Endolysosomal Cation Channels and MITF in Melanocytes and Melanoma
Abstract
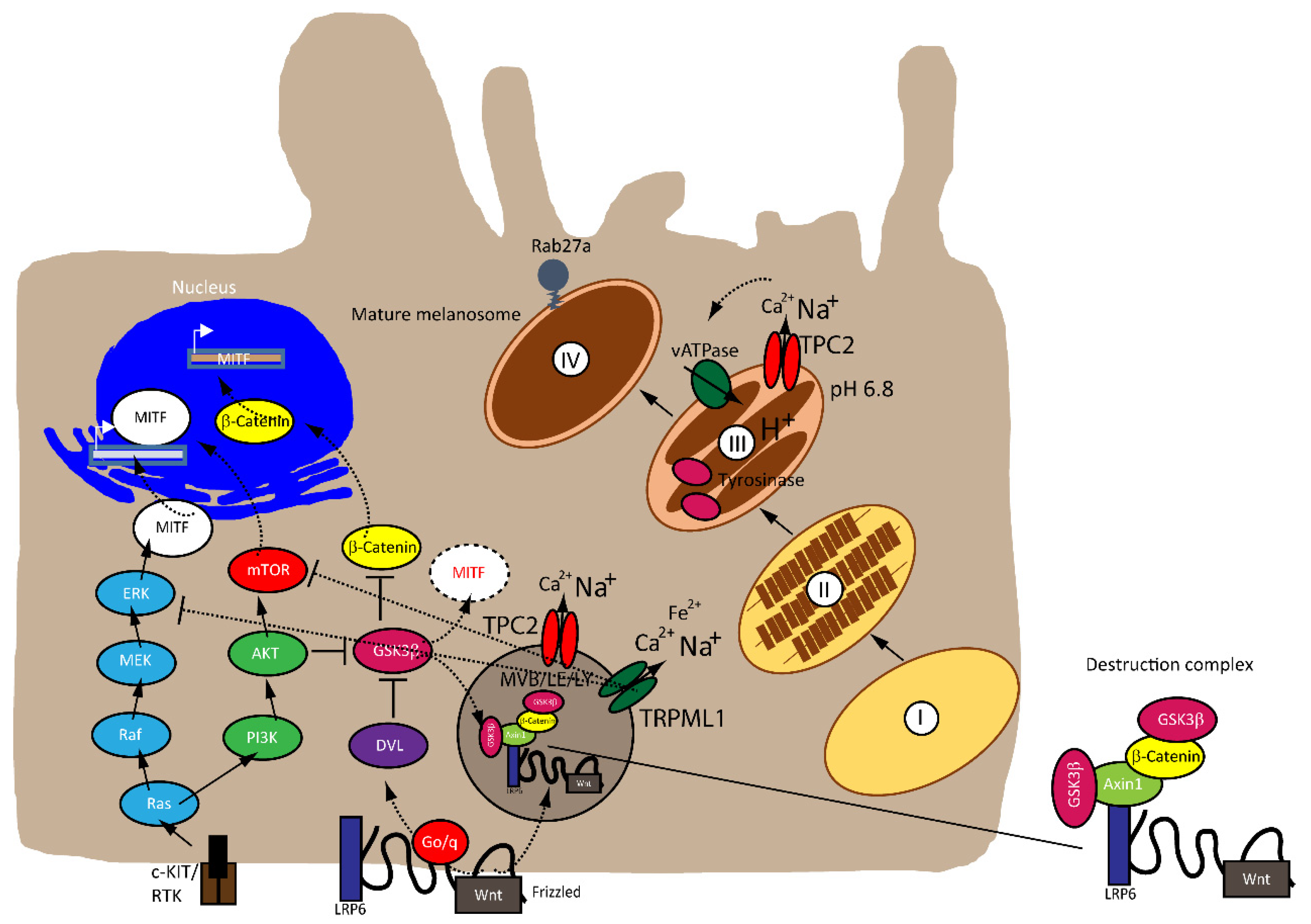
:1. Introduction
2. Role of MITF in Melanoma and Pathways Implicated
2.1. MITF
2.2. Rheostat Model of MITF
2.3. MITF and MAPK
2.4. MITF and Canonical Wnt Pathway
3. Endolysosomal Cation Channels in Melanoma
3.1. TPC2
3.2. TRPML1
4. Summary and Conclusions
Funding
Conflicts of Interest
References
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Hearing, V.J. Melanocytes and their diseases. Cold Spring Harb. Perspect. Med. 2014, 4, a017046. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Marks, M.S. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007, 8, 786–797. [Google Scholar] [CrossRef] [Green Version]
- Wasmeier, C.; Hume, A.N.; Bolasco, G.; Seabra, M.C. Melanosomes at a glance. J. Cell Sci. 2008, 121, 3995–3999. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Wakamatsu, K. Human hair melanins: What we have learned and have not learned from mouse coat color pigmentation. Pigment Cell Melanoma Res. 2011, 24, 63–74. [Google Scholar] [CrossRef]
- Bellono, N.W.; Escobar, I.E.; Oancea, E. A melanosomal two-pore sodium channel regulates pigmentation. Sci. Rep. 2016, 6, 26570. [Google Scholar] [CrossRef]
- Ambrosio, A.L.; Boyle, J.A.; Aradi, A.E.; Christian, K.A.; Di Pietro, S.M. TPC2 controls pigmentation by regulating melanosome pH and size. Proc. Natl. Acad. Sci. USA 2016, 113, 5622–5627. [Google Scholar] [CrossRef] [Green Version]
- Chao, Y.K.; Schludi, V.; Chen, C.C.; Butz, E.; Nguyen, O.N.P.; Muller, M.; Kruger, J.; Kammerbauer, C.; Ben-Johny, M.; Vollmar, A.M.; et al. TPC2 polymorphisms associated with a hair pigmentation phenotype in humans result in gain of channel function by independent mechanisms. Proc. Natl. Acad. Sci. USA 2017, 114, E8595–E8602. [Google Scholar] [CrossRef] [Green Version]
- Lerner, A.B.; Fitzpatrick, T.B.; Calkins, E.; Summerson, W.H. Mammalian tyrosinase; preparation and properties. J. Biol. Chem. 1949, 178, 185–195. [Google Scholar] [CrossRef]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef]
- Schallreuter, K.U.; Kothari, S.; Chavan, B.; Spencer, J.D. Regulation of melanogenesis--controversies and new concepts. Exp. Dermatol. 2008, 17, 395–404. [Google Scholar] [CrossRef]
- Land, E.J.; Ramsden, C.A.; Riley, P.A. Quinone chemistry and melanogenesis. Methods Enzymol. 2004, 378, 88–109. [Google Scholar]
- Sulem, P.; Gudbjartsson, D.F.; Stacey, S.N.; Helgason, A.; Rafnar, T.; Jakobsdottir, M.; Steinberg, S.; Gudjonsson, S.A.; Palsson, A.; Thorleifsson, G.; et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 2008, 40, 835–837. [Google Scholar] [CrossRef]
