Diversity of AMPA Receptor Ligands: Chemotypes, Binding Modes, Mechanisms of Action, and Therapeutic Effects
Abstract
1. Introduction
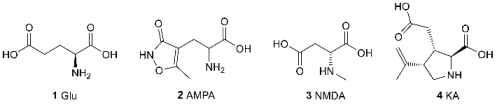
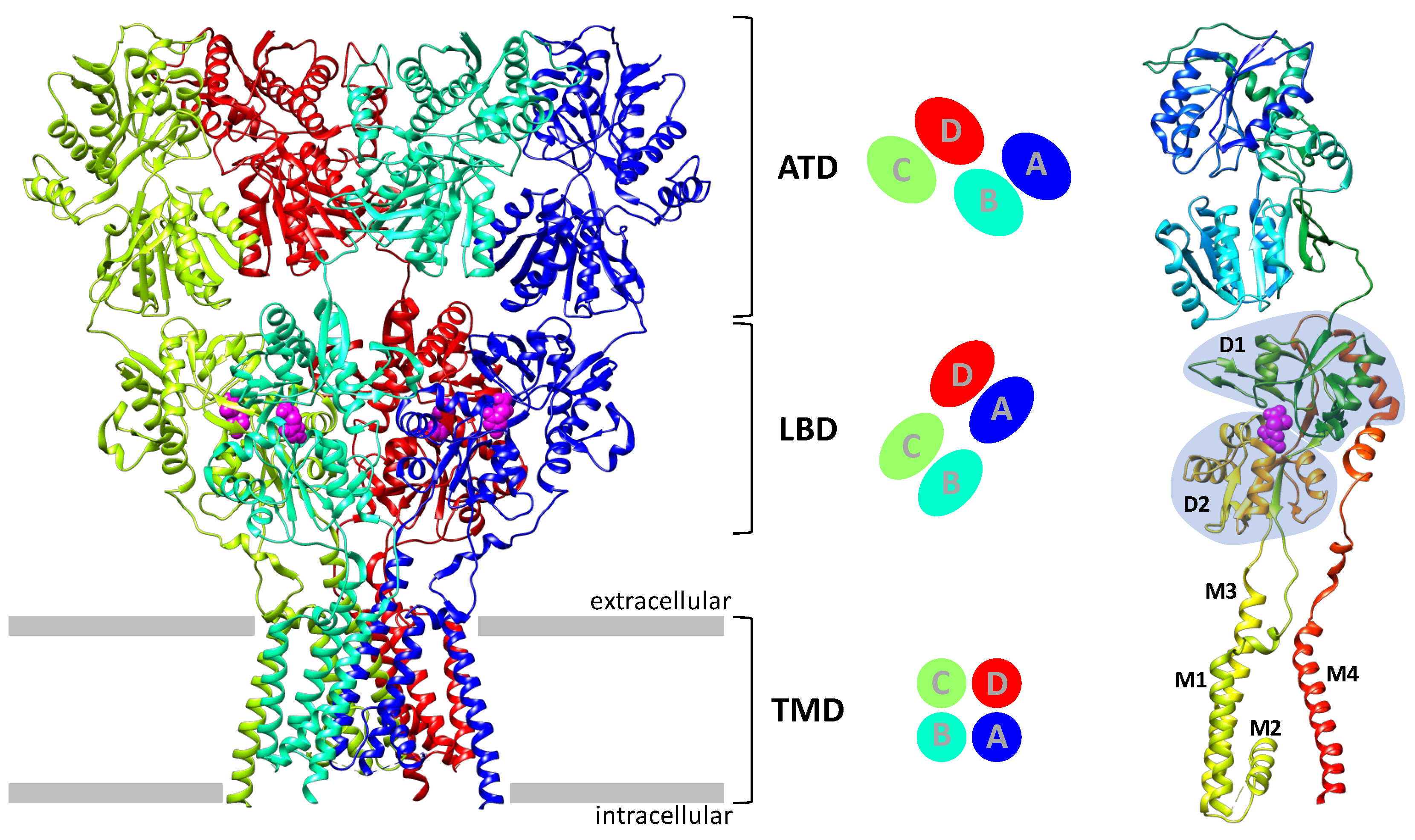
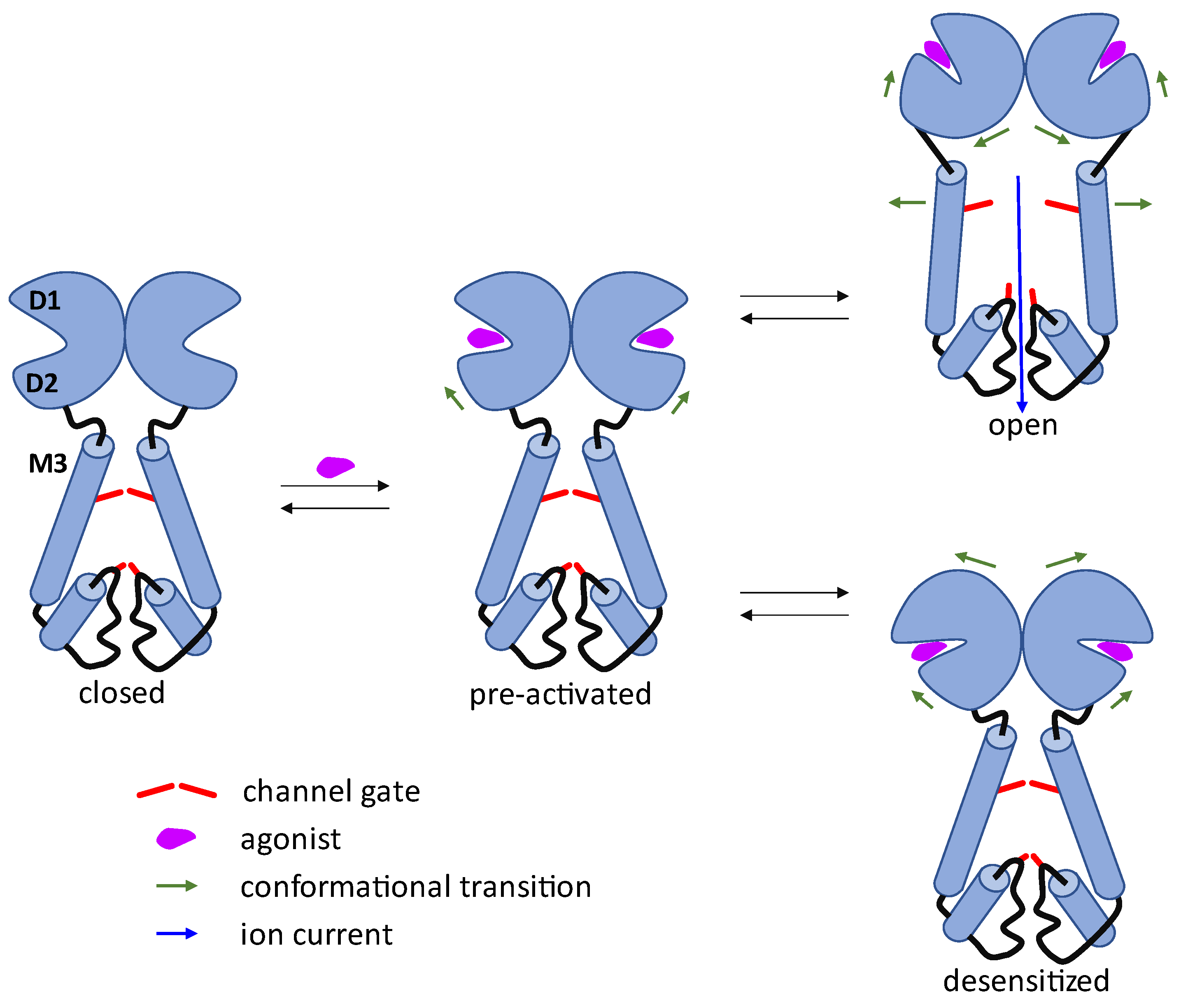
2. AMPA Receptor Architecture and Mechanism of Operation
- Transmembrane AMPA receptor regulatory proteins (TARP), including the first discovered stargazine (γ2), γ3, γ4, γ5, γ7, and γ8. Their structure includes four transmembrane helices and a number of extracellular loops containing β-sheet fragments and disordered regions. The formation of complexes with TARP potentiates the AMPA receptor currents;
- Germ cell-specific gene 1-like protein (GSG1L) is similar to TARP in overall structure (allowing one to assign these two groups to the claudin family) but has an “inhibitory” effect on the receptor;
- Cornichon homolog (CNIH) proteins contain four transmembrane helices, with the spatial structure of their transmembrane domain being very similar to that of TARP (despite the different helix arrangement), but do not contain an extracellular domain;
- Cysteine-knot AMPA receptor modulating proteins (Shisa/CKAMP family) include one transmembrane domain, an extracellular cysteine-knot motif, and a long intracellular C-terminal tail;
- The SynDIG1 and SynDIG4 proteins (Dispanin C family) contain a single transmembrane domain and a long extracellular C-terminal tail with a membrane-associated domain.
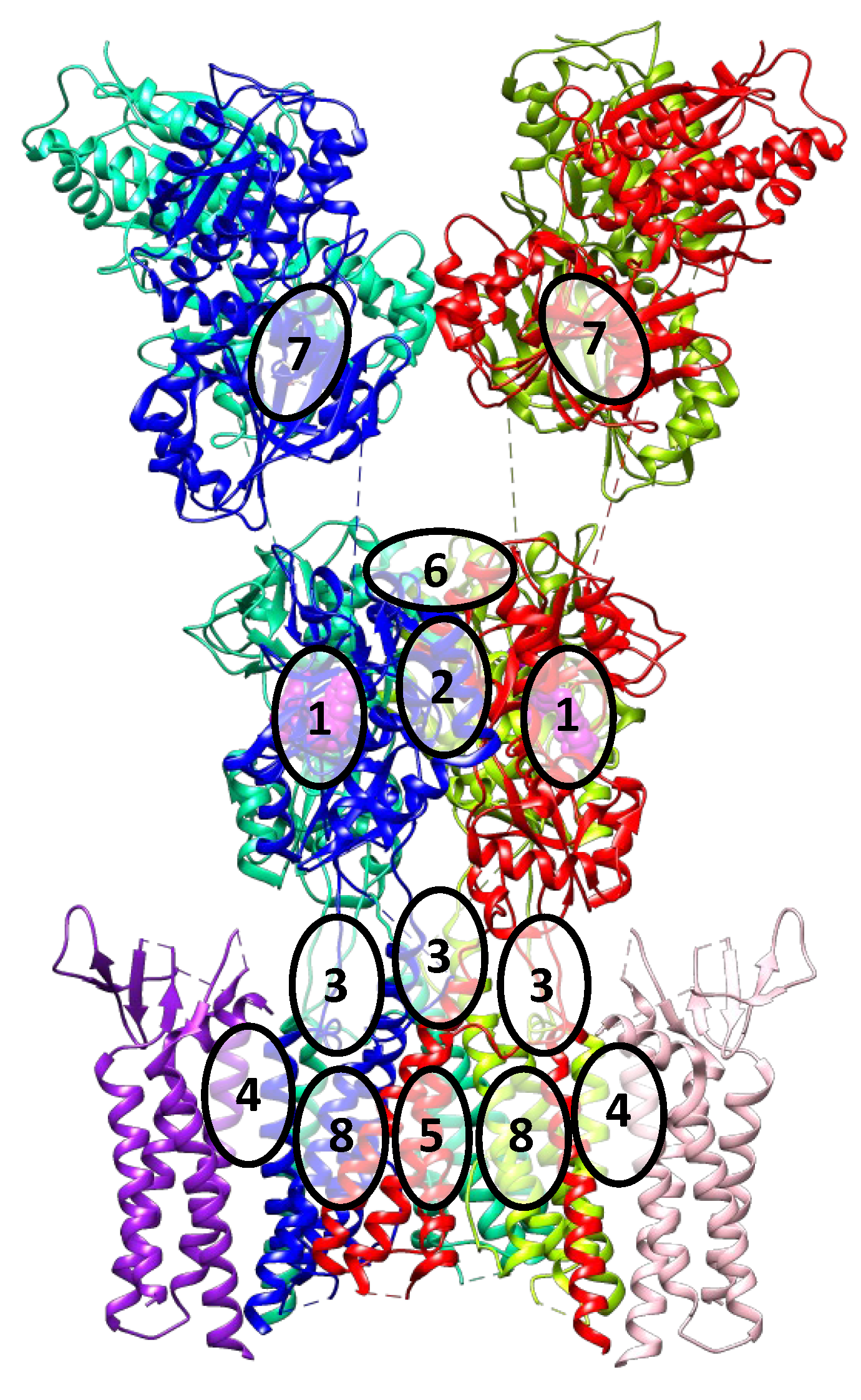
3. Binding Sites and Ligand Types of the AMPA Receptor
- (1).
