Polyphenolic Compounds Inhibit Osteoclast Differentiation While Reducing Autophagy through Limiting ROS and the Mitochondrial Membrane Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Antibodies
2.2. Osteoclast Differentiation
2.3. TRAP Staining
2.4. Acidic Vesicular Organelles Staining
2.5. Western Blot Analysis
2.6. RNA Extraction and Real-Time PCR
2.7. Immunofluorescence Staining
2.8. Detection of Mitochondrial Membrane Potential
2.9. Detection of ROS
2.10. Detection of Mitochondrial ROS
2.11. Intercellular Ca2+ Measurement
2.12. Cell Viability Assay
2.13. Statistical Analysis
3. Results
3.1. Effect of Polyphenolic Compounds on Osteoclast Differentiation
3.2. Effect of Polyphenolic Compounds on Actin Ring Formation
3.3. Effect of Polyphenolic Compounds on Osteoclast-Related Gene Expressions during Differentiation
3.4. Effect of TA, GA, and EA on Protein Levels of Osteoclast Differentiation Marker Molecules during Osteoclastogenesis
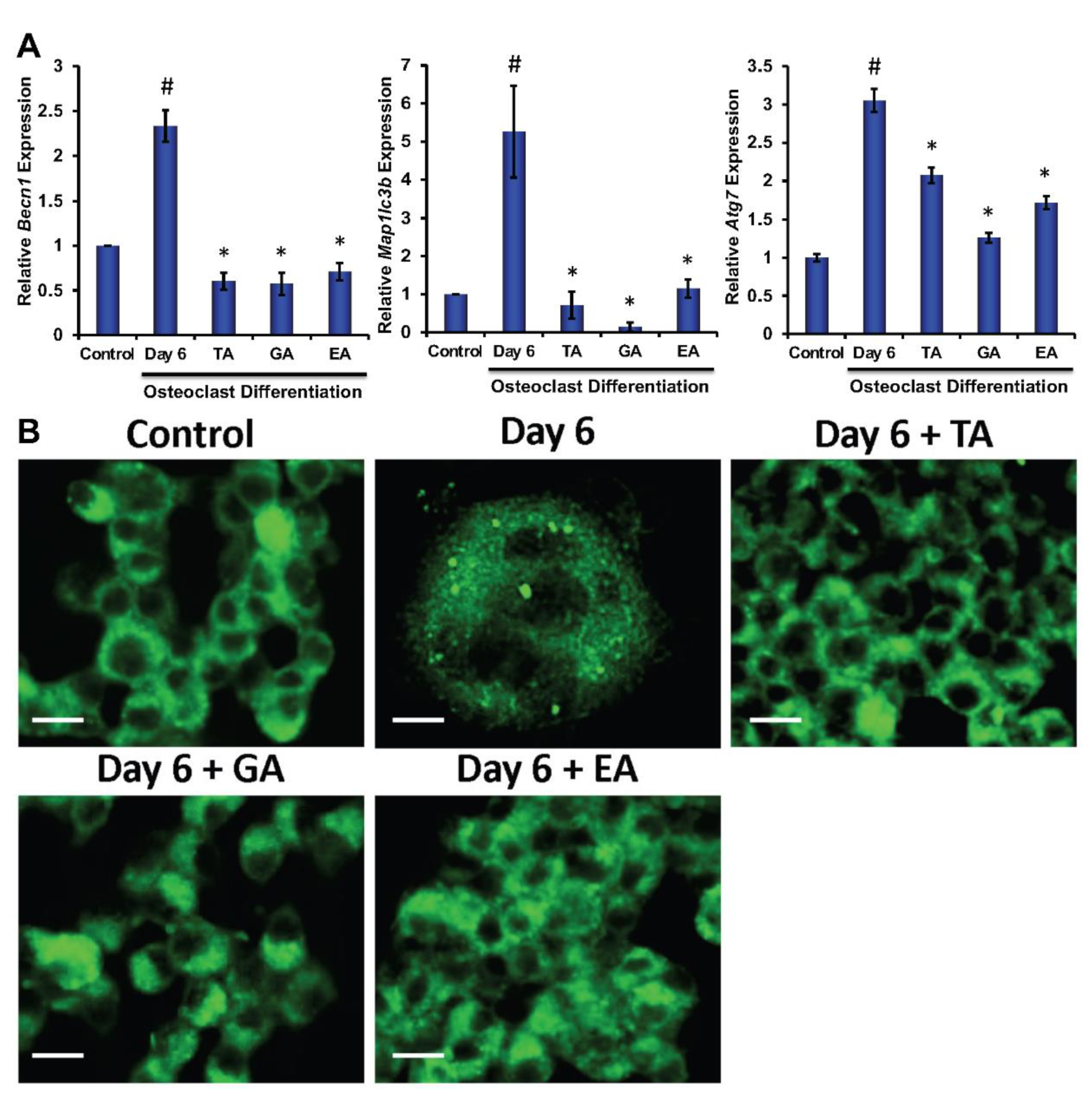
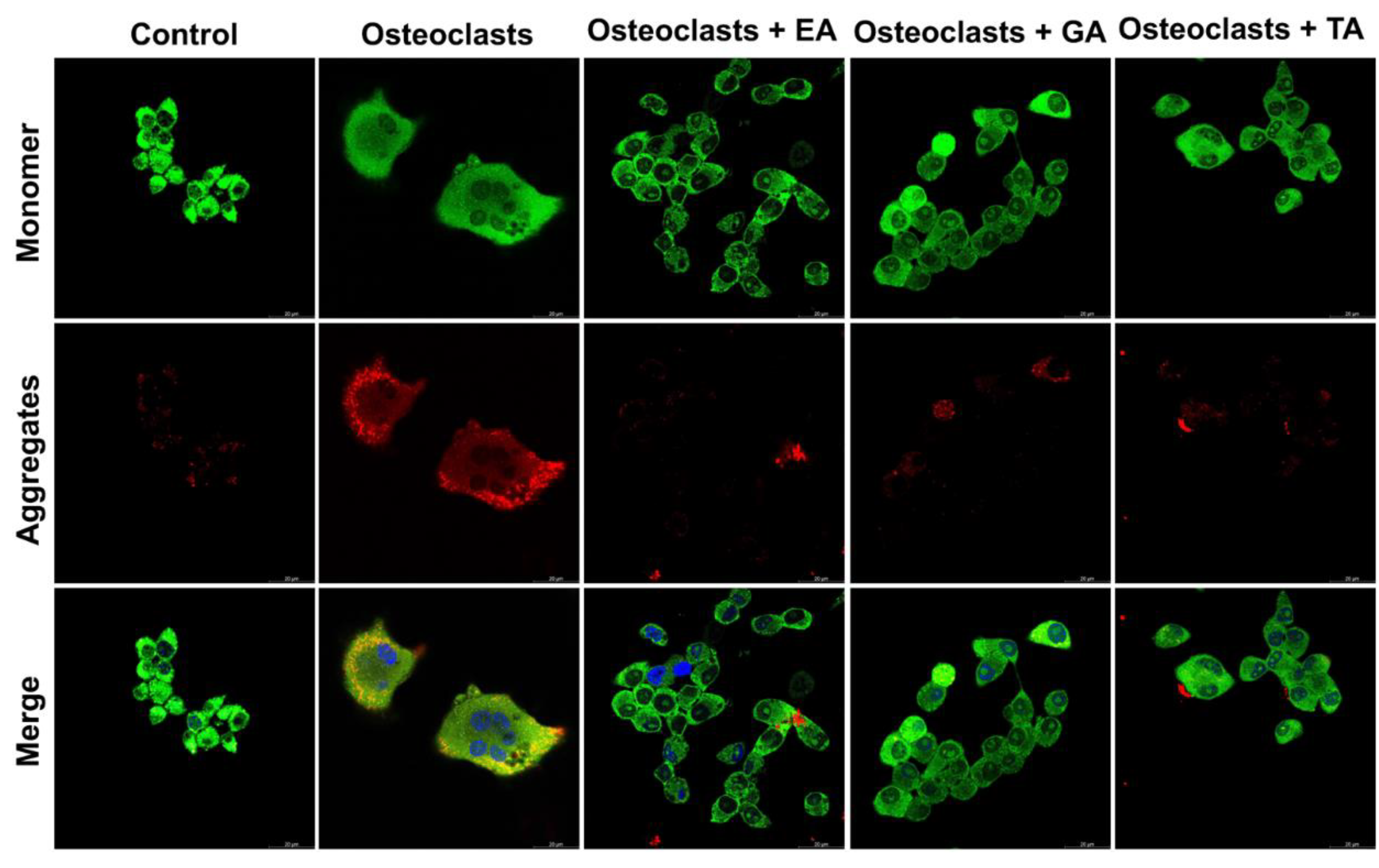
3.5. Effect of Polyphenolic Compounds on the Expression of Autophagic Genes in Myeloid Cells during Osteoclast Differentiation
3.6. Effect of Polyphenolic Compounds on the Expression of Apoptotic and Cell Survival-Related Molecules in Myeloid Cells during Osteoclast Differentiation
3.7. Determination of Downstream Signaling Pathways
3.8. Effect of Polyphenolic Compounds on Production of ROS, and Intracellular Ca2+ during Osteoclast Differentiation
3.9. Effect of Polyphenolic Compounds on Mitochondrial Membrane Potential during Osteoclast Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Scalbert, A.; Manach, C.; Morand, C.; Remesy, C.; Jimenez, L. Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 2005, 45, 287–306. [Google Scholar] [CrossRef] [PubMed]
- Munin, A.; Edwards-Levy, F. Encapsulation of natural polyphenolic compounds; a review. Pharmaceutics 2011, 3, 793–829. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.T.; Wong, T.Y.; Wei, C.I.; Huang, Y.W.; Lin, Y. Tannins and human health: A review. Crit. Rev. Food Sci. Nutr. 1998, 38, 421–464. [Google Scholar] [CrossRef] [PubMed]
- Rosman, R.; Saifullah, B.; Maniam, S.; Dorniani, D.; Hussein, M.Z.; Fakurazi, S. Improved Anticancer Effect of Magnetite Nanocomposite Formulation of GALLIC Acid (Fe(3)O(4)-PEG-GA) Against Lung, Breast and Colon Cancer Cells. Nanomaterials 2018, 8, 83. [Google Scholar] [CrossRef]
