Structural Basis of Botulinum Toxin Type F Binding to Glycosylated Human SV2A: In Silico Studies at the Periphery of a Lipid Raft
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Modeling and Systems Set Up
2.2. Molecular Dynamics Simulation
2.3. Analysis
2.4. Molecular Modeling of BoNT/F Subtypes
3. Results
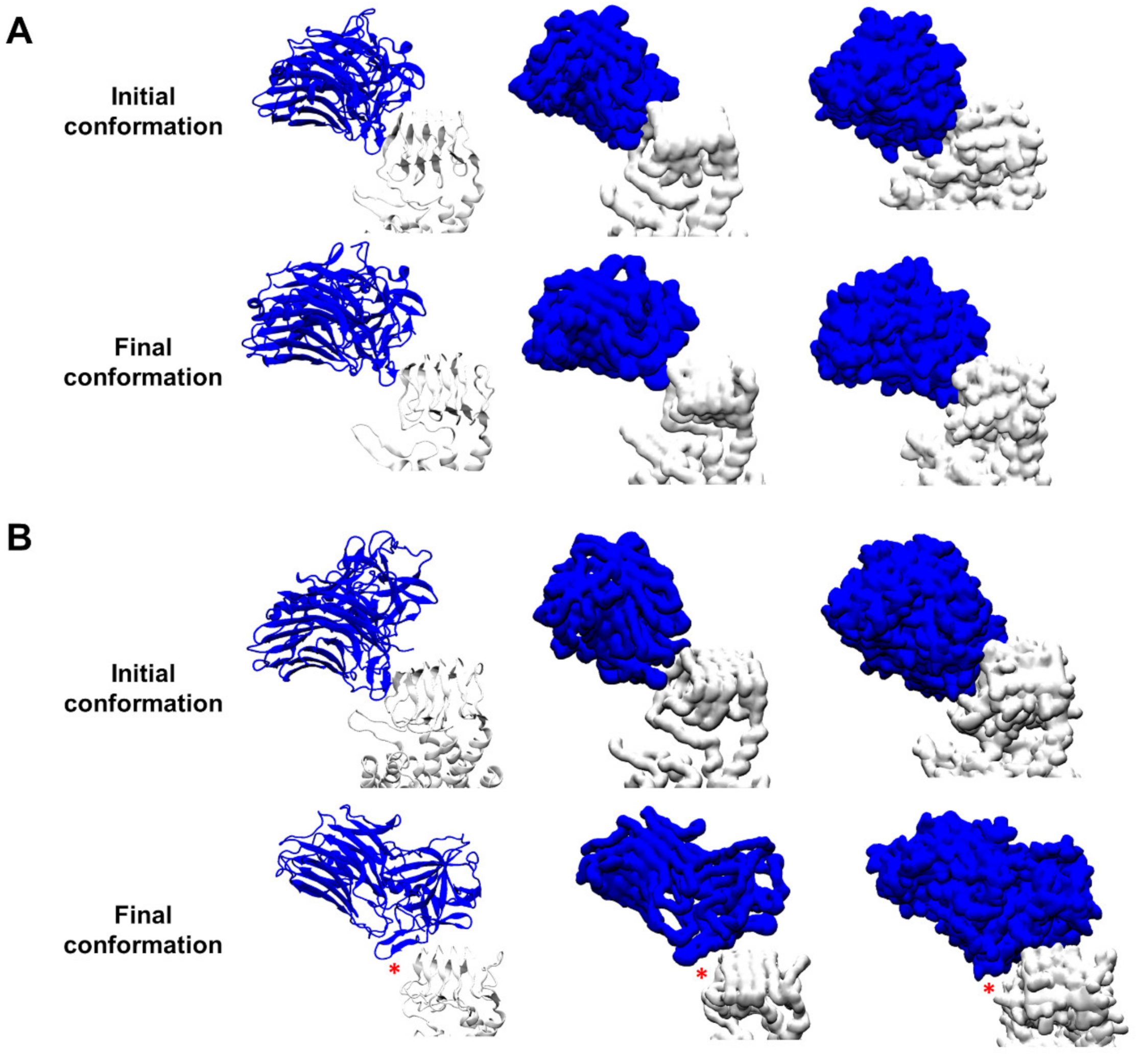
3.1. Protein–Protein Contacts between the Complexes BoNT/F1–hSV2Ag and BoNT/A1–hSV2Ag
3.2. Structural Comparison between BoNT/F1 and the Part of BoNT/A1 That Binds to Protein Moiety of hSV2Ag
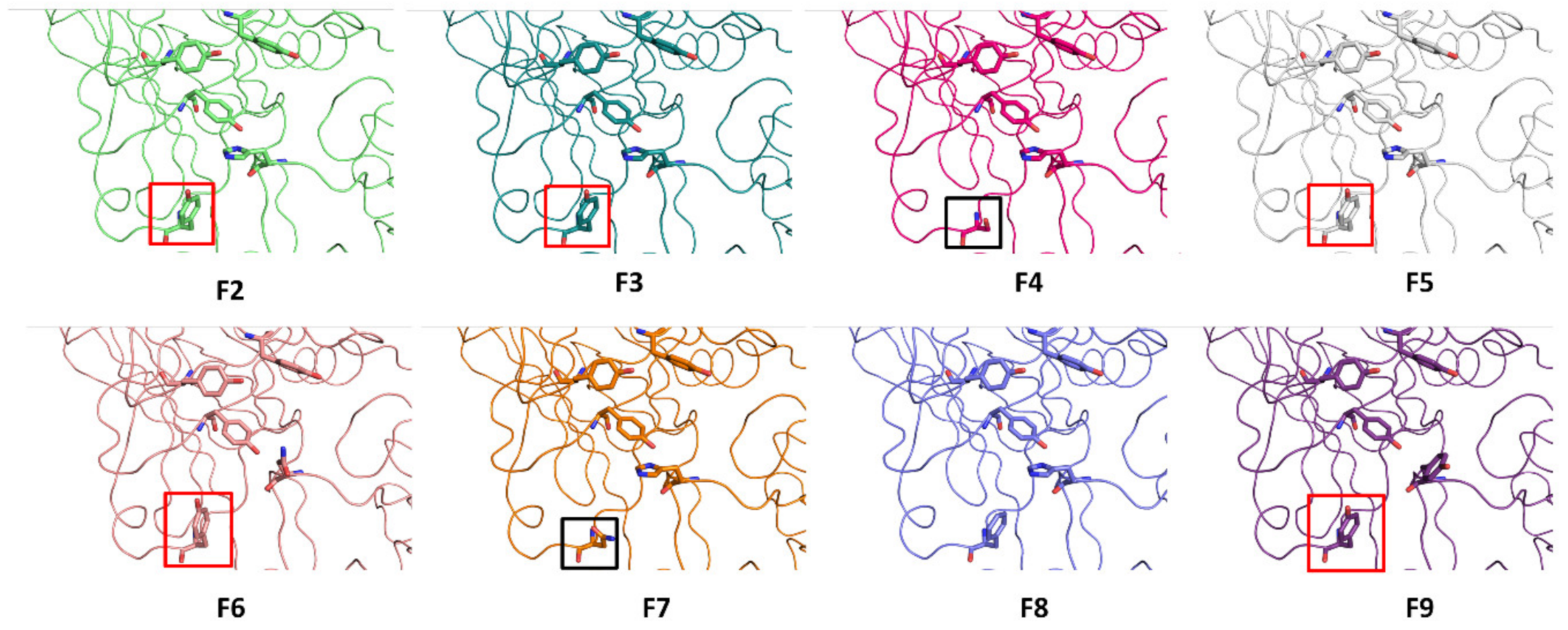
3.3. Surface-Exposed Aromatic Amino Acids of BoNT/F1 Are Key Residues for Binding to N-Glycan of SV2
3.4. Molecular Details Describing Interaction of BoNT/F1 with Lipid-Raft-Associated GT1b
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, A.; Sumner, C.J.; Castor, M.; Maslanka, S.; Sobel, J. Adult botulism type F in the United States, 1981–2002. Neurology 2005, 65, 1694–1700. [Google Scholar] [CrossRef] [PubMed]
- Verderio, C.; Rossetto, O.; Grumelli, C.; Frassoni, C.; Montecucco, C.; Matteoli, M. Entering neurons: Botulinum toxins and synaptic vesicle recycling. EMBO Rep. 2006, 7, 995–999. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Masuyer, G.; Zhang, J.; Shen, Y.; Lundin, D.; Henriksson, L.; Miyashita, S.I.; Martinez-Carranza, M.; Dong, M.; Stenmark, P. Identification and characterization of a novel botulinum neurotoxin. Nat. Commun. 2017, 8, 14130. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Stenmark, P. The Structure and Classification of Botulinum Toxins. Handb. Exp. Pharmacol. 2021, 263, 11–33. [Google Scholar] [CrossRef]
- Eleopra, R.; Rinaldo, S.; Montecucco, C.; Rossetto, O.; Devigili, G. Clinical duration of action of different botulinum toxin types in humans. Toxicon 2020, 179, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Pirazzini, M.; Rossetto, O.; Eleopra, R.; Montecucco, C. Botulinum Neurotoxins: Biology, Pharmacology, and Toxicology. Pharmacol. Rev. 2017, 69, 200–235. [Google Scholar] [CrossRef] [Green Version]