- The orthosteric binding site for agonists and competitive antagonists is located inside the clamshell of the ligand-binding domain;
- (2).
- The positive allosteric modulator binding site is located at the interface between the subunits of the dimeric ligand-binding domain;
- (3).
- The binding site for negative allosteric modulators (non-competitive antagonists, non-competitive inhibitors) is located in the linker region between the ligand-binding and transmembrane domains;
- (4).
- The binding site for TARP-dependent allosteric modulators is located at the interface between the interacting transmembrane segments of TARP and the receptor;
- (5).
- Ion channel blocker binding sites are located in the pore of the ion channel (mainly for the Ca2+-permeable forms of the receptor);
- (6).
- Con-ikot-ikot, a protein toxin from the Conus striatus cone snail, acts as an AMPAR positive allosteric modulator by binding on top of the LBD dimer-of-dimers (in the free space between LBDs and ATDs). This effectively immobilizes the LBD layer of the receptor and prevents desensitization, leading to prolonged receptor activation (although mostly to only partially open states), over-excitation, and toxicity [3,43]. It is not yet clear if this binding site could be exploited by the small molecule ligands.
- (7).
- Based on molecular modeling and structural data, another potential binding site is predicted to be located at the interface between the lower lobes of the subunits of the dimeric amino-terminal domain [44]. Presumably, this site is present (or druggable) only in the GluA3 receptors, while the ligands interacting with it are not known yet. Its position at the dimer interface is similar to that of the binding sites in related NMDA receptors, as well as in metabotropic glutamate receptors. Taking into account the specificity of the amino acid sequence of the AMPA receptor amino-terminal domain, these data make it a promising potential target for the development of new modulators with high selectivity.
- (8).
- Based on the analysis of the structure of the AMPA receptor complex with trans-4-butylcyclohexane carboxylic acid (4-BCCA), new binding sites were found in the transmembrane domain. Driven by the structural data as well as site-directed mutagenesis and molecular modeling, three possible mechanisms of action of 4-BCCA were proposed that involve either direct blocking of the ion channel (interfering with the flow of permeant ions), or influencing the dynamics of the M3 helices, or destabilizing the protein surface through competition with the surrounding membrane lipids [45]. Presumably, other related compounds which are inhibitors of the AMPA receptor also bind at these sites and can serve as promising potential antiepileptic drugs. Moreover, these sites are located in close proximity to those for the negative allosteric modulators, which opens the possibility of interaction of the respective structural fragments and could explain the synergistic effect observed with the simultaneous administration of perampanel and decanoic acid [46].
3.1. The Orthosteric Binding Site for Agonists and Competitive Antagonists
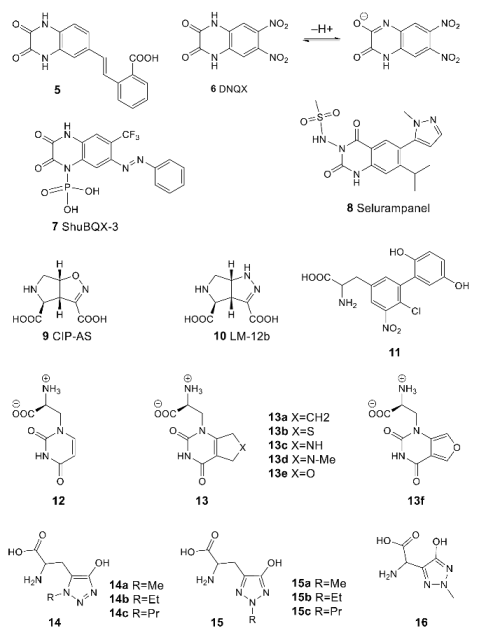
3.2. The Positive Allosteric Modulator (PAM) Binding Site
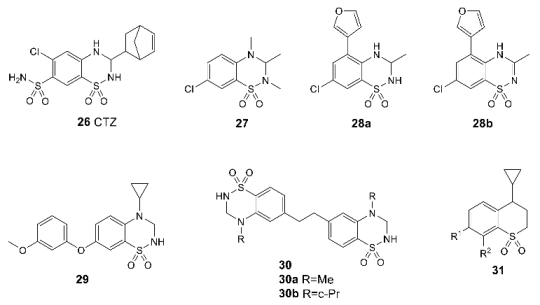

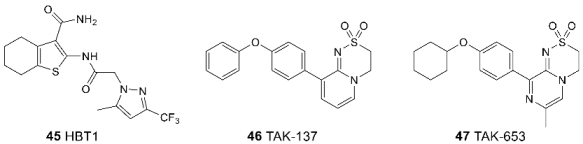
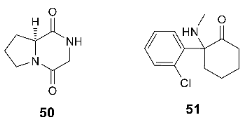
3.3. Binding Site for Negative Allosteric Modulators (Non-competitive Antagonists, Non-competitive Inhibitors)
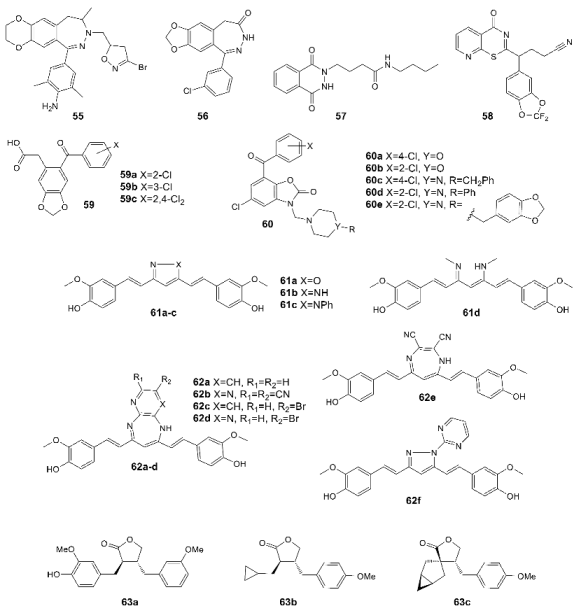
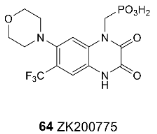
3.4. Chemotypes with Activity Cliffs
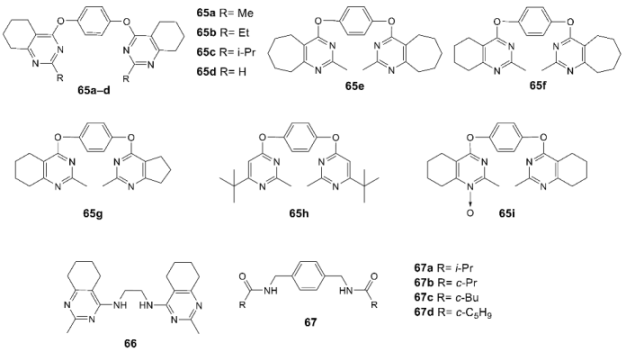
3.5. Binding Site for TARP-Dependent Allosteric Modulators
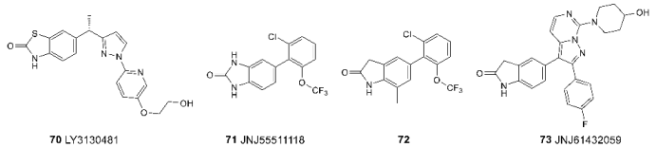
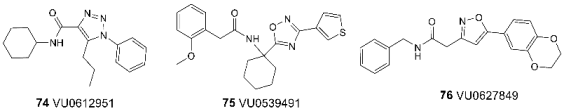
3.6. Binding Sites for Ion Channel Blockers
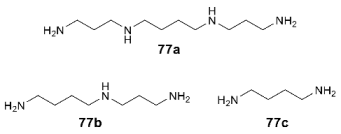
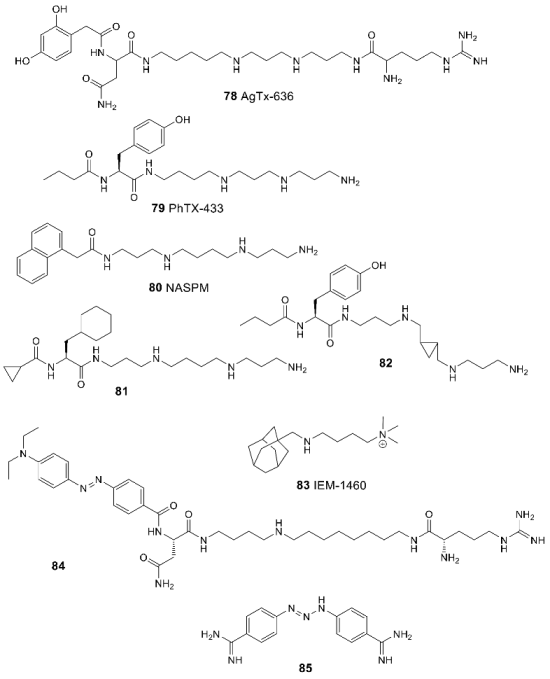
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reiner, A.; Levitz, J. Glutamatergic signaling in the central nervous system: Ionotropic and metabotropic receptors in concert. Neuron 2018, 98, 1080–1098. [Google Scholar] [CrossRef] [PubMed]
- Brogi, S.; Campiani, G.; Brindisi, M.; Butini, S. Allosteric modulation of ionotropic glutamate receptors: An outlook on new therapeutic approaches to treat central nervous system disorders. ACS Med. Chem. Lett. 2019, 10, 228–236. [Google Scholar] [CrossRef]
- Hansen, K.B.; Wollmuth, L.P.; Bowie, D.; Furukawa, H.; Menniti, F.S.; Sobolevsky, A.I.; Swanson, G.T.; Swanger, S.A.; Greger, I.H.; Nakagawa, T.; et al. Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol. Rev. 2021, 73, 298–487. [Google Scholar] [CrossRef] [PubMed]
- Watkins, J.C.; Evans, R.H. Excitatory amino acid transmitters. Annu. Rev. Pharmacol. Toxicol. 1981, 21, 165–204. [Google Scholar] [CrossRef]
- Evans, R.H.; Watkins, J.C. Team Evans and Watkins: Excitatory amino acid research at Bristol University 1973–1981. Neuropharmacology 2021, 198, 108768. [Google Scholar] [CrossRef]
- Honoré, T.; Lauridsen, J.; Krogsgaard-Larsen, P. The binding of [3H]AMPA, a structural analogue of glutamic acid, to rat brain membranes. J. Neurochem. 1982, 38, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Partin, K.M. AMPA receptor potentiators: From drug design to cognitive enhancement. Curr. Opin. Pharmacol. 2015, 20, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Lauterborn, J.C.; Palmer, L.C.; Jia, Y.; Pham, D.T.; Hou, B.; Wang, W.; Trieu, B.H.; Cox, C.D.; Kantorovich, S.; Gall, C.M.; et al. Chronic ampakine treatments stimulate dendritic growth and promote learning in middle-aged rats. J. Neurosci. 2016, 36, 1636–1646. [Google Scholar] [CrossRef]
- Lee, K.; Goodman, L.; Fourie, C.; Schenk, S.; Leitch, B.; Montgomery, J.M. AMPA receptors as therapeutic targets for neurological disorders. In Ion Channels as Therapeutic Targets, Part A; Donev, R., Ed.; Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2016; Volume 103, pp. 203–261. [Google Scholar] [CrossRef]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.; Myers, S.J.; Dingledine, R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef]
- Pirotte, B.; Francotte, P.; Goffin, E.; de Tullio, P. AMPA receptor positive allosteric modulators: A patent review. Expert Opin. Ther. Pat. 2013, 23, 615–628. [Google Scholar] [CrossRef]
- Ward, S.E.; Pennicott, L.E.; Beswick, P. AMPA receptor-positive allosteric modulators for the treatment of schizophrenia: An overview of recent patent applications. Future Med. Chem. 2015, 7, 473–491. [Google Scholar] [CrossRef]
- Reuillon, T.; Ward, S.E.; Beswick, P. AMPA receptor positive allosteric modulators: Potential for the treatment of neuropsychiatric and neurological disorders. Curr. Top. Med. Chem. 2016, 16, 3536–3565. [Google Scholar] [CrossRef] [PubMed]
- Greger, I.H.; Watson, J.F.; Cull-Candy, S.G. Structural and functional architecture of AMPA-type glutamate receptors and their auxiliary proteins. Neuron 2017, 94, 713–730. [Google Scholar] [CrossRef]
- Twomey, E.C.; Sobolevsky, A.I. Structural mechanisms of gating in ionotropic glutamate receptors. Biochemistry 2018, 57, 267–276. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, S.; Swensen, A.C.; Qian, W.-J.; Gouaux, E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 2019, 364, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Sommer, B.; Keinänen, K.; Verdoorn, T.A.; Wisden, W.; Burnashev, N.; Herb, A.; Köhler, M.; Takagi, T.; Sakmann, B.; Seeburg, P.H. Flip and flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science 1990, 249, 1580–1585. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, G.; Regan, P.; Whitcomb, D.J.; Cho, K. Ca2+-permeable AMPA receptor: A new perspective on amyloid-beta mediated pathophysiology of Alzheimer’s disease. Neuropharmacology 2017, 112, 221–227. [Google Scholar] [CrossRef]