- Ismail, T.; Calcabrini, C.; Diaz, A.R.; Fimognari, C.; Turrini, E.; Catanzaro, E.; Akhtar, S.; Sestili, P. Ellagitannins in Cancer Chemoprevention and Therapy. Toxins 2016, 8, 151. [Google Scholar] [CrossRef]
- Prochazkova, D.; Bousova, I.; Wilhelmova, N. Antioxidant and prooxidant properties of flavonoids. Fitoterapia 2011, 82, 513–523. [Google Scholar] [CrossRef]
- Nomura, M.; Kaji, A.; He, Z.; Ma, W.Y.; Miyamoto, K.; Yang, C.S.; Dong, Z. Inhibitory mechanisms of tea polyphenols on the ultraviolet B-activated phosphatidylinositol 3-kinase-dependent pathway. J. Biol. Chem. 2001, 276, 46624–46631. [Google Scholar] [CrossRef]
- Lin, J.K. Cancer chemoprevention by tea polyphenols through modulating signal transduction pathways. Arch. Pharm. Res. 2002, 25, 561–571. [Google Scholar] [CrossRef]
- Khlebnikov, A.I.; Schepetkin, I.A.; Domina, N.G.; Kirpotina, L.N.; Quinn, M.T. Improved quantitative structure-activity relationship models to predict antioxidant activity of flavonoids in chemical, enzymatic, and cellular systems. Bioorg. Med. Chem. 2007, 15, 1749–1770. [Google Scholar] [CrossRef]
- Crozier, A.; Jaganath, I.B.; Clifford, M.N. Dietary phenolics: Chemistry, bioavailability and effects on health. Nat. Prod. Rep. 2009, 26, 1001–1043. [Google Scholar] [CrossRef]
- D’Archivio, M.; Filesi, C.; di Benedetto, R.; Gargiulo, R.; Giovannini, C.; Masella, R. Polyphenols, dietary sources and bioavailability. Ann. Ist. Super. Sanita 2007, 43, 348–361. [Google Scholar]
- Henrotin, Y.; Lambert, C.; Couchourel, D.; Ripoll, C.; Chiotelli, E. Nutraceuticals: Do they represent a new era in the management of osteoarthritis? A narrative review from the lessons taken with five products. Osteoarthr. Cartil. 2011, 19, 1–21. [Google Scholar] [CrossRef]
- Shen, C.L.; Yeh, J.K.; Cao, J.J.; Chyu, M.C.; Wang, J.S. Green tea and bone health: Evidence from laboratory studies. Pharmacol. Res. 2011, 64, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Welch, A.A.; Hardcastle, A.C. The effects of flavonoids on bone. Curr. Osteoporos. Rep. 2014, 12, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Callaway, D.A.; Jiang, J.X. Reactive oxygen species and oxidative stress in osteoclastogenesis, skeletal aging and bone diseases. J. Bone Miner. Metab. 2015, 33, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Bodet, C.; Chandad, F.; Grenier, D. Cranberry components inhibit interleukin-6, interleukin-8, and prostaglandin E production by lipopolysaccharide-activated gingival fibroblasts. Eur. J. Oral Sci. 2007, 115, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, P.; Persson, E.; Conaway, H.H.; Lerner, U.H. IL-6, leukemia inhibitory factor, and oncostatin M stimulate bone resorption and regulate the expression of receptor activator of NF-kappa B ligand, osteoprotegerin, and receptor activator of NF-kappa B in mouse calvariae. J. Immunol. 