlo, S.; et al. Disruption of ionic interactions between the nucleotide binding domain 1 (NBD1) and middle (M) domain in Hsp100 disaggregase unleashes toxic hyperactivity and partial independence from Hsp70. J. Biol. Chem. 2013, 288, 2857–2869. [Google Scholar] [CrossRef]
- Haslberger, T.; Zdanowicz, A.; Brand, I.; Kirstein, J.; Turgay, K.; Mogk, A.; Bukau, B. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat. Struct. Mol. Biol. 2008, 15, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Franke, K.B.; Kamal, S.M.; Kim, H.; Lünsdorf, H.; Jäger, J.; Nimtz, M.; Trcek, J.; Jänsch, L.; Bukau, B.; et al. Stand-alone ClpG disaggregase confers superior heat tolerance to bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E273–E282. [Google Scholar] [CrossRef] [PubMed]
- Bojer, M.S.; Struve, C.; Ingmer, H.; Hansen, D.S.; Krogfelt, K.A. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS ONE 2010, 5, e15467. [Google Scholar] [CrossRef] [PubMed]
- Boll, E.J.; Marti, R.; Hasman, H.; Overballe-Petersen, S.; Stegger, M.; Ng, K.; Knochel, S.; Krogfelt, K.A.; Hummerjohann, J.; Struve, C. Turn Up the Heat-Food and Clinical Escherichia coli Isolates Feature Two Transferrable Loci of Heat Resistance. Front. Microbiol. 2017, 8, 579. [Google Scholar] [CrossRef]
- Lee, C.; Wigren, E.; Trcek, J.; Peters, V.; Kim, J.; Hasni, M.S.; Nimtz, M.; Lindqvist, Y.; Park, C.; Curth, U.; et al. A novel protein quality control mechanism contributes to heat shock resistance of worldwide-distributed Pseudomonas aeruginosa clone C strains. Environ. Microbiol. 2015, 17, 4511–4526. [Google Scholar] [CrossRef]
- Mercer, R.G.; Zheng, J.; Garcia-Hernandez, R.; Ruan, L.; Ganzle, M.G.; McMullen, L.M. Genetic determinants of heat resistance in Escherichia coli. Front. Microbiol. 2015, 6, 932. [Google Scholar] [CrossRef]
- Lee, C.; Wigren, E.; Lünsdorf, H.; Römling, U. Protein homeostasis-more than resisting a hot bath. Curr. Opin. Microbiol. 2016, 30, 147–154. [Google Scholar] [CrossRef]
- Marti, R.; Muniesa, M.; Schmid, M.; Ahrens, C.H.; Naskova, J.; Hummerjohann, J. Short communication: Heat-resistant Escherichia coli as potential persistent reservoir of extended-spectrum beta-lactamases and Shiga toxin-encoding phages in dairy. J. Dairy Sci. 2016, 99, 8622–8632. [Google Scholar] [CrossRef]
- Boll, E.J.; Frimodt-Moller, J.; Olesen, B.; Krogfelt, K.A.; Struve, C. Heat resistance in extended-spectrum beta-lactamase-producing Escherichia coli may favor environmental survival in a hospital setting. Res. Microbiol. 2016, 167, 345–349. [Google Scholar] [CrossRef]
- Bojer, M.S.; Hammerum, A.M.; Jorgensen, S.L.; Hansen, F.; Olsen, S.S.; Krogfelt, K.A.; Struve, C. Concurrent emergence of multidrug resistance and heat resistance by CTX-M-15-encoding conjugative plasmids in Klebsiella pneumoniae. APMIS 2012, 120, 699–705. [Google Scholar] [CrossRef]
- Bojer, M.S.; Struve, C.; Ingmer, H.; Krogfelt, K.A. ClpP-dependent and -independent activities encoded by the polycistronic clpK-encoding locus contribute to heat shock survival in Klebsiella pneumoniae. Res. Microbiol. 2013, 164, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Seyffer, F.; Kummer, E.; Oguchi, Y.; Winkler, J.; Kumar, M.; Zahn, R.; Sourjik, V.; Bukau, B.; Mogk, A. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat. Struct. Mol. Biol. 2012, 19, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Diamant, S.; Ben-Zvi, A.P.; Bukau, B.; Goloubinoff, P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem. 2000, 275, 21107–21113. [Google Scholar] [CrossRef]
- Franke, K.B.; Bukau, B.; Mogk, A. Mutant Analysis Reveals Allosteric Regulation of ClpB Disaggregase. Front. Mol. Biosci. 2017, 4, 6. [Google Scholar] [CrossRef]
- Deville, C.; Carroni, M.; Franke, K.B.; Topf, M.; Bukau, B.; Mogk, A.; Saibil, H.R. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv. 2017, 3, e1701726. [Google Scholar] [CrossRef]
- Deville, C.; Franke, K.; Mogk, A.; Bukau, B.; Saibil, H.R. Two-Step Activation Mechanism of the ClpB Disaggregase for Sequential Substrate Threading by the Main ATPase Motor. Cell Rep. 2019, 27, 3433–3446.e3434. [Google Scholar] [CrossRef]
- Mogk, A.; Deuerling, E.; Vorderwulbecke, S.; Vierling, E.; Bukau, B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 2003, 50, 585–595. [Google Scholar] [CrossRef]
- Cherkasov, V.; Hofmann, S.; Druffel-Augustin, S.; Mogk, A.; Tyedmers, J.; Stoecklin, G.; Bukau, B. Coordination of translational control and protein homeostasis during severe heat stress. Curr. Biol. 2013, 23, 2452–2462. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katikaridis, P.; Meins, L.; Kamal, S.M.; Römling, U.; Mogk, A. ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase. Biomolecules 2019, 9, 815. https://doi.org/10.3390/biom9120815
Katikaridis P, Meins L, Kamal SM, Römling U, Mogk A. ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase. Biomolecules. 2019; 9(12):815. https://doi.org/10.3390/biom9120815
Chicago/Turabian StyleKatikaridis, Panagiotis, Lena Meins, Shady Mansour Kamal, Ute Römling, and Axel Mogk. 2019. "ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase" Biomolecules 9, no. 12: 815. https://doi.org/10.3390/biom9120815
APA StyleKatikaridis, P., Meins, L., Kamal, S. M., Römling, U., & Mogk, A. (2019). ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase. Biomolecules, 9(12), 815. https://doi.org/10.3390/biom9120815