- Twomey, E.C.; Yelshanskaya, M.V.; Vassilevski, A.A.; Sobolevsky, A.I. Mechanisms of channel block in calcium-permeable AMPA receptors. Neuron 2018, 99, 956–968. [Google Scholar] [CrossRef]
- Umino, M.; Umino, A.; Nishikawa, T. Effects of selective calcium-permeable AMPA receptor blockade by IEM 1460 on psychotomimetic-induced hyperactivity in the mouse. J. Neural Transm. 2018, 125, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Rozov, A.; Burnashev, N. Fast interaction between AMPA and NMDA receptors by intracellular calcium. Cell Calcium 2016, 60, 407–414. [Google Scholar] [CrossRef]
- Guo, C.; Ma, Y.-Y. Calcium permeable AMPA receptors and excitotoxicity in neurological disorders. Front. Neural Circuits 2021, 15, 711564. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Guntupalli, S.; Jang, S.E.; Anggono, V. Regulation of AMPA receptor trafficking by protein ubiquitination. Front. Mol. Neurosci. 2017, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T. Post-translational palmitoylation of ionotropic glutamate receptors in excitatory synaptic functions. Br. J. Pharmacol. 2021, 178, 784–797. [Google Scholar] [CrossRef]
- Chen, S.; Gouaux, E. Structure and mechanism of AMPA receptor—Auxiliary protein complexes. Curr. Opin. Struct. Biol. 2019, 54, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Coombs, I.D.; Soto, D.; McGee, T.P.; Gold, M.G.; Farrant, M.; Cull-Candy, S.G. Homomeric GluA2(R) AMPA receptors can conduct when desensitized. Nat. Commun. 2019, 10, 4312. [Google Scholar] [CrossRef] [PubMed]
- Salazar, H.; Mischke, S.; Plested, A.J.R. Measurements of the timescale and conformational space of AMPA receptor desensitization. Biophys. J. 2020, 119, 206–218. [Google Scholar] [CrossRef]
- Kamalova, A.; Nakagawa, T. AMPA receptor structure and auxiliary subunits. J. Physiol. 2021, 599, 453–469. [Google Scholar] [CrossRef]
- Jacobi, E.; von Engelhardt, J. Modulation of information processing by AMPA receptor auxiliary subunits. J. Physiol. 2021, 599, 471–483. [Google Scholar] [CrossRef]
- Abdollahi Nejat, M.; Klaassen, R.V.; Spijker, S.; Smit, A.B. Auxiliary subunits of the AMPA receptor: The Shisa family of proteins. Curr. Opin. Pharmacol. 2021, 58, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Radin, D.P.; Li, Y.-X.; Rogers, G.; Purcell, R.; Lippa, A. Tarps differentially affect the pharmacology of ampakines. Biochem. Pharmacol. 2018, 154, 446–451. [Google Scholar] [CrossRef]
- Ishii, T.; Stolz, J.R.; Swanson, G.T. Auxiliary proteins are the predominant determinants of differential efficacy of clinical candidates acting as AMPA receptor positive allosteric modulators. Mol. Pharmacol. 2020, 97, 336–350. [Google Scholar] [CrossRef]
- Kato, A.S.; Witkin, J.M. Auxiliary subunits of AMPA receptors: The discovery of a forebrain-selective antagonist, LY3130481/CERC-611. Biochem. Pharmacol. 2018, 147, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Knopp, K.L.; Simmons, R.M.A.; Guo, W.; Adams, B.L.; Gardinier, K.M.; Gernert, D.L.; Ornstein, P.L.; Porter, W.; Reel, J.; Ding, C.; et al. Modulation of TARP γ8-containing AMPA receptors as a novel therapeutic approach for chronic pain. J. Pharmacol. Exp. Ther. 2019, 369, 345–363. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Vicente, D.; Bayés, À. AMPA receptor auxiliary subunits emerged during early vertebrate evolution by neo/subfunctionalization of unrelated proteins. Open Biol. 2020, 10, 200234. [Google Scholar] [CrossRef] [PubMed]
- Riva, I.; Eibl, C.; Volkmer, R.; Carbone, A.L.; Plested, A.J. Control of AMPA receptor activity by the extracellular loops of auxiliary proteins. eLife 2017, 6, e28680. [Google Scholar] [CrossRef]
- Carrillo, E.; Shaikh, S.A.; Berka, V.; Durham, R.J.; Litwin, D.B.; Lee, G.; MacLean, D.M.; Nowak, L.M.; Jayaraman, V. Mechanism of modulation of AMPA receptors by TARP-γ8. J. Gen. Physiol. 2020, 152, e201912451. [Google Scholar] [CrossRef] [PubMed]
- Cais, O.; Herguedas, B.; Krol, K.; Cull-Candy, S.G.; Farrant, M.; Greger, I.H. Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating. Cell Rep. 2014, 9, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Baranovic, J. AMPA receptors in the synapse: Very little space and even less time. Neuropharmacology 2021, 196, 108711. [Google Scholar] [CrossRef]
- Matthews, P.M.; Pinggera, A.; Kampjut, D.; Greger, I.H. Biology of AMPA receptor interacting proteins—From biogenesis to synaptic plasticity. Neuropharmacology 2021, 197, 108709. [Google Scholar] [CrossRef]
- Díaz-Alonso, J.; Nicoll, R.A. AMPA receptor trafficking and LTP: Carboxy-termini, amino-termini and TARPs. Neuropharmacology 2021, 197, 108710. [Google Scholar] [CrossRef]
- Wu, Q.-L.; Gao, Y.; Li, J.-T.; Ma, W.-Y.; Chen, N.-H. The role of AMPARs composition and trafficking in synaptic plasticity and diseases. Cell. Mol. Neurobiol. 2022, 42, 2489–2504. [Google Scholar] [CrossRef] [PubMed]
- Baranovic, J.; Braunbeck, S.; Zaki, N.; Minniberger, S.; Chebli, M.; Plested, A.J.R. The action of Con-ikot-ikot toxin on single AMPA-type glutamate receptors. J. Gen. Physiol. 2022, 154, e202112912. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Krieger, J.; Herguedas, B.; García-Nafría, J.; Dutta, A.; Shaikh, S.A.; Greger, I.H.; Bahar, I. Druggability simulations and X-ray crystallography reveal a ligand-binding site in the GluA3 AMPA receptor N-terminal domain. Structure 2019, 27, 241–252. [Google Scholar] [CrossRef]
- Yelshanskaya, M.V.; Singh, A.K.; Narangoda, C.; Williams, R.S.B.; Kurnikova, M.G.; Sobolevsky, A.I. Structural basis of AMPA receptor inhibition by trans-4-butylcyclohexane carboxylic acid. Br. J. Pharmacol. 2022, 179, 3628–3644. [Google Scholar] [CrossRef]
- Augustin, K.; Williams, S.; Cunningham, M.; Devlin, A.M.; Friedrich, M.; Jayasekera, A.; Hussain, M.A.; Holliman, D.; Mitchell, P.; Jenkins, A.; et al. Perampanel and decanoic acid show synergistic action against AMPA receptors and seizures. Epilepsia 2018, 59, e172–e178. [Google Scholar] [CrossRef]
- Blakemore, L.J.; Trombley, P.Q. Mechanisms of zinc modulation of olfactory bulb AMPA receptors. Neuroscience 2019, 410, 160–175. [Google Scholar] [CrossRef]
- Carrillo, E.; Bhatia, N.K.; Akimzhanov, A.M.; Jayaraman, V. Activity dependent inhibition of AMPA receptors by Zn2+. J. Neurosci. 2020, 40, 8629–8636. [Google Scholar] [CrossRef]
- Ivanova, V.O.; Balaban, P.M.; Bal, N.V. Modulation of AMPA receptors by nitric oxide in nerve cells. Int. J. Mol. Sci. 2020, 21, 981. [Google Scholar] [CrossRef] [PubMed]
- Krintel, C.; Dorosz, J.; Larsen, A.H.; Thorsen, T.S.; Venskutonytė, R.; Mirza, O.; Gajhede, M.; Boesen, T.; Kastrup, J.S. Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2. FEBS J. 2021, 288, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Shi, E.Y.; Yuan, C.L.; Sipple, M.T.; Srinivasan, J.; Ptak, C.P.; Oswald, R.E.; Nowak, L.M. Noncompetitive antagonists induce cooperative AMPA receptor channel gating. J. Gen. Physiol. 2019, 151, 156–173. [Google Scholar] [CrossRef]
- Coombs, I.D.; Cull-Candy, S.G. Single-channel mechanisms underlying the function, diversity and plasticity of AMPA receptors. Neuropharmacology 2021, 198, 108781. [Google Scholar] [CrossRef] [PubMed]
- Carannante, I.; Johansson, Y.; Silberberg, G.; Hellgren Kotaleski, J. Data-driven model of postsynaptic currents mediated by NMDA or AMPA receptors in striatal neurons. Front. Comput. Neurosci. 2022, 16, 806086. [Google Scholar] [CrossRef] [PubMed]