2002, 169, 3353–3362. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Xing, L.; Qin, C.; Schwarz, E.M.; Boyce, B.F. Osteoclast precursor interaction with bone matrix induces osteoclast formation directly by an interleukin-1-mediated autocrine mechanism. J. Biol. Chem. 2008, 283, 9917–9924. [Google Scholar] [CrossRef] [PubMed]
- Imboden, J.B. The immunopathogenesis of rheumatoid arthritis. Annu. Rev. Pathol. 2009, 4, 417–434. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Das, H. KLF2 in Regulation of NF-kappaB-Mediated Immune Cell Function and Inflammation. Int. J. Mol. Sci. 2017, 18, 2383. [Google Scholar] [CrossRef]
- Das, M.; Lu, J.; Joseph, M.; Aggarwal, R.; Kanji, S.; McMichael, B.K.; Lee, B.S.; Agarwal, S.; Ray-Chaudhury, A.; Iwenofu, O.H.; et al. Kruppel-like factor 2 (KLF2) regulates monocyte differentiation and functions in mBSA and IL-1beta-induced arthritis. Curr. Mol. Med. 2012, 12, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Deb, M.; Laha, D.; Joseph, M.; Kanji, S.; Aggarwal, R.; Iwenofu, O.H.; Pompili, V.J.; Jarjour, W.; Das, H. Myeloid Kruppel-Like Factor 2 Critically Regulates K/BxN Serum-Induced Arthritis. Cells 2019, 8, 908. [Google Scholar] [CrossRef]
- Das, M.; Laha, D.; Kanji, S.; Joseph, M.; Aggarwal, R.; Iwenofu, O.H.; Pompili, V.J.; Jain, M.K.; Das, H. Induction of Kruppel-like factor 2 reduces K/BxN serum-induced arthritis. J. Cell. Mol. Med. 2019, 23, 1386–1395. [Google Scholar] [CrossRef] [PubMed]
- Deb, M.; Laha, D.; Maity, J.; Das, H. SETD2-mediated epigenetic regulation of noncanonical Wnt5A during osteoclastogenesis. Clin. Epigenetics 2021, 13, 192. [Google Scholar] [CrossRef] [PubMed]
- Kanji, S.; Sarkar, R.; Pramanik, A.; Kshirsagar, S.; Greene, C.J.; Das, H. Dental pulp-derived stem cells inhibit osteoclast differentiation by secreting osteoprotegerin and deactivating AKT signalling in myeloid cells. J. Cell. Mole. Med. 2021, 25, 2390–2403. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Takahashi, N. Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis. 2002, 8, 147–159. [Google Scholar] [CrossRef]
- Rolph, D.; Das, H. Transcriptional Regulation of Osteoclastogenesis: The Emerging Role of KLF2. Front. Immunol. 2020, 11, 937. [Google Scholar] [CrossRef]
