Crystal Structure of the Multidomain Pectin Methylesterase PmeC5 from Butyrivibrio fibrisolvens D1T
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Modelling
2.2. Protein Expression and Purification
2.3. Crystallization
2.4. Data Collection and Structure Determination
3. Results and Discussion
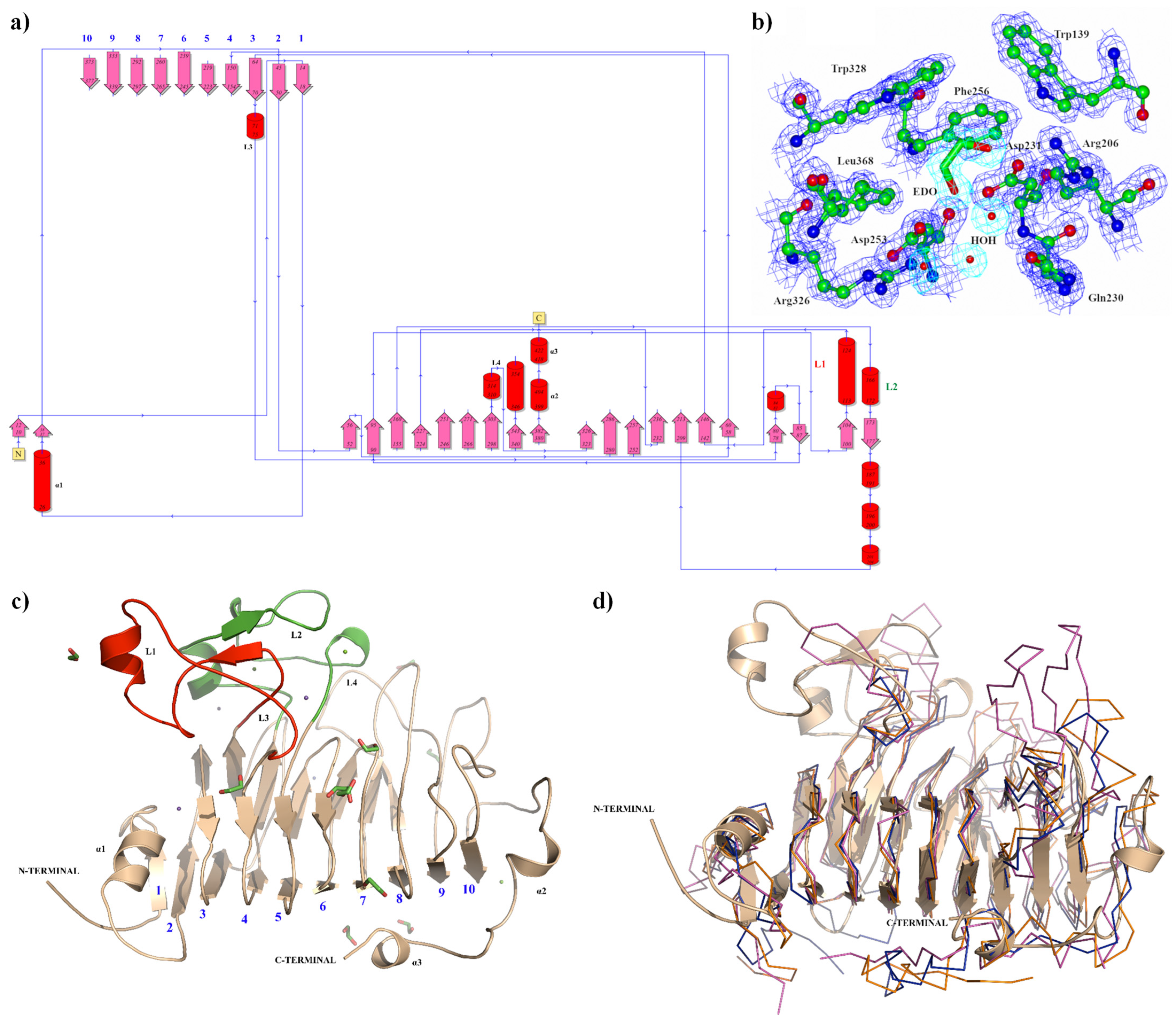
3.1. The Pectin Methylesterase Pmec5 Structure from B. Fibrisolvens D1T
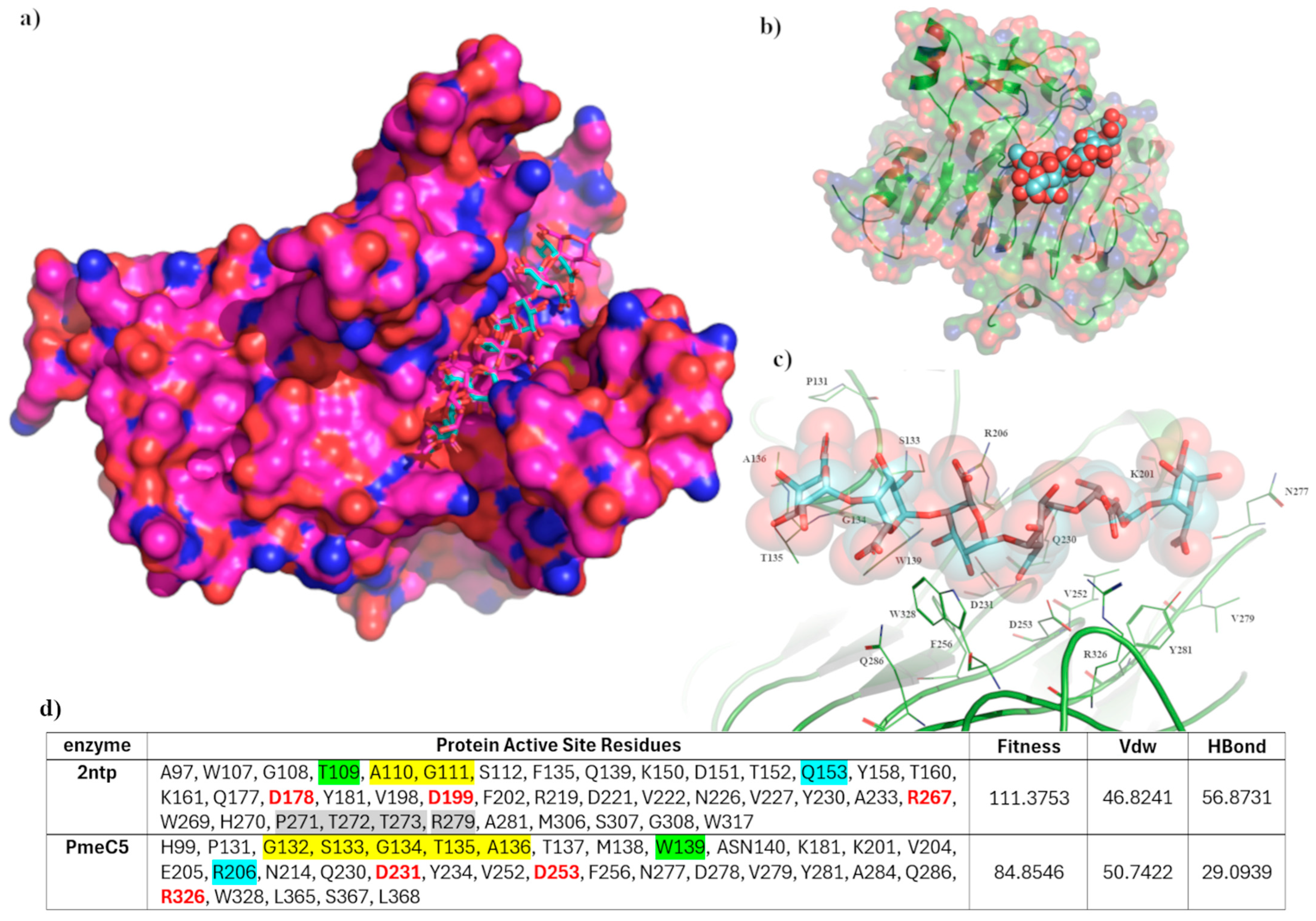
3.2. The Substrate Binding Domain of Pmec5
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeyanathan, J.; Palevich, N.; Reilly, K.; Palevich, F.P.; Maclean, P.H.; Li, D.; Altermann, E.; Kim, C.C.; van Scheepstal, I.M.; Hoskin, S.O.; et al. Isolation and characterization of Methanosphaera sp. ISO3-F5, a member of a novel and widespread species of rumen methanogens growing with methanol plus hydrogen. Microbe 2024, 5, 100210. [Google Scholar] [CrossRef]
- Palevich, N.; Jeyanathan, J.; Reilly, K.; Palevich, F.P.; Maclean, P.H.; Li, D.; Altermann, E.; Kelly, W.J.; Leahy, S.C.; Attwood, G.T.; et al. Complete genome sequence of Methanosphaera sp. ISO3-F5, a rumen methylotrophic methanogen. Microbiol. Resour. Announc. 2024, 13, e0004324. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed. Sci. Technol. 2010, 160, 1–22. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants. Animal 2010, 4, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Czerkawski, J.W. Methane production in ruminants and its significance. World Rev. Nutr. Diet. 1969, 11, 240–282. [Google Scholar] [CrossRef] [PubMed]
- Kelly, W.J.; Mackie, R.I.; Attwood, G.T.; Janssen, P.H.; McAllister, T.A.; Leahy, S.C. Hydrogen and formate production and utilisation in the rumen and the human colon. Anim. Microbiome 2022, 4, 22. [Google Scholar] [CrossRef]
- Leahy, S.C.; Janssen, P.H.; Attwood, G.T.; Mackie, R.I.; McAllister, T.A.; Kelly, W.J. Electron flow: Key to mitigating ruminant methanogenesis. Trends Microbiol. 2022, 30, 209–212. [Google Scholar] [CrossRef]