- Rummel, A.; Hafner, K.; Mahrhold, S.; Darashchonak, N.; Holt, M.; Jahn, R.; Beermann, S.; Karnath, T.; Bigalke, H.; Binz, T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J. Neurochem. 2009, 110, 1942–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Z.; Chen, C.; Barbieri, J.T.; Kim, J.J.; Baldwin, M.R. Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry 2009, 48, 5631–5641. [Google Scholar] [CrossRef] [Green Version]
- Eleopra, R.; Tugnoli, V.; Montecucco, C. 6-Biology and Clinical Pharmacology of Botulinum Neurotoxin Type C and Other Non-A/Non-B Botulinum Neurotoxins. In Botulinum Toxin; Jankovic, J., Albanese, A., Atassi, M.Z., Dolly, J.O., Hallett, M., Mayer, N.H., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2009; pp. 77–84. [Google Scholar]
- Cenciarelli, O.; Riley, P.W.; Baka, A. Biosecurity Threat Posed by Botulinum Toxin. Toxins 2019, 11, 681. [Google Scholar] [CrossRef] [Green Version]
- Yao, G.; Zhang, S.; Mahrhold, S.; Lam, K.-h.; Stern, D.; Bagramyan, K.; Perry, K.; Kalkum, M.; Rummel, A.; Dong, M.; et al. N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A. Nat. Struct. Mol. Biol. 2016, 23, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Azzaz, F.; Yahi, N.; Chahinian, H.; Fantini, J. The Epigenetic Dimension of Protein Structure Is an Intrinsic Weakness of the AlphaFold Program. Biomolecules 2022, 12, 1527. [Google Scholar] [CrossRef]
- Azzaz, F.; Fantini, J. The epigenetic dimension of protein structure. Biomol. Concepts 2022, 13, 55–60. [Google Scholar] [CrossRef]
- Dong, M.; Liu, H.; Tepp, W.H.; Johnson, E.A.; Janz, R.; Chapman, E.R. Glycosylated SV2A and SV2B Mediate the Entry of Botulinum Neurotoxin E into Neurons. Mol. Biol. Cell 2008, 19, 5226–5237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Patel, D.S.; Ståhle, J.; Park, S.J.; Kern, N.R.; Kim, S.; Lee, J.; Cheng, X.; Valvano, M.A.; Holst, O.; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput. 2019, 15, 775–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzaz, F.; Hilaire, D.; Fantini, J. Structural basis of botulinum neurotoxin serotype A1 binding to human SV2A or SV2C receptors. bioRxiv 2022. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Benoit, R.M.; Frey, D.; Hilbert, M.; Kevenaar, J.T.; Wieser, M.M.; Stirnimann, C.U.; McMillan, D.; Ceska, T.; Lebon, F.; Jaussi, R.; et al. Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A. Nature 2014, 505, 108–111. [Google Scholar] [CrossRef]