- Pollok, S.; Reiner, A. Subunit-selective iGluR antagonists can potentiate heteromeric receptor responses by blocking desensitization. Proc. Natl. Acad. Sci. USA 2020, 117, 25851–25858. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Kunugi, A.; Tajima, Y.; Suzuki, N.; Suzuki, M.; Toyofuku, M.; Kuno, H.; Sogabe, S.; Kosugi, Y.; Awasaki, Y.; et al. Strictly regulated agonist-dependent activation of AMPA-R is the key characteristic of TAK-653 for robust synaptic responses and cognitive improvement. Sci. Rep. 2021, 11, 14532. [Google Scholar] [CrossRef] [PubMed]
- Demmer, C.S.; Rombach, D.; Liu, N.; Nielsen, B.; Pickering, D.S.; Bunch, L. Revisiting the quinoxalinedione scaffold in the construction of new ligands for the ionotropic glutamate receptors. ACS Chem. Neurosci. 2017, 8, 2477–2495. [Google Scholar] [CrossRef]
- Dudić, A.; Reiner, A. Quinoxalinedione deprotonation is important for glutamate receptor binding. Biol. Chem. 2019, 400, 927–938. [Google Scholar] [CrossRef]
- Barber, D.M.; Liu, S.-A.; Gottschling, K.; Sumser, M.; Hollmann, M.; Trauner, D. Optical control of AMPA receptors using a photoswitchable quinoxaline-2,3-dione antagonist. Chem. Sci. 2017, 8, 611–615. [Google Scholar] [CrossRef]
- Orain, D.; Tasdelen, E.; Haessig, S.; Koller, M.; Picard, A.; Dubois, C.; Lingenhoehl, K.; Desrayaud, S.; Floersheim, P.; Carcache, D.; et al. Design and synthesis of Selurampanel, a novel orally active and competitive AMPA receptor antagonist. ChemMedChem 2017, 12, 197–201. [Google Scholar] [CrossRef]
- Møllerud, S.; Pinto, A.; Marconi, L.; Frydenvang, K.; Thorsen, T.S.; Laulumaa, S.; Venskutonytė, R.; Winther, S.; Moral, A.M.C.; Tamborini, L.; et al. Structure and affinity of two bicyclic glutamate analogues at AMPA and kainate receptors. ACS Chem. Neurosci. 2017, 8, 2056–2064. [Google Scholar] [CrossRef]
- Szymańska, E.; Chałupnik, P.; Johansen, T.N.; Nielsen, B.; Cuñado Moral, A.M.; Pickering, D.S.; Więckowska, A.; Kieć-Kononowicz, K. Aryl- and heteroaryl-substituted phenylalanines as AMPA receptor ligands. Chem. Biol. Drug. Des. 2017, 90, 1271–1281. [Google Scholar] [CrossRef]
- Szymańska, E.; Nielsen, B.; Johansen, T.N.; Cuñado Moral, A.M.; Pickering, D.S.; Szczepańska, K.; Mickowska, A.; Kieć-Kononowicz, K. Pharmacological characterization and binding modes of novel racemic and optically active phenylalanine-based antagonists of AMPA receptors. Eur. J. Med. Chem. 2017, 138, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Latacz, G.; Sałat, K.; Furgała-Wojas, A.; Martyniak, A.; Olejarz-Maciej, A.; Honkisz-Orzechowska, E.; Szymańska, E. Phenylalanine-based AMPA receptor antagonist as the anticonvulsant agent with neuroprotective activity—In vitro and in vivo studies. Molecules 2022, 27, 875. [Google Scholar] [CrossRef] [PubMed]
- Brogi, S.; Brindisi, M.; Butini, S.; Kshirsagar, G.U.; Maramai, S.; Chemi, G.; Gemma, S.; Campiani, G.; Novellino, E.; Fiorenzani, P.; et al. (S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic acid (AMPA) and kainate receptor ligands: Further exploration of bioisosteric replacements and structural and biological investigation. J. Med. Chem. 2018, 61, 2124–2130. [Google Scholar] [CrossRef]
- Frydenvang, K.; Pickering, D.S.; Kshirsagar, G.U.; Chemi, G.; Gemma, S.; Sprogøe, D.; Kærn, A.M.; Brogi, S.; Campiani, G.; Butini, S.; et al. Ionotropic glutamate receptor GluA2 in complex with bicyclic pyrimidinedione-based compounds: When small compound modifications have distinct effects on binding interactions. ACS Chem. Neurosci. 2020, 11, 1791–1800. [Google Scholar] [CrossRef]
- Sainas, S.; Temperini, P.; Farnsworth, J.C.; Yi, F.; Møllerud, S.; Jensen, A.A.; Nielsen, B.; Passoni, A.; Kastrup, J.S.; Hansen, K.B.; et al. Use of the 4-hydroxytriazole moiety as a bioisosteric tool in the development of ionotropic glutamate receptor ligands. J. Med. Chem. 2019, 62, 4467–4482. [Google Scholar] [CrossRef]
- Xiao, D.; Xie, F.; Xu, X.; Zhou, X. The impact and mechanism of ampakine CX1739 on protection against respiratory depression in rats. Future Med. Chem. 2020, 12, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Hara, H.; Kimura, H. Role of the AMPA receptor in antidepressant effects of ketamine and potential of AMPA receptor potentiators as a novel antidepressant. Neuropharmacology 2022, 222, 109308. [Google Scholar] [CrossRef]
- Kadriu, B.; Musazzi, L.; Johnston, J.N.; Kalynchuk, L.E.; Caruncho, H.J.; Popoli, M.; Zarate, C.A. Positive AMPA receptor modulation in the treatment of neuropsychiatric disorders: A long and winding road. Drug Discov. Today 2021, 26, 2816–2838. [Google Scholar] [CrossRef]
- Cetin, S.; Knez, D.; Gobec, S.; Kos, J.; Pišlar, A. Cell models for Alzheimer’s and Parkinson’s disease: At the interface of biology and drug discovery. Biomed. Pharmacother. 2022, 149, 112924. [Google Scholar] [CrossRef]
- Banerjee, R.; Rai, A.; Iyer, S.M.; Narwal, S.; Tare, M. Animal models in the study of Alzheimer’s disease and Parkinson’s disease: A historical perspective. Anim. Model. Exp. Med. 2022, 5, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chen, X.; Li, S.; Li, X.-J. Genetically modified large animal models for investigating neurodegenerative diseases. Cell Biosci. 2021, 11, 218. [Google Scholar] [CrossRef] [PubMed]
- Rapaka, D.; Adiukwu, P.C.; Bitra, V.R. Experimentally induced animal models for cognitive dysfunction and Alzheimer’s disease. MethodsX 2022, 9, 101933. [Google Scholar] [CrossRef] [PubMed]
- Białoń, M.; Wąsik, A. Advantages and limitations of animal schizophrenia models. Int. J. Mol. Sci. 2022, 23, 5968. [Google Scholar] [CrossRef] [PubMed]
- Frydenvang, K.; Pickering, D.S.; Kastrup, J.S. Structural basis for positive allosteric modulation of AMPA and kainate receptors. J. Physiol. 2022, 600, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Drapier, T.; Geubelle, P.; Bouckaert, C.; Nielsen, L.; Laulumaa, S.; Goffin, E.; Dilly, S.; Francotte, P.; Hanson, J.; Pochet, L.; et al. Enhancing action of positive allosteric modulators through the design of dimeric compounds. J. Med. Chem. 2018, 61, 5279–5291. [Google Scholar] [CrossRef]
- Karlov, D.S.; Lavrov, M.I.; Palyulin, V.A.; Zefirov, N.S. Pharmacophore analysis of positive allosteric modulators of AMPA receptors. Russ. Chem. Bull. 2016, 65, 581–587. [Google Scholar] [CrossRef]
- Radchenko, E.V.; Karlov, D.S.; Lavrov, M.I.; Palyulin, V.A. Structural requirements for molecular design of positive allosteric modulators of AMPA receptor. Mendeleev Commun. 2017, 27, 623–625. [Google Scholar] [CrossRef]
- Karlov, D.S.; Lavrov, M.I.; Palyulin, V.A.; Zefirov, N.S. MM-GBSA and MM-PBSA performance in activity evaluation of AMPA receptor positive allosteric modulators. J. Biomol. Struct. Dyn. 2018, 36, 2508–2516. [Google Scholar] [CrossRef]
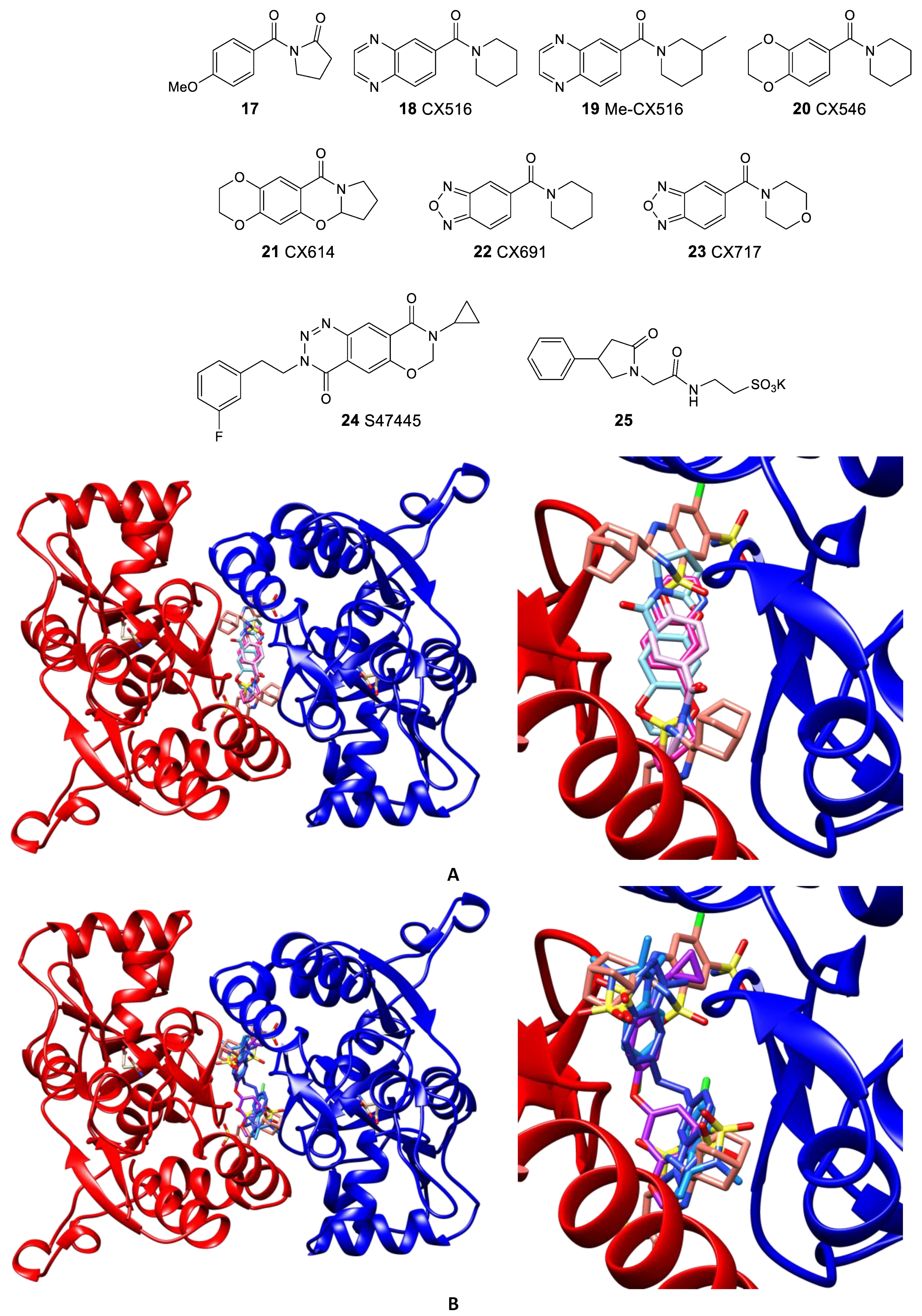
- Krintel, C.; Harpsøe, K.; Zachariassen, L.G.; Peters, D.; Frydenvang, K.; Pickering, D.S.; Gajhede, M.; Kastrup, J.S. Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1645–1652. [Google Scholar] [CrossRef]
- Grove, S.J.A.; Jamieson, C.; Maclean, J.K.F.; Morrow, J.A.; Rankovic, Z. Positive allosteric modulators of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor. J. Med. Chem. 2010, 53, 7271–7279. [Google Scholar] [CrossRef]
- Giralt, A.; Gómez-Climent, M.Á.; Alcalá, R.; Bretin, S.; Bertrand, D.; María Delgado-García, J.; Pérez-Navarro, E.; Alberch, J.; Gruart, A. The AMPA receptor positive allosteric modulator S 47445 rescues in vivo CA3-CA1 long-term potentiation and structural synaptic changes in old mice. Neuropharmacology 2017, 123, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Borozdenko, D.A.; Ezdoglian, A.A.; Shmigol, T.A.; Gonchar, D.I.; Lyakhmun, D.N.; Tarasenko, D.V.; Golubev, Y.V.; Cherkashova, E.A.; Namestnikova, D.D.; Gubskiy, I.L.; et al. A novel phenylpyrrolidine derivative: Synthesis and effect on cognitive functions in rats with experimental ishemic stroke. Molecules 2021, 26, 6124. [Google Scholar] [CrossRef] [PubMed]