- Valenti, M.T.; Carbonare, L.D.; Mottes, M. Role of autophagy in bone and muscle biology. World J. Stem Cells 2016, 8, 396–398. [Google Scholar] [CrossRef]
- Shi, J.; Wang, L.; Zhang, H.; Jie, Q.; Li, X.; Shi, Q.; Huang, Q.; Gao, B.; Han, Y.; Guo, K.; et al. Glucocorticoids: Dose-related effects on osteoclast formation and function via reactive oxygen species and autophagy. Bone 2015, 79, 222–232. [Google Scholar] [CrossRef]
- Lin, N.Y.; Stefanica, A.; Distler, J.H. Autophagy: A key pathway of TNF-induced inflammatory bone loss. Autophagy 2013, 9, 1253–1255. [Google Scholar] [CrossRef]
- Das, G.; Shravage, B.V.; Baehrecke, E.H. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb. Perspect. Biol. 2012, 4, a008813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef]
- Maity, J.; Bohr, V.A.; Laskar, A.; Karmakar, P. Transient overexpression of Werner protein rescues starvation induced autophagy in Werner syndrome cells. Biochim. Biophys. Acta 2014, 1842, 2387–2394. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Deb, M.; Das, H. KLF2 (kruppel-like factor 2 [lung]) regulates osteoclastogenesis by modulating autophagy. Autophagy 2019, 15, 2063–2075. [Google Scholar] [CrossRef] [PubMed]
- Teitelbaum, S.L. The osteoclast and its unique cytoskeleton. Ann. N. Y. Acad. Sci. 2011, 1240, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Eisner, D.A.; Caldwell, J.L.; Kistamás, K.; Trafford, A.W. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 2017, 121, 181–195. [Google Scholar] [CrossRef]
- Zeb, A. Ellagic acid in suppressing in vivo and in vitro oxidative stresses. Mol. Cell. Biochem. 2018, 448, 27–41. [Google Scholar] [CrossRef]
- Hasima, N.; Ozpolat, B. Regulation of autophagy by polyphenolic compounds as a potential therapeutic strategy for cancer. Cell Death Dis. 2014, 5, e1509. [Google Scholar] [CrossRef]
- Aoki, S.; Shimizu, K.; Ito, K. Autophagy-dependent mitochondrial function regulates osteoclast differentiation and maturation. Biochem. Biophys. Res. Commun. 2020, 527, 874–880. [Google Scholar] [CrossRef]
- Doss, H.M.; Samarpita, S.; Ganesan, R.; Rasool, M. Ferulic acid, a dietary polyphenol suppresses osteoclast differentiation and bone erosion via the inhibition of RANKL dependent NF-kappaB signalling pathway. Life Sci. 2018, 207, 284–295. [Google Scholar] [CrossRef]
- Graef, J.L.; Rendina-Ruedy, E.; Crockett, E.K.; Ouyang, P.; Wu, L.; King, J.B.; Cichewicz, R.H.; Lin, D.; Lucas, E.A.; Smith, B.J. Osteoclast Differentiation is Downregulated by Select Polyphenolic Fractions from Dried Plum via Suppression of MAPKs and Nfatc1 in Mouse C57BL/6 Primary Bone Marrow Cells. Curr. Dev. Nutr. 2017, 1, e000406. [Google Scholar] [CrossRef]