- Böck, J.; Krogsaeter, E.; Passon, M.; Chao, Y.K.; Sharma, S.; Grallert, H.; Peters, A.; Grimm, C. Human genome diversity data reveal that L564P is the predominant TPC2 variant and a prerequisite for the blond hair associated M484L gain-of-function effect. PLoS Genet. 2021, 17, e1009236. [Google Scholar] [CrossRef]
- Sample, A.; He, Y.Y. Mechanisms and prevention of UV-induced melanoma. Photodermatol. Photoimmunol. Photomed. 2018, 34, 13–24. [Google Scholar] [CrossRef]
- Premi, S.; Wallisch, S.; Mano, C.M.; Weiner, A.B.; Bacchiocchi, A.; Wakamatsu, K.; Bechara, E.J.; Halaban, R.; Douki, T.; Brash, D.E. Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science 2015, 347, 842–847. [Google Scholar] [CrossRef]
- Williams, P.F.; Olsen, C.M.; Hayward, N.K.; Whiteman, D.C. Melanocortin 1 receptor and risk of cutaneous melanoma: A meta-analysis and estimates of population burden. Int. J. Cancer 2011, 129, 1730–1740. [Google Scholar] [CrossRef]
- Favia, A.; Desideri, M.; Gambara, G.; D’Alessio, A.; Ruas, M.; Esposito, B.; Del Bufalo, D.; Parrington, J.; Ziparo, E.; Palombi, F.; et al. VEGF-induced neoangiogenesis is mediated by NAADP and two-pore channel-2-dependent Ca2+ signaling. Proc. Natl. Acad. Sci. USA 2014, 111, E4706–E4715. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, O.N.; Grimm, C.; Schneider, L.S.; Chao, Y.K.; Atzberger, C.; Bartel, K.; Watermann, A.; Ulrich, M.; Mayr, D.; Wahl-Schott, C.; et al. Two-Pore Channel Function Is Crucial for the Migration of Invasive Cancer Cells. Cancer Res. 2017, 77, 1427–1438. [Google Scholar] [CrossRef] [Green Version]
- Grimm, C.; Bartel, K.; Vollmar, A.M.; Biel, M. Endolysosomal Cation Channels and Cancer-A Link with Great Potential. Pharmaceuticals 2018, 11, 4. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Yue, J. TPC2 mediates autophagy progression and extracellular vesicle secretion in cancer cells. Exp. Cell. Res. 2018, 370, 478–489. [Google Scholar] [CrossRef]
- Alharbi, A.F.; Parrington, J. Endolysosomal Ca(2+) Signaling in Cancer: The Role of TPC2, From Tumorigenesis to Metastasis. Front. Cell Dev. Biol. 2019, 7, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faris, P.; Pellavio, G.; Ferulli, F.; Di Nezza, F.; Shekha, M.; Lim, D.; Maestri, M.; Guerra, G.; Ambrosone, L.; Pedrazzoli, P.; et al. Nicotinic Acid Adenine Dinucleotide Phosphate (NAADP) Induces Intracellular Ca(2+) Release through the Two-Pore Channel TPC1 in Metastatic Colorectal Cancer Cells. Cancers 2019, 11, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Amore, A.; Hanbashi, A.A.; Di Agostino, S.; Palombi, F.; Sacconi, A.; Voruganti, A.; Taggi, M.; Canipari, R.; Blandino, G.; Parrington, J.; et al. Loss of Two-Pore Channel 2 (TPC2) Expression Increases the Metastatic Traits of Melanoma Cells by a Mechanism Involving the Hippo Signalling Pathway and Store-Operated Calcium Entry. Cancers 2020, 12, 2391. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Gerndt, S.; Chao, Y.K.; Zisis, T.; Nguyen, O.N.P.; Gerwien, A.; Urban, N.; Müller, C.; Gegenfurtner, F.A.; Geisslinger, F.; et al. Gene editing and synthetically accessible inhibitors reveal role for TPC2 in HCC cell proliferation and tumor growth. Cell Chem. Biol. 2021. [Google Scholar] [CrossRef]
- Netcharoensirisuk, P.; Abrahamian, C.; Tang, R.; Chen, C.C.; Rosato, A.S.; Beyers, W.; Chao, Y.K.; Filippini, A.; Di Pietro, S.; Bartel, K.; et al. Flavonoids increase melanin production and reduce proliferation, migration and invasion of melanoma cells by blocking endolysosomal/melanosomal TPC2. Sci. Rep. 2021, 11, 8515. [Google Scholar] [CrossRef] [PubMed]
- Vachtenheim, J.; Borovanský, J. “Transcription physiology” of pigment formation in melanocytes: Central role of MITF. Exp. Dermatol. 2010, 19, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Steingrímsson, E.; Copeland, N.G.; Jenkins, N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004, 38, 365–411. [Google Scholar] [CrossRef]
- Slade, L.; Pulinilkunnil, T. The MiTF/TFE Family of Transcription Factors: Master Regulators of Organelle Signaling, Metabolism, and Stress Adaptation. Mol. Cancer Res. 2017, 15, 1637–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Spina, M.; Contreras, P.S.; Rissone, A.; Meena, N.K.; Jeong, E.; Martina, J.A. MiT/TFE Family of Transcription Factors: An Evolutionary Perspective. Front. Cell Dev. Biol. 2021, 8, 1580. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Zhang, D.; Beyer, L.A.; Halsey, K.E.; Fukui, H.; Raphael, Y.; Dolan, D.F.; Hornyak, T.J. Hearing dysfunction in heterozygous Mitf(Mi-wh)/+ mice, a model for Waardenburg syndrome type 2 and Tietz syndrome. Pigment Cell Melanoma Res. 2013, 26, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauschendorf, M.A.; Zimmer, A.D.; Laut, A.; Demmer, P.; Rösler, B.; Happle, R.; Sartori, S.; Fischer, J. Homozygous intronic MITF mutation causes severe Waardenburg syndrome type 2A. Pigment Cell Melanoma Res. 2019, 32, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, G.M.; Aithal, C.; Kraechan, A.; Heise, C.; Cadilha, B.L.; Zhang, J.; Duewell, P.; Ballotti, R.; Endres, S.; Bertolotto, C.; et al. Microphthalmia-Associated Transcription Factor (MITF) Regulates Immune Cell Migration into Melanoma. Transl. Oncol. 2019, 12, 350–360. [Google Scholar] [CrossRef]