- Murru, L.; Vezzoli, E.; Longatti, A.; Ponzoni, L.; Falqui, A.; Folci, A.; Moretto, E.; Bianchi, V.; Braida, D.; Sala, M.; et al. Pharmacological modulation of AMPAR rescues intellectual disability-like phenotype in Tm4sf2-/y mice. Cereb. Cortex. 2017, 27, 5369–5384. [Google Scholar] [CrossRef] [PubMed]
- Tatsukawa, T.; Raveau, M.; Ogiwara, I.; Hattori, S.; Miyamoto, H.; Mazaki, E.; Itohara, S.; Miyakawa, T.; Montal, M.; Yamakawa, K. Scn2a haploinsufficient mice display a spectrum of phenotypes affecting anxiety, sociability, memory flexibility and ampakine CX516 rescues their hyperactivity. Mol. Autism 2019, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Rhine, M.A.; Parrott, J.M.; Schultz, M.N.; Kazdoba, T.M.; Crawley, J.N. Hypothesis-driven investigations of diverse pharmacological targets in two mouse models of autism. Autism Res. 2019, 12, 401–421. [Google Scholar] [CrossRef]
- Guo, B.; Chen, J.; Chen, Q.; Ren, K.; Feng, D.; Mao, H.; Yao, H.; Yang, J.; Liu, H.; Liu, Y.; et al. Anterior cingulate cortex dysfunction underlies social deficits in Shank3 mutant mice. Nat. Neurosci. 2019, 22, 1223–1234. [Google Scholar] [CrossRef]
- Cavalleri, L.; Merlo Pich, E.; Millan, M.J.; Chiamulera, C.; Kunath, T.; Spano, P.F.; Collo, G. Ketamine enhances structural plasticity in mouse mesencephalic and human iPSC-derived dopaminergic neurons via AMPAR-driven BDNF and mTOR signaling. Mol. Psychiatry 2018, 23, 812–823. [Google Scholar] [CrossRef]
- Mozafari, N.; Shamsizadeh, A.; Fatemi, I.; Allahtavakoli, M.; Moghadam-Ahmadi, A.; Kaviani, E.; Kaeidi, A. CX691, as an AMPA receptor positive modulator, improves the learning and memory in a rat model of Alzheimer’s disease. Iran. J. Basic Med. Sci. 2018, 21, 724–730. [Google Scholar] [CrossRef]
- Gordillo-Salas, M.; Pascual-Antón, R.; Ren, J.; Greer, J.; Adell, A. Antidepressant-like effects of CX717, a positive allosteric modulator of AMPA receptors. Mol. Neurobiol. 2020, 57, 3498–3507. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.; Savino, E.; Mocaer, E.; Bretin, S.; Racagni, G.; Riva, M.A. Upregulation of neurotrophins by S47445, a novel positive allosteric modulator of AMPA receptors in aged rats. Pharmacol. Res. 2017, 121, 59–69. [Google Scholar] [CrossRef]
- Mendez-David, I.; Guilloux, J.-P.; Papp, M.; Tritschler, L.; Mocaer, E.; Gardier, A.M.; Bretin, S.; David, D.J. S47445 produces antidepressant- and anxiolytic-like effects through neurogenesis dependent and independent mechanisms. Front. Pharmacol. 2017, 8, 462. [Google Scholar] [CrossRef] [PubMed]
- Pilar-Cuellar, F.; Castro, E.; Bretin, S.; Mocaer, E.; Pazos, Á.; Díaz, Á. S 47445 counteracts the behavioral manifestations and hippocampal neuroplasticity changes in bulbectomized mice. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 93, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Bretin, S.; Louis, C.; Seguin, L.; Wagner, S.; Thomas, J.-Y.; Challal, S.; Rogez, N.; Albinet, K.; Iop, F.; Villain, N.; et al. Pharmacological characterisation of S 47445, a novel positive allosteric modulator of AMPA receptors. PLoS ONE 2017, 12, e0184429. [Google Scholar] [CrossRef]
- Morley-Fletcher, S.; Zuena, A.R.; Mairesse, J.; Gatta, E.; Van Camp, G.; Bouwalerh, H.; Riozzi, B.; Battaglia, G.; Pittaluga, A.; Olivero, G.; et al. The reduction in glutamate release is predictive of cognitive and emotional alterations that are corrected by the positive modulator of AMPA receptors S 47445 in perinatal stressed rats. Neuropharmacology 2018, 135, 284–296. [Google Scholar] [CrossRef]
- Louis, C.; Llopis, K.; Danober, L.; Panayi, F.; Lestage, P.; Beracochea, D. New procognitive enhancers acting at the histamine H3 and AMPA receptors reverse natural forgetting in mice: Comparisons with donepezil and memantine in the object recognition task. Behav. Pharmacol. 2019, 30, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Bernard, K.; Gouttefangeas, S.; Bretin, S.; Galtier, S.; Robert, P.; Holthoff-Detto, V.; Cummings, J.; Pueyo, M. A 24-week double-blind placebo-controlled study of the efficacy and safety of the AMPA modulator S47445 in patients with mild to moderate Alzheimer’s disease and depressive symptoms. Alzheimer’s Dement. 2019, 5, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.P.; Francotte, P.; Frydenvang, K.; Tapken, D.; Goffin, E.; Fraikin, P.; Caignard, D.-H.; Lestage, P.; Danober, L.; Pirotte, B.; et al. Synthesis and pharmacology of mono-, di-, and trialkyl-substituted 7-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxides combined with X-ray structure analysis to understand the unexpected structure-activity relationship at AMPA receptors. ACS Chem. Neurosci. 2016, 7, 378–390. [Google Scholar] [CrossRef]
- Citti, C.; Battisti, U.M.; Cannazza, G.; Jozwiak, K.; Stasiak, N.; Puja, G.; Ravazzini, F.; Ciccarella, G.; Braghiroli, D.; Parenti, C.; et al. 7-Chloro-5-(furan-3-yl)-3-methyl-4H-benzo[e][1,2,4]thiadiazine 1,1-dioxide as positive allosteric modulator of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor. The end of the unsaturated-inactive paradigm? ACS Chem. Neurosci. 2016, 7, 149–160. [Google Scholar] [CrossRef]
- Krintel, C.; Francotte, P.; Pickering, D.S.; Juknaitė, L.; Pøhlsgaard, J.; Olsen, L.; Frydenvang, K.; Goffin, E.; Pirotte, B.; Kastrup, J.S. Enthalpy-entropy compensation in the binding of modulators at ionotropic glutamate receptor GluA2. Biophys. J. 2016, 110, 2397–2406. [Google Scholar] [CrossRef]
- Battisti, U.M.; Citti, C.; Rastelli, G.; Pinzi, L.; Puja, G.; Ravazzini, F.; Ciccarella, G.; Braghiroli, D.; Cannazza, G. An unexpected reversal in the pharmacological stereoselectivity of benzothiadiazine AMPA positive allosteric modulators. Med. Chem. Commun. 2016, 7, 2410–2417. [Google Scholar] [CrossRef]
- Goffin, E.; Drapier, T.; Larsen, A.P.; Geubelle, P.; Ptak, C.P.; Laulumaa, S.; Rovinskaja, K.; Gilissen, J.; de Tullio, P.; Olsen, L.; et al. 7-Phenoxy-substituted 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxides as positive allosteric modulators of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors with nanomolar potency. J. Med. Chem. 2018, 61, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Etsè, K.S.; Dorosz, J.; McLain Christensen, K.; Thomas, J.-Y.; Botez Pop, I.; Goffin, E.; Colson, T.; Lestage, P.; Danober, L.; Pirotte, B.; et al. Development of thiochroman dioxide analogues of benzothiadiazine dioxides as new positive allosteric modulators of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. ACS Chem. Neurosci. 2021, 12, 2679–2692. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, J.T.; Fitzpatrick, C.M.; Larsen, M.; Skovgaard, L.; Nielsen, S.D.; Clausen, R.P.; Troelsen, K.; Pickering, D.S. Differential role of AMPA receptors in mouse tests of antidepressant and anxiolytic action. Brain. Res. 2015, 1601, 117–126. [Google Scholar] [CrossRef]
- Fitzpatrick, C.M.; Larsen, M.; Madsen, L.H.; Caballero-Puntiverio, M.; Pickering, D.S.; Clausen, R.P.; Andreasen, J.T. Positive allosteric modulation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid glutamate receptors differentially modulates the behavioural effects of citalopram in mouse models of antidepressant and anxiolytic action. Behav. Pharmacol. 2016, 27, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.E.; Bax, B.D.; Harries, M. Challenges for and current status of research into positive modulators of AMPA receptors. Br. J. Pharmacol. 2010, 160, 181–190. [Google Scholar] [CrossRef]