- Mahrhold, S.; Bergström, T.; Stern, D.; Dorner, B.G.; Åstot, C.; Rummel, A. Only the complex N559-glycan in the synaptic vesicle glycoprotein 2C mediates high affinity binding to botulinum neurotoxin serotype A1. Biochem. J. 2016, 473, 2645–2654. [Google Scholar] [CrossRef]
- Nishio, M.; Umezawa, Y.; Fantini, J.; Weiss, M.S.; Chakrabarti, P. CH-π hydrogen bonds in biological macromolecules. Phys. Chem. Chem. Phys. PCCP 2014, 16, 12648–12683. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Yahi, N. The driving force of alpha-synuclein insertion and amyloid channel formation in the plasma membrane of neural cells: Key role of ganglioside- and cholesterol-binding domains. Adv. Exp. Med. Biol. 2013, 991, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Chahinian, H.; Yahi, N. Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Chahinian, H.; Yahi, N. A Vaccine Strategy Based on the Identification of an Annular Ganglioside Binding Motif in Monkeypox Virus Protein E8L. Viruses 2022, 14, 2531. [Google Scholar] [CrossRef]
- Dong, M.; Yeh, F.; Tepp, W.H.; Dean, C.; Johnson, E.A.; Janz, R.; Chapman, E.R. SV2 is the protein receptor for botulinum neurotoxin A. Science 2006, 312, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Eleopra, R.; Tugnoli, V.; Quatrale, R.; Rossetto, O.; Montecucco, C.; Dressler, D. Clinical use of non-A botulinum toxins: Botulinum toxin type C and botulinum toxin type F. Neurotox. Res. 2006, 9, 127–131. [Google Scholar] [CrossRef]
- Houser, M.K.; Sheean, G.L.; Lees, A.J. Further studies using higher doses of botulinum toxin type F for torticollis resistant to botulinum toxin type A. J. Neurol. Neurosurg. Psychiatry 1998, 64, 577–580. [Google Scholar] [CrossRef] [Green Version]