- Montaseri, A.; Giampietri, C.; Rossi, M.; Riccioli, A.; del Fattore, A.; Filippini, A. The Role of Autophagy in Osteoclast Differentiation and Bone Resorption Function. Biomolecules 2020, 10, 1398. [Google Scholar] [CrossRef]
- Park-Min, K.H. Metabolic reprogramming in osteoclasts. Semin. Immunopathol. 2019, 41, 565–572. [Google Scholar] [CrossRef]
- Rantlha, M.; Sagar, T.; Kruger, M.C.; Coetzee, M.; Deepak, V. Ellagic acid inhibits RANKL-induced osteoclast differentiation by suppressing the p38 MAP kinase pathway. Arch. Pharm. Res. 2017, 40, 79–87. [Google Scholar] [CrossRef]
- Shim, K.S.; Kim, T.; Ha, H.; Lee, C.J.; Lee, B.; Kim, H.S.; Park, J.H.; Ma, J.Y. Water extract of Magnolia officinalis cortex inhibits osteoclastogenesis and bone resorption by downregulation of nuclear factor of activated T cells cytoplasmic 1. Integr. Med. Res. 2015, 4, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Koenigstein, A.; Joseph, J.; Sun, L.; Kalyanaraman, B.; Zaidi, M.; Avadhani, N.G. Role of mitochondrial reactive oxygen species in osteoclast differentiation. Ann. N. Y. Acad. Sci. 2010, 1192, 245–252. [Google Scholar] [CrossRef]
- Moradi, A.; Abolfathi, M.; Javadian, M.; Heidarian, E.; Roshanmehr, H.; Khaledi, M.; Nouri, A. Gallic Acid Exerts Nephroprotective, Anti-Oxidative Stress, and Anti-Inflammatory Effects Against Diclofenac-Induced Renal Injury in Malerats. Arch. Med. Res. 2021, 52, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.Y.; Xue, Y.R.; Xue, Y.C.; Han, X.; Song, Q.T.; Zhang, J.P.; Li, Z.L.; Cheng, J.; Guan, S.J.; Sun, S.J.; et al. Tannic acid ameliorates arsenic trioxide-induced nephrotoxicity, contribution of NF-kappa B and Nrf2 pathways. Biomed. Pharmacother. 2020, 126, 110047. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Zerin, T.; Song, H.Y. Antioxidant Action of Ellagic Acid Ameliorates Paraquat-Induced A549 Cytotoxicity. Biol. Pharm. Bull. 2013, 36, 609–615. [Google Scholar] [CrossRef]
- Nouri, A.; Heibati, F.; Heidarian, E. Gallic acid exerts anti-inflammatory, anti-oxidative stress, and nephroprotective effects against paraquat-induced renal injury in male rats. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 394, 1–9. [Google Scholar] [CrossRef]