- Ward, S.E.; Beswick, P.; Calcinaghi, N.; Dawson, L.A.; Gartlon, J.; Graziani, F.; Jones, D.N.C.; Lacroix, L.; Selina Mok, M.H.; Oliosi, B.; et al. Pharmacological characterization of N-[(2S)-5-(6-fluoro-3-pyridinyl)-2, 3-dihydro-1H-inden-2-yl]-2-propanesulfonamide: A novel, clinical AMPA receptor positive allosteric modulator. Br. J. Pharmacol. 2017, 174, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, C.L.; Patel, N.C.; Schwarz, J.; Scialis, R.J.; Wei, Y.; Hou, X.J.; Xie, L.; Karki, K.; Bryce, D.K.; Osgood, S.M.; et al. The discovery and characterization of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor potentiator N-{(3S,4S)-4-[4-(5-cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242). J. Med. Chem. 2015, 58, 4291–4308. [Google Scholar] [CrossRef]
- Jamieson, C.; Basten, S.; Campbell, R.A.; Cumming, I.A.; Gillen, K.J.; Gillespie, J.; Kazemier, B.; Kiczun, M.; Lamont, Y.; Lyons, A.J.; et al. A novel series of positive modulators of the AMPA receptor: Discovery and structure based hit-to-lead studies. Bioorg. Med. Chem. Lett. 2010, 20, 5753–5756. [Google Scholar] [CrossRef]
- Jamieson, C.; Campbell, R.A.; Cumming, I.A.; Gillen, K.J.; Gillespie, J.; Kazemier, B.; Kiczun, M.; Lamont, Y.; Lyons, A.J.; Maclean, J.K.F.; et al. A novel series of positive modulators of the AMPA receptor: Structure-based lead optimization. Bioorg. Med. Chem. Lett. 2010, 20, 6072–6075. [Google Scholar] [CrossRef]
- Harms, J.E.; Benveniste, M.; Maclean, J.K.F.; Partin, K.M.; Jamieson, C. Functional analysis of a novel positive allosteric modulator of AMPA receptors derived from a structure-based drug design strategy. Neuropharmacology 2013, 64, 45–52. [Google Scholar] [CrossRef]
- Ward, S.E.; Harries, M.H.; Aldegheri, L.; Bradford, A.M.; Ballini, E.; Dawson, L.; Lacroix, L.; Pardoe, J.; Starr, K.; Weil, A.; et al. Pharmacological characterisation of MDI-222, a novel AMPA receptor positive allosteric modulator with an improved safety profile. J. Psychopharmacol. 2020, 34, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, C.; Maclean, J.K.F.; Brown, C.I.; Campbell, R.A.; Gillen, K.J.; Gillespie, J.; Kazemier, B.; Kiczun, M.; Lamont, Y.; Lyons, A.J.; et al. Structure based evolution of a novel series of positive modulators of the AMPA receptor. Bioorg. Med. Chem. Lett. 2011, 21, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Kunugi, A.; Tajima, Y.; Kuno, H.; Sogabe, S.; Kimura, H. HBT1, a novel AMPA receptor potentiator with lower agonistic effect, avoided bell-shaped response in in vitro BDNF production. J. Pharmacol. Exp. Ther. 2018, 364, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Murakami, K.; Tajima, Y.; Hara, H.; Kunugi, A.; Kimura, H. TAK-137, an AMPA receptor potentiator with little agonistic effect, produces antidepressant-like effect without causing psychotomimetic effects in rats. Pharmacol. Biochem. Behav. 2019, 183, 80–86. [Google Scholar] [CrossRef]
- Suzuki, A.; Tajima, Y.; Kunugi, A.; Kimura, H. Electrophysiological characterization of a novel AMPA receptor potentiator, TAK-137, in rat hippocampal neurons. Neurosci. Lett. 2019, 712, 134488. [Google Scholar] [CrossRef]
- Kunugi, A.; Tanaka, M.; Suzuki, A.; Tajima, Y.; Suzuki, N.; Suzuki, M.; Nakamura, S.; Kuno, H.; Yokota, A.; Sogabe, S.; et al. TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window. Neuropsychopharmacology 2019, 44, 961–970. [Google Scholar] [CrossRef]
- Witkin, J.M.; Martin, A.E.; Golani, L.K.; Xu, N.Z.; Smith, J.L. Rapid-acting antidepressants. Adv. Pharmacol. 2019, 86, 47–96. [Google Scholar] [CrossRef]
- Tanaka, M.; Kunugi, A.; Suzuki, A.; Suzuki, N.; Suzuki, M.; Kimura, H. Preclinical characterization of AMPA receptor potentiator TAK-137 as a therapeutic drug for schizophrenia. Pharmacol. Res. Perspect. 2019, 7, e00479. [Google Scholar] [CrossRef]
- Lavrov, M.I.; Grigor’ev, V.V.; Bachurin, S.O.; Palyulin, V.A.; Zefirov, N.S. Novel bivalent positive allosteric modulators of AMPA receptor. Dokl. Biochem. Biophys. 2015, 464, 322–324. [Google Scholar] [CrossRef]
- Grigoriev, V.V.; Lavrov, M.I.; Zamoyski, V.L.; Garibova, T.L.; Palyulin, V.A.; Bachurin, S.O. New positive allosteric modulator of AMPA receptors: In vitro and in vivo studies. Dokl. Biochem. Biophys. 2019, 488, 304–306. [Google Scholar] [CrossRef]
- Lavrov, M.I.; Karlov, D.S.; Voronina, T.A.; Grigoriev, V.V.; Ustyugov, A.A.; Bachurin, S.O.; Palyulin, V.A. Novel positive allosteric modulators of AMPA receptors based on 3,7-diazabicyclo[3.3.1]nonane scaffold. Mol. Neurobiol. 2020, 57, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Vyunova, T.V.; Andreeva, L.A.; Shevchenko, K.V.; Grigoriev, V.V.; Palyulin, V.A.; Lavrov, M.I.; Bondarenko, E.V.; Kalashnikova, E.E.; Myasoedov, N.F. Characterization of a new positive allosteric modulator of AMPA receptors—PAM-43: Specific binding of the ligand and its ability to potentiate AMPAR currents. Curr. Mol. Pharmacol. 2020, 13, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Vasilenko, D.A.; Sadovnikov, K.S.; Sedenkova, K.N.; Karlov, D.S.; Radchenko, E.V.; Grishin, Y.K.; Rybakov, V.B.; Kuznetsova, T.S.; Zamoyski, V.L.; Grigoriev, V.V.; et al. A facile approach to bis(isoxazoles), promising ligands of the AMPA receptor. Molecules 2021, 26, 6411. [Google Scholar] [CrossRef] [PubMed]
- Gudasheva, T.A.; Grigoriev, V.V.; Koliasnikova, K.N.; Zamoyski, V.L.; Seredenin, S.B. Neuropeptide cycloprolylglycine is an endogenous positive modulator of AMPA receptors. Dokl. Biochem. Biophys. 2016, 471, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Gudasheva, T.A.; Povarnina, P.Y.; Koliasnikova, K.N.; Alyaeva, A.G.; Vorontsova, O.N.; Seredenin, S.B. The anxiolytic effect of the neuropeptide cycloprolylglycine Is mediated by AMPA and TrkB receptors. Dokl. Biochem. Biophys. 2020, 493, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrova, L.R.; Phillips, A.G.; Wang, Y.T. Antidepressant effects of ketamine and the roles of AMPA glutamate receptors and other mechanisms beyond NMDA receptor antagonism. J. Psychiatry Neurosci. 2017, 42, 222–229. [Google Scholar] [CrossRef]
- Llamosas, N.; Perez-Caballero, L.; Berrocoso, E.; Bruzos-Cidon, C.; Ugedo, L.; Torrecilla, M. Ketamine promotes rapid and transient activation of AMPA receptor-mediated synaptic transmission in the dorsal raphe nucleus. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 88, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Matveychuk, D.; Thomas, R.K.; Swainson, J.; Khullar, A.; MacKay, M.-A.; Baker, G.B.; Dursun, S.M. Ketamine as an antidepressant: Overview of its mechanisms of action and potential predictive biomarkers. Ther. Adv. Psychopharmacol. 2020, 10, 2045125320916657. [Google Scholar] [CrossRef]
- Kang, M.J.Y.; Hawken, E.; Vazquez, G.H. The mechanisms behind rapid antidepressant effects of Ketamine: A systematic review with a focus on molecular neuroplasticity. Front. Psychiatry 2022, 13, 860882. [Google Scholar] [CrossRef]
- Hess, E.M.; Riggs, L.M.; Michaelides, M.; Gould, T.D. Mechanisms of ketamine and its metabolites as antidepressants. Biochem. Pharmacol. 2022, 197, 114892. [Google Scholar] [CrossRef]
- Freudenberg, F.; Celikel, T.; Reif, A. The role of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors in depression: Central mediators of pathophysiology and antidepressant activity? Neurosci. Biobehav. Rev. 2015, 52, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Hanada, T. The discovery and development of perampanel for the treatment of epilepsy. Expert Opin. Drug Discov. 2014, 9, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Potschka, H.; Trinka, E. Perampanel: Does it have broad-spectrum potential? Epilepsia 2019, 60 (Suppl. S1), 22–36. [Google Scholar] [CrossRef] [PubMed]