- Tao, L.; Peng, L.; Berntsson, R.P.; Liu, S.M.; Park, S.; Yu, F.; Boone, C.; Palan, S.; Beard, M.; Chabrier, P.E.; et al. Engineered botulinum neurotoxin B with improved efficacy for targeting human receptors. Nat. Commun. 2017, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Elliott, M.; Favre-Guilmard, C.; Liu, S.M.; Maignel, J.; Masuyer, G.; Beard, M.; Boone, C.; Carré, D.; Kalinichev, M.; Lezmi, S.; et al. Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models. Sci. Adv. 2019, 5, eaau7196. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Masuyer, G.; Zhang, S.; Zhang, J.; Miyashita, S.I.; Burgin, D.; Lovelock, L.; Coker, S.F.; Fu, T.M.; Stenmark, P.; et al. Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy. PLoS Biol. 2020, 18, e3000618. [Google Scholar] [CrossRef]
- Mahrhold, S.; Strotmeier, J.; Garcia-Rodriguez, C.; Lou, J.; Marks, J.D.; Rummel, A.; Binz, T. Identification of the SV2 protein receptor-binding site of botulinum neurotoxin type E. Biochem. J. 2013, 453, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Stern, D.; Weisemann, J.; Le Blanc, A.; von Berg, L.; Mahrhold, S.; Piesker, J.; Laue, M.; Luppa, P.B.; Dorner, M.B.; Dorner, B.G.; et al. A lipid-binding loop of botulinum neurotoxin serotypes B, DC and G is an essential feature to confer their exquisite potency. PLoS Pathog. 2018, 14, e1007048. [Google Scholar] [CrossRef] [Green Version]
- Di Scala, C.; Fantini, J. Hybrid in silico/in vitro approaches for the identification of functional cholesterol-binding domains in membrane proteins. In Cholesterol Homeostasis; Springer: Berlin/Heidelberg, Germany, 2017; pp. 7–19. [Google Scholar]
- Di Scala, C.; Yahi, N.; Boutemeur, S.; Flores, A.; Rodriguez, L.; Chahinian, H.; Fantini, J. Common molecular mechanism of amyloid pore formation by Alzheimer’s β-amyloid peptide and α-synuclein. Sci. Rep. 2016, 6, 28781. [Google Scholar] [CrossRef] [Green Version]
- Di Scala, C.; Yahi, N.; Flores, A.; Boutemeur, S.; Kourdougli, N.; Chahinian, H.; Fantini, J. Broad neutralization of calcium-permeable amyloid pore channels with a chimeric Alzheimer/Parkinson peptide targeting brain gangliosides. Biochim. Biophys. Acta 2016, 1862, 213–222. [Google Scholar] [CrossRef]