- Thomas, A.; South, S.; Vijayagopal, P.; Juma, S. Effect of Tart Cherry Polyphenols on Osteoclast Differentiation and Activity. J. Med. Food 2020, 23, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Nicolin, V.; de Tommasi, N.; Nori, S.L.; Costantinides, F.; Berton, F.; di Lenarda, R. Modulatory Effects of Plant Polyphenols on Bone Remodeling: A Prospective View From the Bench to Bedside. Front. Endocrinol. 2019, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.C.; Wei, Y.J.; An, Z.Y.; Zou, Z.J.; Xiao, G.H.; Bhagat, G.; White, M.; Reichelt, J.; Levine, B. Akt-Mediated Regulation of Autophagy and Tumorigenesis Through Beclin 1 Phosphorylation. Science 2012, 338, 956–959. [Google Scholar] [CrossRef]
- Kajiya, H. Calcium Signaling in Osteoclast Differentiation and Bone Resorption. Calcium Signal. 2012, 740, 917–932. [Google Scholar]
- Feno, S.; Butera, G.; Reane, D.V.; Rizzuto, R.; Raffaello, A. Crosstalk between Calcium and ROS in Pathophysiological Conditions. Oxidative Med. Cell. Longev. 2019, 2019, 9324018. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Affourtit, C.; Esteves, T.C.; Green, K.; Lambert, A.J.; Miwa, S.; Pakay, J.L.; Parker, N. Mitochondrial superoxide: Production, biological effects, and activation of uncoupling proteins. Free Radic. Biol. Med. 2004, 37, 755–767. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laha, D.; Sarkar, J.; Maity, J.; Pramanik, A.; Howlader, M.S.I.; Barthels, D.; Das, H. Polyphenolic Compounds Inhibit Osteoclast Differentiation While Reducing Autophagy through Limiting ROS and the Mitochondrial Membrane Potential. Biomolecules 2022, 12, 1220. https://doi.org/10.3390/biom12091220
Laha D, Sarkar J, Maity J, Pramanik A, Howlader MSI, Barthels D, Das H. Polyphenolic Compounds Inhibit Osteoclast Differentiation While Reducing Autophagy through Limiting ROS and the Mitochondrial Membrane Potential. Biomolecules. 2022; 12(9):1220. https://doi.org/10.3390/biom12091220
Chicago/Turabian StyleLaha, Dipranjan, Jaganmay Sarkar, Jyotirindra Maity, Asmita Pramanik, Md Sariful Islam Howlader, Derek Barthels, and Hiranmoy Das. 2022. "Polyphenolic Compounds Inhibit Osteoclast Differentiation While Reducing Autophagy through Limiting ROS and the Mitochondrial Membrane Potential" Biomolecules 12, no. 9: 1220. https://doi.org/10.3390/biom12091220
APA StyleLaha, D., Sarkar, J., Maity, J., Pramanik, A., Howlader, M. S. I., Barthels, D., & Das, H. (2022). Polyphenolic Compounds Inhibit Osteoclast Differentiation While Reducing Autophagy through Limiting ROS and the Mitochondrial Membrane Potential. Biomolecules, 12(9), 1220. https://doi.org/10.3390/biom12091220