- Bailey, R. Pasture quality and ruminant nutrition: I. Carbohydrate composition of ryegrass varieties grown as sheep pastures. N. Z. J. Agric. Res. 1964, 7, 497–507. [Google Scholar] [CrossRef]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell. Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Jarvis, M.C. Structure and properties of pectin gels in plant cell walls. Plant Cell Environ. 1984, 7, 153–164. [Google Scholar] [CrossRef]
- Ralet, M.-C.; Lerouge, P.; Quéméner, B. Mass spectrometry for pectin structure analysis. Carbohydr. Res. 2009, 344, 1798–1807. [Google Scholar] [CrossRef]
- Carpita, N.C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 445–476. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Pelloux, J.; Rusterucci, C.; Mellerowicz, E. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Seedorf, H.; Kittelmann, S.; Janssen, P.H. Few Highly Abundant Operational Taxonomic Units Dominate within Rumen Methanogenic Archaeal Species in New Zealand Sheep and Cattle. Appl. Environ. Microbiol. 2015, 81, 986–995. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; A Eloe-Fadrosh, E.; A Pavlopoulos, G.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef]
- Palevich, N.; Kelly, W.J.; Leahy, S.C.; Denman, S.; Altermann, E.; Rakonjac, J.; Attwood, G.T. Comparative Genomics of Rumen Butyrivibrio spp. Uncovers a Continuum of Polysaccharide-Degrading Capabilities. Appl. Environ. Microbiol. 2019, 86. [Google Scholar] [CrossRef]
- Palevich, N.; Maclean, P.H.; Kelly, W.J.; Leahy, S.C.; Rakonjac, J.; Attwood, G.T. Complete Genome Sequence of the Polysaccharide-Degrading Rumen Bacterium Pseudobutyrivibrio xylanivorans MA3014 Reveals an Incomplete Glycolytic Pathway. Genome Biol. Evol. 2020, 12, 1566–1572. [Google Scholar] [CrossRef]
- Blackburn, T.H.; Hobson, P.N. Further Studies on the Isolation of Proteolytic Bacteria from the Sheep Rumen. J. Gen. Microbiol. 1962, 29, 69–81. [Google Scholar] [CrossRef]
- Maia, M.R.; Chaudhary, L.C.; Figueres, L.; Wallace, R.J. Metabolism of polyunsaturated fatty acids and their toxicity to the mi-croflora of the rumen. Antonie Van Leeuwenhoek 2007, 91, 303–314. [Google Scholar] [CrossRef]