- Lavu, A.; Aboulatta, L.; Abou-Setta, A.M.; Aloud, B.; Askin, N.; Rabbani, R.; Shouman, W.; Zarychanski, R.; Eltonsy, S. Efficacy and safety of perampanel in epilepsy: A systematic review and meta-analysis of randomised controlled trials. Seizure 2022, 102, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Suda, S.; Sowa, K.; Sakamoto, Y.; Nito, C.; Nishiyama, Y.; Aoki, J.; Ueda, M.; Yokobori, S.; Yamada, M.; et al. AMPA receptor antagonist Perampanel ameliorates post-stroke functional and cognitive impairments. Neuroscience 2018, 386, 256–264. [Google Scholar] [CrossRef]
- Suda, S.; Kimura, K. Therapeutic potential of AMPA receptor antagonist perampanel against cerebral ischemia: Beyond epileptic disorder. Neural Regen. Res. 2019, 14, 1525–1526. [Google Scholar] [CrossRef]
- Lange, F.; Weßlau, K.; Porath, K.; Hörnschemeyer, J.; Bergner, C.; Krause, B.J.; Mullins, C.S.; Linnebacher, M.; Köhling, R.; Kirschstein, T. AMPA receptor antagonist perampanel affects glioblastoma cell growth and glutamate release in vitro. PLoS ONE 2019, 14, e0211644. [Google Scholar] [CrossRef]
- Nozawa, A.; Ozeki, M.; Matsuoka, M.; Nakama, M.; Yasue, S.; Endo, S.; Kawamoto, N.; Ohnishi, H.; Fukao, T. Perampanel inhibits neuroblastoma cell proliferation through down-regulation of AKT and ERK pathways. Anticancer Res. 2019, 39, 3595–3599. [Google Scholar] [CrossRef]
- Ribeiro, M.P.C.; Custódio, J.B.A.; Santos, A.E. Ionotropic glutamate receptor antagonists and cancer therapy: Time to think out of the box? Cancer Chemother. Pharmacol. 2017, 79, 219–225. [Google Scholar] [CrossRef]
- Yelshanskaya, M.V.; Singh, A.K.; Sampson, J.M.; Narangoda, C.; Kurnikova, M.; Sobolevsky, A.I. Structural bases of noncompetitive inhibition of AMPA-subtype ionotropic glutamate receptors by antiepileptic drugs. Neuron 2016, 91, 1305–1315. [Google Scholar] [CrossRef]
- Stenum-Berg, C.; Musgaard, M.; Chavez-Abiega, S.; Thisted, C.L.; Barrella, L.; Biggin, P.C.; Kristensen, A.S. Mutational analysis and modeling of negative allosteric modulator binding sites in AMPA receptors. Mol. Pharmacol. 2019, 96, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Guseynov, A.-A.D.; Pisarev, S.A.; Shulga, D.A.; Palyulin, V.A.; Fedorov, M.V.; Karlov, D.S. Computational characterization of the glutamate receptor antagonist perampanel and its close analogs: Density functional exploration of conformational space and molecular docking study. J. Mol. Model. 2019, 25, 312. [Google Scholar] [CrossRef] [PubMed]
- Narangoda, C.; Sakipov, S.N.; Kurnikova, M.G. AMPA receptor noncompetitive inhibitors occupy a promiscuous binding site. ACS Chem. Neurosci. 2019, 10, 4511–4521. [Google Scholar] [CrossRef] [PubMed]
- Espahbodinia, M.; Ettari, R.; Wen, W.; Wu, A.; Shen, Y.-C.; Niu, L.; Grasso, S.; Zappalà, M. Development of novel N-3-bromoisoxazolin-5-yl substituted 2,3-benzodiazepines as noncompetitive AMPAR antagonists. Bioorg. Med. Chem. 2017, 25, 3631–3637. [Google Scholar] [CrossRef]
- Qneibi, M.; Jaradat, N.; Hawash, M.; Olgac, A.; Emwas, N. Ortho versus meta chlorophenyl-2,3-benzodiazepine analogues: Synthesis, molecular modeling, and biological activity as AMPAR antagonists. ACS Omega 2020, 5, 3588–3595. [Google Scholar] [CrossRef]
- El-Helby, A.-G.A.; Ayyad, R.R.A.; El-Adl, K.; Elkady, H. Phthalazine-1,4-dione derivatives as non-competitive AMPA receptor antagonists: Design, synthesis, anticonvulsant evaluation, ADMET profile and molecular docking. Mol. Divers. 2019, 23, 283–298. [Google Scholar] [CrossRef]
- Inami, H.; Shishikura, J.-I.; Yasunaga, T.; Hirano, M.; Kimura, T.; Yamashita, H.; Ohno, K.; Sakamoto, S. Synthesis and pharmacological evaluation of 3-[(4-oxo-4H-pyrido[3,2-e][1,3]thiazin-2-yl)(phenyl)amino]propanenitrile derivatives as orally active AMPA receptor antagonists. Chem. Pharm. Bull. 2019, 67, 699–706. [Google Scholar] [CrossRef]
- Qneibi, M.; Hawash, M.; Jaradat, N.; Bdir, S. Affecting AMPA receptor biophysical gating properties with negative allosteric modulators. Mol. Neurobiol. 2022, 59, 5264–5275. [Google Scholar] [CrossRef]
- Qneibi, M.; Hawash, M.; Bdir, S.; Nacak Baytas, S. Targeting the kinetics mechanism of AMPA receptor inhibition by 2-oxo-3H-benzoxazole derivatives. Bioorg. Chem. 2022, 129, 106163. [Google Scholar] [CrossRef]
- Qneibi, M.; Hamed, O.; Fares, O.; Jaradat, N.; Natsheh, A.-R.; AbuHasan, Q.; Emwas, N.; Al-Kerm, R.; Al-Kerm, R. The inhibitory role of curcumin derivatives on AMPA receptor subunits and their effect on the gating biophysical properties. Eur. J. Pharm. Sci. 2019, 136, 104951. [Google Scholar] [CrossRef]
- Qneibi, M.; Hamed, O.; Natsheh, A.-R.; Fares, O.; Jaradat, N.; Emwas, N.; AbuHasan, Q.; Al-Kerm, R.; Al-Kerm, R. Inhibition and assessment of the biophysical gating properties of GluA2 and GluA2/A3 AMPA receptors using curcumin derivatives. PLoS ONE 2019, 14, e0221132. [Google Scholar] [CrossRef] [PubMed]
- Qneibi, M.; Hamed, O.; Jaradat, N.; Hawash, M.; Al-Kerm, R.; Al-Kerm, R.; Sobuh, S.; Tarazi, S. The AMPA receptor biophysical gating properties and binding site: Focus on novel curcumin-based diazepines as non-competitive antagonists. Bioorg. Chem. 2021, 116, 105406. [Google Scholar] [CrossRef] [PubMed]
- Rečnik, L.-M.; Thatcher, R.J.; Mallah, S.; Butts, C.P.; Collingridge, G.L.; Molnár, E.; Jane, D.E.; Willis, C.L. Synthesis and pharmacological characterisation of arctigenin analogues as antagonists of AMPA and kainate receptors. Org. Biomol. Chem. 2021, 19, 9154–9162. [Google Scholar] [CrossRef] [PubMed]
- Jaremko, W.J.; Huang, Z.; Wen, W.; Wu, A.; Karl, N.; Niu, L. Identification and characterization of RNA aptamers: A long aptamer blocks the AMPA receptor and a short aptamer blocks both AMPA and kainate receptors. J. Biol. Chem. 2017, 292, 7338–7347. [Google Scholar] [CrossRef] [PubMed]
- Jaremko, W.; Huang, Z.; Karl, N.; Pierce, V.D.; Lynch, J.; Niu, L. A kainate receptor-selective RNA aptamer. J. Biol. Chem. 2020, 295, 6280–6288. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Niu, L. RNA aptamers for AMPA receptors. Neuropharmacology 2021, 199, 108761. [Google Scholar] [CrossRef]
- Nazarova, A.A.; Sedenkova, K.N.; Karlov, D.S.; Lavrov, M.I.; Grishin, Y.K.; Kuznetsova, T.S.; Zamoyski, V.L.; Grigoriev, V.V.; Averina, E.B.; Palyulin, V.A. Bivalent AMPA receptor positive allosteric modulators of the bis(pyrimidine) series. Med. Chem. Commun. 2019, 10, 1615–1619. [Google Scholar] [CrossRef]
- Sedenkova, K.N.; Averina, E.B.; Nazarova, A.A.; Grishin, Y.K.; Karlov, D.S.; Zamoyski, V.L.; Grigoriev, V.V.; Kuznetsova, T.S.; Palyulin, V.A. The first AMPA receptor negative modulators based on the tetrahydroquinazoline scaffold. Mendeleev Commun. 2018, 28, 423–425. [Google Scholar] [CrossRef]
- Sedenkova, K.N.; Zverev, D.V.; Nazarova, A.A.; Lavrov, M.I.; Radchenko, E.V.; Grishin, Y.K.; Gabrel’yan, A.V.; Zamoyski, V.L.; Grigoriev, V.V.; Averina, E.B.; et al. Novel nanomolar allosteric modulators of AMPA receptor of bis(pyrimidine) series: Synthesis, biotesting and SAR analysis. Molecules 2022, 27, 8252. [Google Scholar] [CrossRef]
- Temnyakova, N.S.; Vasilenko, D.A.; Lavrov, M.I.; Karlov, D.S.; Grishin, Y.K.; Zamoyski, V.L.; Grigoriev, V.V.; Averina, E.B.; Palyulin, V.A. Novel bivalent positive allosteric AMPA receptor modulator of bis-amide series. Mendeleev Commun. 2021, 31, 216–218. [Google Scholar] [CrossRef]
- Temnyakova, N.S.; Vasilenko, D.A.; Lavrov, M.I.; Karlov, D.S.; Grishin, Y.K.; Zamoyski, V.L.; Grigoriev, V.V.; Averina, E.B.; Palyulin, V.A. New nanomolar negative modulators of AMPA receptors. Mendeleev Commun. 2021, 31, 219–220. [Google Scholar] [CrossRef]
- Lavrov, M.I.; Veremeeva, P.N.; Karlov, D.S.; Zamoyski, V.L.; Grigoriev, V.V.; Palyulin, V.A. Tricyclic derivatives of bispidine as AMPA receptor allosteric modulators. Mendeleev Commun. 2019, 29, 619–621. [Google Scholar] [CrossRef]