- Yahi, N.; Fantini, J. Deciphering the glycolipid code of Alzheimer’s and Parkinson’s amyloid proteins allowed the creation of a universal ganglioside-binding peptide. PLoS ONE 2014, 9, e104751. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Garmy, N.; Yahi, N. Prediction of glycolipid-binding domains from the amino acid sequence of lipid raft-associated proteins: Application to HpaA, a protein involved in the adhesion of Helicobacter pylori to gastrointestinal cells. Biochemistry 2006, 45, 10957–10962. [Google Scholar] [CrossRef]
- Benson, M.A.; Fu, Z.; Kim, J.J.; Baldwin, M.R. Unique ganglioside recognition strategies for clostridial neurotoxins. J. Biol. Chem. 2011, 286, 34015–34022. [Google Scholar] [CrossRef] [Green Version]
- Chai, Q.; Arndt, J.W.; Dong, M.; Tepp, W.H.; Johnson, E.A.; Chapman, E.R.; Stevens, R.C. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature 2006, 444, 1096–1100. [Google Scholar] [CrossRef]
- Jin, R.; Rummel, A.; Binz, T.; Brunger, A.T. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 2006, 444, 1092–1095. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzaz, F.; Hilaire, D.; Fantini, J. Structural Basis of Botulinum Toxin Type F Binding to Glycosylated Human SV2A: In Silico Studies at the Periphery of a Lipid Raft. Biomolecules 2022, 12, 1821. https://doi.org/10.3390/biom12121821
Azzaz F, Hilaire D, Fantini J. Structural Basis of Botulinum Toxin Type F Binding to Glycosylated Human SV2A: In Silico Studies at the Periphery of a Lipid Raft. Biomolecules. 2022; 12(12):1821. https://doi.org/10.3390/biom12121821
Chicago/Turabian StyleAzzaz, Fodil, Didier Hilaire, and Jacques Fantini. 2022. "Structural Basis of Botulinum Toxin Type F Binding to Glycosylated Human SV2A: In Silico Studies at the Periphery of a Lipid Raft" Biomolecules 12, no. 12: 1821. https://doi.org/10.3390/biom12121821
APA StyleAzzaz, F., Hilaire, D., & Fantini, J. (2022). Structural Basis of Botulinum Toxin Type F Binding to Glycosylated Human SV2A: In Silico Studies at the Periphery of a Lipid Raft. Biomolecules, 12(12), 1821. https://doi.org/10.3390/biom12121821