- McKain, N.; Shingfield, K.J.; Wallace, R.J. Metabolism of conjugated linoleic acids and 18: 1 fatty acids by ruminal bacteria: Products and mechanisms. Microbiology 2010, 156, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Marounek, M.; Duskova, D. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett. Appl. Microbiol. 1999, 29, 429–433. [Google Scholar] [CrossRef]
- Gradel, C.M.; Dehority, B.A. Fermentation of Isolated Pectin and Pectin from Intact Forages by Pure Cultures of Rumen Bacteria. Appl. Microbiol. 1972, 23, 332–340. [Google Scholar] [CrossRef]
- Dehority, B.A. Pectin-fermenting Bacteria Isolated from the Bovine Rumen. J. Bacteriol. 1969, 99, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Palevich, N.; Palevich, F.P.; Attwood, G.T.; Kelly, W.J. Complete genome sequence of the rumen bacterium Butyrivibrio fibrisol-vens D1T. Microbiol. Resour. Announc. 2024, 13, e00267-24. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, K.; Hivarkar, S.S.; Palevich, N.; Chaudhary, P.P.; Dhakephalkar, P.K.; Dagar, S.S. Genomic architecture of three newly isolated unclassified Butyrivibrio species elucidate their potential role in the rumen ecosystem. Genomics 2022, 114, 110281. [Google Scholar] [CrossRef]
- Pidcock, S.E.; Skvortsov, T.; Santos, F.G.; Courtney, S.J.; Sui-Ting, K.; Creevey, C.J.; Huws, S.A. Phylogenetic systematics of Butyrivibrio and Pseudobutyrivibrio genomes illustrate vast taxonomic diversity, open genomes and an abundance of carbohydrate-active enzyme family isoforms. Microb. Genom. 2021, 7, 000638. [Google Scholar] [CrossRef]
- Palevich, N.; Kelly, W.J.; Ganesh, S.; Rakonjac, J.; Attwood, G.T. Butyrivibrio hungatei MB2003 Competes Effectively for Soluble Sugars Released by Butyrivibrio proteoclasticus B316T during Growth on Xylan or Pectin. Appl. Environ. Microbiol. 2019, 85, e02056-18. [Google Scholar] [CrossRef]
- Carbone, V.; Reilly, K.; Sang, C.; Schofield, L.R.; Ronimus, R.S.; Kelly, W.J.; Attwood, G.T.; Palevich, N. Crystal Structures of Bacterial Pectin Methylesterases Pme8A and PmeC2 from Rumen Butyrivibrio. Int. J. Mol. Sci. 2023, 24, 13738. [Google Scholar] [CrossRef]