- Lavrov, M.I.; Karlov, D.S.; Palyulin, V.A.; Grigoriev, V.V.; Zamoyski, V.L.; Brkich, G.E.; Pyatigorskaya, N.V.; Zapolskiy, M.E. Novel positive allosteric modulator of AMPA-receptors based on tricyclic scaffold. Mendeleev Commun. 2018, 28, 311–313. [Google Scholar] [CrossRef]
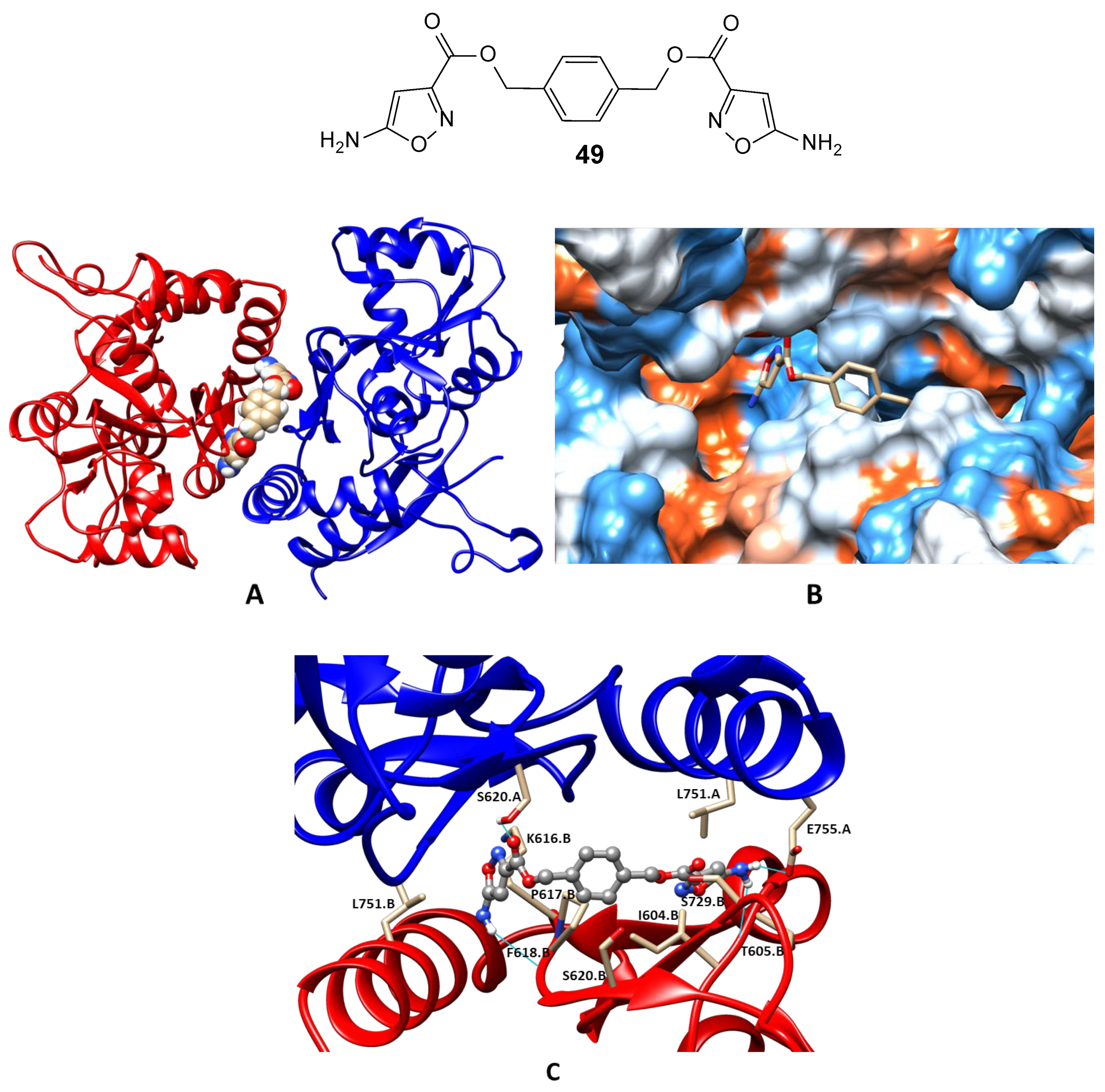
- Lavrov, M.I.; Stroganov, O.V.; Zamoyski, V.L.; Grigoriev, V.V.; Zapolskiy, M.E.; Sysolyatin, S.V.; Malykhin, V.V.; Surmachev, V.N.; Palyulin, V.A. Synthesis of an allosteric modulator of ionotropic glutamate receptors. Mendeleev Commun. 2020, 30, 156–158. [Google Scholar] [CrossRef]
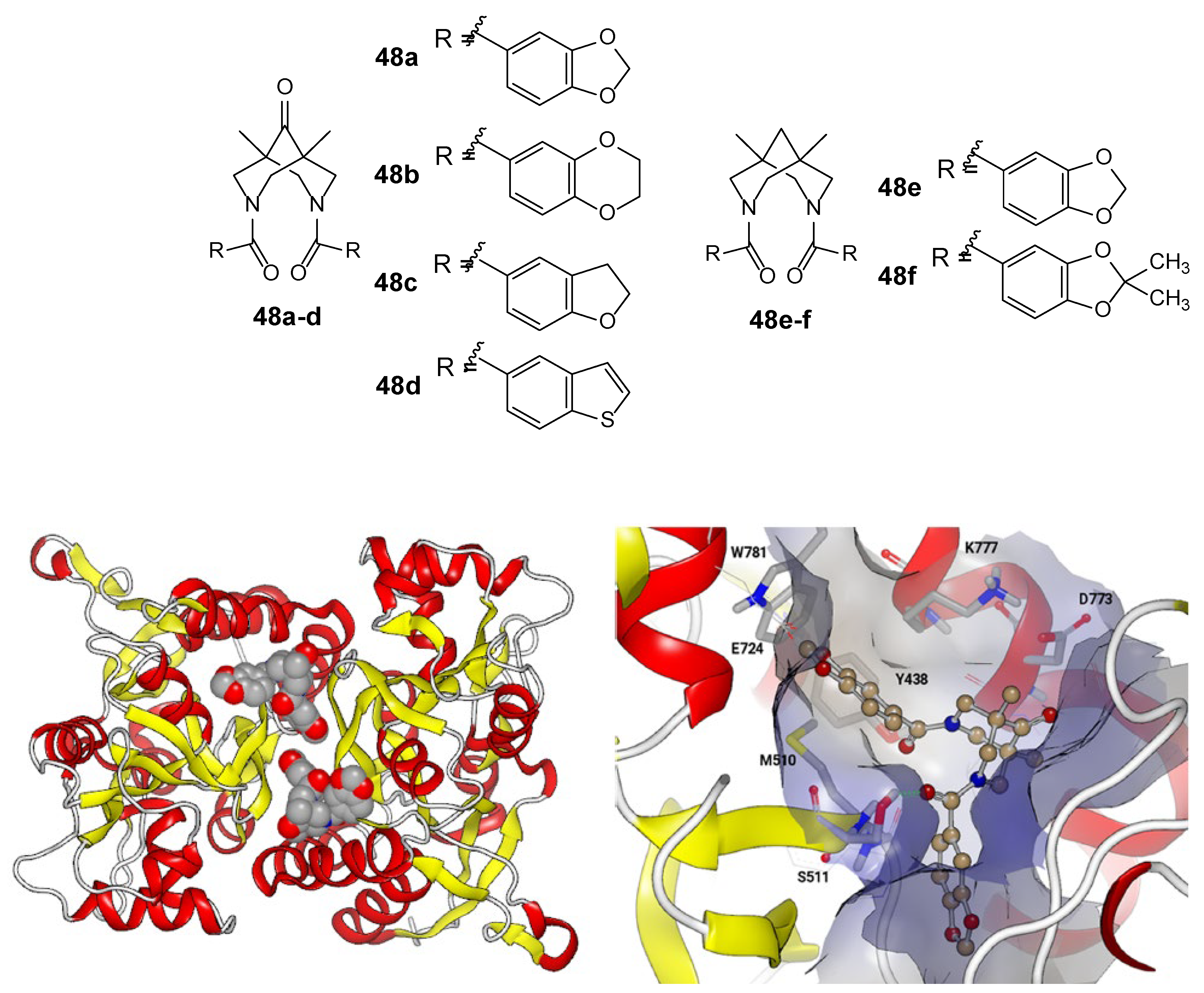
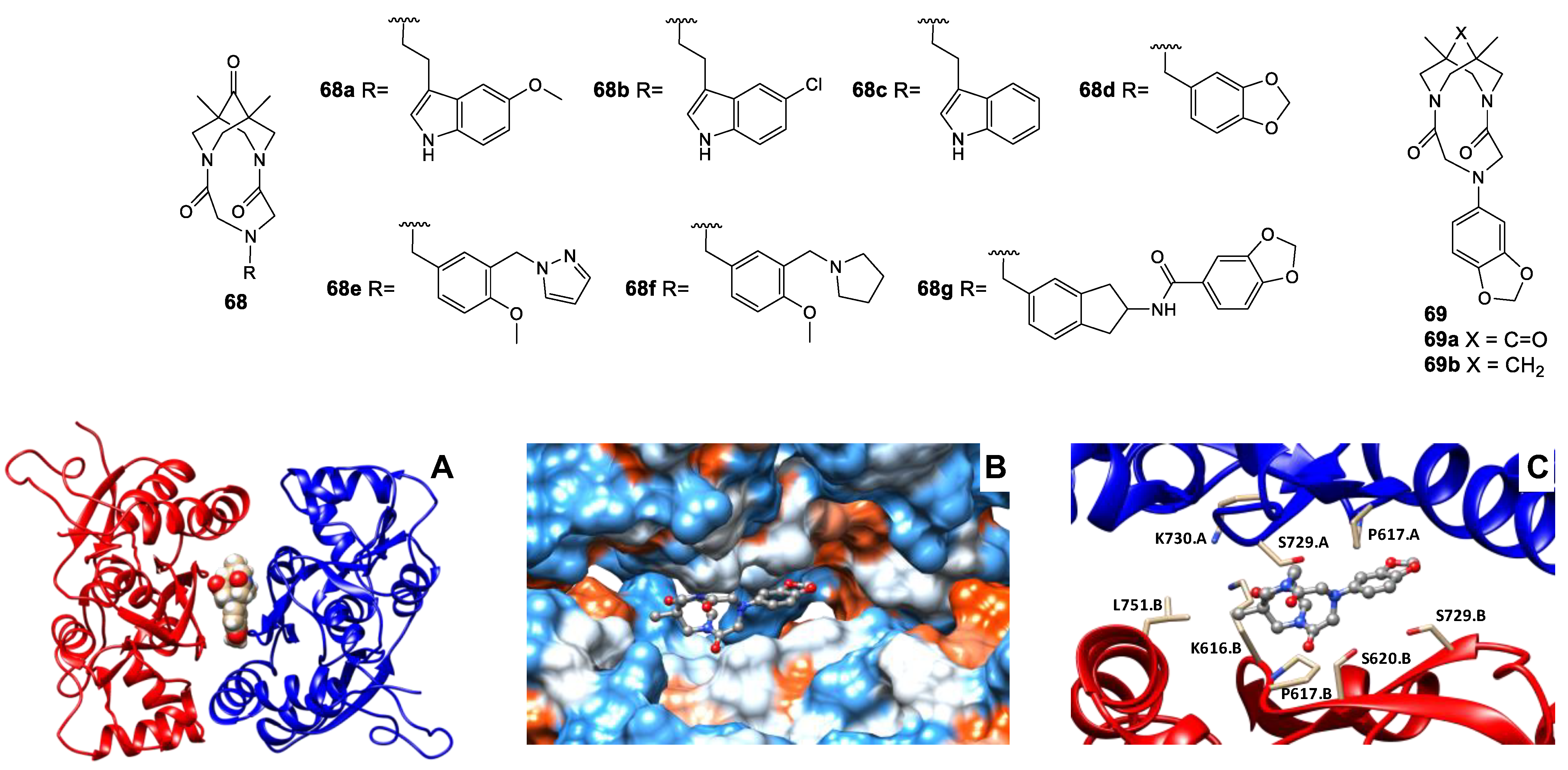
- Golubeva, E.A.; Lavrov, M.I.; Veremeeva, P.N.; Bovina, E.M.; Radchenko, E.V.; Topchiy, M.A.; Asachenko, A.F.; Zamoyski, V.L.; Grigoriev, V.V.; Palyulin, V.A. New 1,11-dimethyl-3,6,9-triazatricyclo[7.3.1.13,11]tetradecane-4,8,12-trione derivative as an allosteric modulator of the glutamatergic system. Mendeleev Commun. 2023, 33, 70–72. [Google Scholar] [CrossRef]
- Lavrov, M.I.; Veremeeva, P.N.; Golubeva, E.A.; Radchenko, E.V.; Zamoyski, V.L.; Grigoriev, V.V.; Palyulin, V.A. Positive and negative AMPA receptor modulators based on tricyclic bispidine derivative: Minor structural change inverts the type of activity. Mendeleev Commun. 2022, 32, 360–363. [Google Scholar] [CrossRef]
- Twomey, E.C.; Yelshanskaya, M.V.; Grassucci, R.A.; Frank, J.; Sobolevsky, A.I. Structural bases of desensitization in AMPA receptor-auxiliary subunit complexes. Neuron 2017, 94, 569–580.e5. [Google Scholar] [CrossRef]
- Dawe, G.B.; Kadir, M.F.; Venskutonytė, R.; Perozzo, A.M.; Yan, Y.; Alexander, R.P.D.; Navarrete, C.; Santander, E.A.; Arsenault, M.; Fuentes, C.; et al. Nanoscale mobility of the apo state and TARP stoichiometry dictate the gating behavior of alternatively spliced AMPA receptors. Neuron 2019, 102, 976–992.e5. [Google Scholar] [CrossRef]
- Herguedas, B.; Watson, J.F.; Ho, H.; Cais, O.; García-Nafría, J.; Greger, I.H. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Science 2019, 364, eaav9011. [Google Scholar] [CrossRef]
- Yelshanskaya, M.V.; Patel, D.S.; Kottke, C.M.; Kurnikova, M.G.; Sobolevsky, A.I. Opening of glutamate receptor channel to subconductance levels. Nature 2022, 605, 172–178. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, Y.; Wang, Y.; Shekhar, M.; Tajkhorshid, E.; Gouaux, E. Activation and desensitization mechanism of AMPA receptor-TARP complex by Cryo-EM. Cell 2017, 170, 1234–1246.e14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Watson, J.F.; Matthews, P.M.; Cais, O.; Greger, I.H. Gating and modulation of a hetero-octameric AMPA glutamate receptor. Nature 2021, 594, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Rao, P.; Clark, S.; Mitra, J.; Ha, T.; Gouaux, E. Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition. Nature 2021, 594, 448–453. [Google Scholar] [CrossRef]
- Herguedas, B.; Kohegyi, B.K.; Dohrke, J.-N.; Watson, J.F.; Zhang, D.; Ho, H.; Shaikh, S.A.; Lape, R.; Krieger, J.M.; Greger, I.H. Mechanisms underlying TARP modulation of the GluA1/2-γ8 AMPA receptor. Nat. Commun. 2022, 13, 734. [Google Scholar] [CrossRef] [PubMed]
- Dohrke, J.-N.; Watson, J.F.; Birchall, K.; Greger, I.H. Characterizing the binding and function of TARP γ8-selective AMPA receptor modulators. J. Biol. Chem. 2020, 295, 14565–14577. [Google Scholar] [CrossRef]
- Coombs, I.D.; Sexton, C.A.; Cull-Candy, S.G.; Farrant, M. Influence of the TARP γ8-selective negative allosteric modulator JNJ-55511118 on AMPA receptor gating and channel conductance. Mol. Pharmacol. 2022, 101, 343–356. [Google Scholar] [CrossRef]
- Gardinier, K.M.; Gernert, D.L.; Porter, W.J.; Reel, J.K.; Ornstein, P.L.; Spinazze, P.; Stevens, F.C.; Hahn, P.; Hollinshead, S.P.; Mayhugh, D.; et al. Discovery of the first α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor antagonist dependent upon transmembrane AMPA receptor regulatory protein (TARP) γ-8. J. Med. Chem. 2016, 59, 4753–4768. [Google Scholar] [CrossRef]
- Maher, M.P.; Wu, N.; Ravula, S.; Ameriks, M.K.; Savall, B.M.; Liu, C.; Lord, B.; Wyatt, R.M.; Matta, J.A.; Dugovic, C.; et al. Discovery and characterization of AMPA receptor modulators selective for TARP-γ8. J. Pharmacol. Exp. Ther. 2016, 357, 394–414. [Google Scholar] [CrossRef]
- Ravula, S.; Savall, B.M.; Wu, N.; Lord, B.; Coe, K.; Wang, K.; Seierstad, M.; Swanson, D.M.; Ziff, J.; Nguyen, M.; et al. Lead optimization of 5-aryl benzimidazolone- and oxindole-based AMPA receptor modulators selective for TARP γ-8. ACS Med. Chem. Lett. 2018, 9, 821–826. [Google Scholar] [CrossRef]
- Savall, B.M.; Wu, D.; Swanson, D.M.; Seierstad, M.; Wu, N.; Vives Martinez, J.; García Olmos, B.; Lord, B.; Coe, K.; Koudriakova, T.; et al. Discovery of imidazo[1,2-a]pyrazines and pyrazolo[1,5-c]pyrimidines as TARP γ-8 selective AMPAR negative modulators. ACS Med. Chem. Lett. 2019, 10, 267–272. [Google Scholar] [CrossRef]
- Azumaya, C.M.; Days, E.L.; Vinson, P.N.; Stauffer, S.; Sulikowski, G.; Weaver, C.D.; Nakagawa, T. Screening for AMPA receptor auxiliary subunit specific modulators. PLoS ONE 2017, 12, e0174742. [Google Scholar] [CrossRef] [PubMed]
- Bowie, D. Polyamine-mediated channel block of ionotropic glutamate receptors and its regulation by auxiliary proteins. J. Biol. Chem. 2018, 293, 18789–18802. [Google Scholar] [CrossRef] [PubMed]
- Hackos, D.H.; Hanson, J.E. Diverse modes of NMDA receptor positive allosteric modulation: Mechanisms and consequences. Neuropharmacology 2017, 112, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Frølund, S.; Bella, A.; Kristensen, A.S.; Ziegler, H.L.; Witt, M.; Olsen, C.A.; Strømgaard, K.; Franzyk, H.; Jaroszewski, J.W. Assessment of structurally diverse philanthotoxin analogues for inhibitory activity on ionotropic glutamate receptor subtypes: Discovery of nanomolar, nonselective, and use-dependent antagonists. J. Med. Chem. 2010, 53, 7441–7451. [Google Scholar] [CrossRef]
- Franzyk, H.; Grzeskowiak, J.W.; Tikhonov, D.B.; Jaroszewski, J.W.; Mellor, I.R. The effects of conformational constraints in the polyamine moiety of philanthotoxins on AMPAR inhibition. ChemMedChem 2014, 9, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Nørager, N.G.; Poulsen, M.H.; Strømgaard, K. Controlling Ca2+ permeable α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors with photochromic ion channel blockers. J. Med. Chem. 2018, 61, 8048–8053. [Google Scholar] [CrossRef] [PubMed]
- Zhigulin, A.S.; Dron, M.Y.; Barygin, O.I. Mechanisms of AMPA receptor inhibition by diminazene. Neurosci. Behav. Physiol. 2022, 52, 308–314. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golubeva, E.A.; Lavrov, M.I.; Radchenko, E.V.; Palyulin, V.A. Diversity of AMPA Receptor Ligands: Chemotypes, Binding Modes, Mechanisms of Action, and Therapeutic Effects. Biomolecules 2023, 13, 56. https://doi.org/10.3390/biom13010056
Golubeva EA, Lavrov MI, Radchenko EV, Palyulin VA. Diversity of AMPA Receptor Ligands: Chemotypes, Binding Modes, Mechanisms of Action, and Therapeutic Effects. Biomolecules. 2023; 13(1):56. https://doi.org/10.3390/biom13010056
Chicago/Turabian StyleGolubeva, Elena A., Mstislav I. Lavrov, Eugene V. Radchenko, and Vladimir A. Palyulin. 2023. "Diversity of AMPA Receptor Ligands: Chemotypes, Binding Modes, Mechanisms of Action, and Therapeutic Effects" Biomolecules 13, no. 1: 56. https://doi.org/10.3390/biom13010056
APA StyleGolubeva, E. A., Lavrov, M. I., Radchenko, E. V., & Palyulin, V. A. (2023). Diversity of AMPA Receptor Ligands: Chemotypes, Binding Modes, Mechanisms of Action, and Therapeutic Effects. Biomolecules, 13(1), 56. https://doi.org/10.3390/biom13010056