- Giovane, A.; Servillo, L.; Balestrieri, C.; Raiola, A.; D’Avino, R.; Tamburrini, M.; Ciardiello, M.A.; Camardella, L. Pectin methylesterase inhibitor. Biochim. Biophys. Acta 2004, 1696, 245–252. [Google Scholar] [CrossRef]
- D’Avino, R.; Camardella, L.; Christensen, T.M.; Giovane, A.; Servillo, L. Tomato pectin methylesterase: Modeling, fluorescence, and inhibitor interaction studies-comparison with the bacterial (Erwinia chrysanthemi) enzyme. Proteins 2003, 53, 830–839. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Camardella, L.; Giovane, A.; Obel, N.; Pauly, M.; Favaron, F.; Cervone, F.; Bellincampi, D. Overexpression of Pectin Methylesterase Inhibitors in Arabidopsis Restricts Fungal Infection by Botrytis cinerea. Plant Physiol. 2007, 143, 1871–1880. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Discovery, C.; Boitreaud, J.; Dent, J.; McPartlon, M.; Meier, J.; Reis, V.; Rogozhnikov, A.; Wu, K. Chai-1: Decoding the molecular interactions of life. bioRxiv 2024. [Google Scholar] [CrossRef]
- Holm, L. Dali server: Structural unification of protein families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Fries, M.; Ihrig, J.; Brocklehurst, K.; Shevchik, V.E.; Pickersgill, R.W. Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J. 2007, 26, 3879–3887. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Cowieson, N.P.; Aragao, D.; Clift, M.; Ericsson, D.J.; Gee, C.; Harrop, S.J.; Mudie, N.; Panjikar, S.; Price, J.R.; Riboldi-Tunnicliffe, A.; et al. MX1: A bending-magnet crystallography beamline serving both chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2015, 22, 187–190. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. Sect. D Struct. Biol. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.; Mayans, O.; Smith, D.; Worboys, K.; Pickersgill, R.W. Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J. Mol. Biol. 2001, 305, 951–960. [Google Scholar] [CrossRef]
- Jenkins, J.; Pickersgill, R. The architecture of parallel β-helices and related folds. Prog. Biophys. Mol. Biol. 2001, 77, 111–175. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Kääriäinen, S.; Wilton, C.; Plewczynski, D. Using Dali for Structural Comparison of Proteins. Curr. Protoc. Bioinform. 2006, 14, 5.5.1–5.5.24. [Google Scholar] [CrossRef]
- Kent, L.M.; Loo, T.S.; Melton, L.D.; Mercadante, D.; Williams, M.A.; Jameson, G.B. Structure and properties of a non-processive, salt-requiring, and acidophilic pectin methylesterase from Aspergillus niger provide insights into the key determinants of proces-sivity control. J. Biol. Chem. 2016, 291, 1289–1306. [Google Scholar] [CrossRef]
- Johansson, K.; El-Ahmad, M.; Friemann, R.; Jörnvall, H.; Markovič, O.; Eklund, H. Crystal structure of plant pectin methylesterase. FEBS Lett. 2002, 514, 243–249. [Google Scholar] [CrossRef]
- Eklöf, J.M.; Tan, T.; Divne, C.; Brumer, H. The crystal structure of the outer membrane lipoprotein YbhC from Escherichia coli sheds new light on the phylogeny of carbohydrate esterase family 8. Proteins Struct. Funct. Bioinform. 2009, 76, 1029–1036. [Google Scholar] [CrossRef]


| Data Collection Statistics * | Items |
|---|---|
| Space group | P1211 |
| Unit cell parameters: a, b, c (Å) α, β, γ (°) | 43.30, 61.95, 77.37 90.00, 105.39, 90.00 |
| Wavelength (Å) | 0.953722 |
| Temperature (K) | 100 |
| Resolution Range (Å) | 47.66–1.33 |
| No. of observed ref. | 624,009 (30,348) |
| No. of unique ref. | 89,955 (4440) |
| Rsym a | 0.045 (0.274) |
| Rpim b | 0.028 (0.171) |
| Completeness (%) | 99.4 (98.4) |
| Multiplicity | 6.9 (6.8) |
| I/σ(I) | 23.3 (5.8) |
| CC1/2 | 1.000 (0.958) |
| Refinement statistics | |
| Resolution range (Å) | 47.66–1.33 |
| All reflections used | 90,509 |
| Size Rfree set (%) | 5 |
| All reflections (Rfree) | 4472 |
| R-values | |
| Rcryst (%) | 11.97 |
| Rfree (%) | 14.01 |
| Matthews coefficient (Å3 Da−1) | 1.74 |
| Solvent content (%) | 28.70 |
| RMSD ** | |
| Rms Bond Length (Å) | 0.0127 |
| Rms Bond Angle (°) | 1.8987 |
| Ramachandran plot | |
| Residues in favored regions (%) | 96.9 |
| Residues in allowed regions (%) | 3.1 |
| Average B factors (Å2) | |
| Protein | 9.055 |
| Water (HOH) | 23.282 |
| Mg2+ Na | 20.333 18.173 |
| Organism and Gene | Class | PDB Monomer | Z-Score a | RMSD b | Lali c | %id d |
|---|---|---|---|---|---|---|
| Dickeya dadantii 3937 Gene Name: PEMA, PEM EC: 3.1.1.11 | Pectinesterase | 2NTP-A | 33.2 | 2.4 | 289 | 27 |
| Aspergillus niger ATCC 1015 Gene Name: ASPNIDRAFT_214857 EC: 3.1.1.11 | Pectinesterase | 5C1C-A | 32.7 | 2.1 | 271 | 30 |
| Erwinia chrysanthemi Gene Name: PEMA EC: 3.1.1.11 | Pectin methylesterase | 1QJV-A | 32.7 | 2.4 | 290 | 27 |
| Daucus carota Protein sequence: P83218, PME EC: 3.1.1.11 | Pectinesterase | 1GQ8-A | 31.5 | 2.4 | 284 | 27 |
| Escherichia coli K-12 Gene Names: b0772, JW0755, ybhC EC: 3.1.2 | Acyl-CoA thioester hydrolase | 3GRH-A | 28.9 | 2.4 | 269 | 19 |
| Sitophilus oryzae Gene Name: CE8-1 EC: 3.1.1.11 | Pectinesterase | 4PMH-A | 28.6 | 2.2 | 261 | 26 |
| Butyrivibrio fibrisolvens Gene Name: SAMN02745229_01989 EC: 3.1.1.11 | Pectinesterase | 8TMS-A | 25.0 | 2.9 | 229 | 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carbone, V.; Reilly, K.; Sang, C.; Schofield, L.R.; Kelly, W.J.; Ronimus, R.S.; Attwood, G.T.; Palevich, N. Crystal Structure of the Multidomain Pectin Methylesterase PmeC5 from Butyrivibrio fibrisolvens D1T. Biomolecules 2025, 15, 720. https://doi.org/10.3390/biom15050720
Carbone V, Reilly K, Sang C, Schofield LR, Kelly WJ, Ronimus RS, Attwood GT, Palevich N. Crystal Structure of the Multidomain Pectin Methylesterase PmeC5 from Butyrivibrio fibrisolvens D1T. Biomolecules. 2025; 15(5):720. https://doi.org/10.3390/biom15050720
Chicago/Turabian StyleCarbone, Vincenzo, Kerri Reilly, Carrie Sang, Linley R. Schofield, William J. Kelly, Ron S. Ronimus, Graeme T. Attwood, and Nikola Palevich. 2025. "Crystal Structure of the Multidomain Pectin Methylesterase PmeC5 from Butyrivibrio fibrisolvens D1T" Biomolecules 15, no. 5: 720. https://doi.org/10.3390/biom15050720
APA StyleCarbone, V., Reilly, K., Sang, C., Schofield, L. R., Kelly, W. J., Ronimus, R. S., Attwood, G. T., & Palevich, N. (2025). Crystal Structure of the Multidomain Pectin Methylesterase PmeC5 from Butyrivibrio fibrisolvens D1T. Biomolecules, 15(5), 720. https://doi.org/10.3390/biom15050720