- Cheli, Y.; Ohanna, M.; Ballotti, R.; Bertolotto, C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010, 23, 27–40. [Google Scholar] [CrossRef]
- Wellbrock, C.; Arozarena, I. Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment Cell Melanoma Res. 2015, 28, 390–406. [Google Scholar] [CrossRef] [Green Version]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; Larue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes. Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef] [Green Version]
- Hoek, K.S.; Goding, C.R. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010, 23, 746–759. [Google Scholar] [CrossRef]
- Miller, A.J.; Levy, C.; Davis, I.J.; Razin, E.; Fisher, D.E. Sumoylation of MITF and its related family members TFE3 and TFEB. J. Biol. Chem. 2005, 280, 146–155. [Google Scholar] [CrossRef] [Green Version]
- Haass, N.K.; Beaumont, K.A.; Hill, D.S.; Anfosso, A.; Mrass, P.; Munoz, M.A.; Kinjyo, I.; Weninger, W. Real-time cell cycle imaging during melanoma growth, invasion, and drug response. Pigment Cell Melanoma Res. 2014, 27, 764–776. [Google Scholar] [CrossRef]
- Barysch, M.J.; Mangana, J.; Dummer, R. A new B-Raf inhibitor combo for advanced melanoma. Oncotarget 2018, 9, 34457–34458. [Google Scholar] [CrossRef]
- Greaves, W.O.; Verma, S.; Patel, K.P.; Davies, M.A.; Barkoh, B.A.; Galbincea, J.M.; Yao, H.; Lazar, A.J.; Aldape, K.D.; Medeiros, L.J.; et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J. Mol. Diagn. 2013, 15, 220–226. [Google Scholar] [CrossRef]
- Heppt, M.V.; Siepmann, T.; Engel, J.; Schubert-Fritschle, G.; Eckel, R.; Mirlach, L.; Kirchner, T.; Jung, A.; Gesierich, A.; Ruzicka, T.; et al. Prognostic significance of BRAF and NRAS mutations in melanoma: A German study from routine care. BMC Cancer 2017, 17, 536. [Google Scholar] [CrossRef]
- Zhou, A.Y.; Johnson, D.B. Combinatorial Therapies in Melanoma: MAPK Inhibitors and Beyond. Am. J. Clin. Dermatol. 2018, 19, 181–193. [Google Scholar] [CrossRef]
- Inamdar, G.S.; Madhunapantula, S.V.; Robertson, G.P. Targeting the MAPK pathway in melanoma: Why some approaches succeed and other fail. Biochem. Pharmacol. 2010, 80, 624–637. [Google Scholar] [CrossRef] [Green Version]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef] [Green Version]
- Lister, J.A.; Capper, A.; Zeng, Z.; Mathers, M.E.; Richardson, J.; Paranthaman, K.; Jackson, I.J.; Elizabeth Patton, E. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J. Investig. Dermatol. 2014, 134, 133–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemesath, T.J.; Price, E.R.; Takemoto, C.; Badalian, T.; Fisher, D.E. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature 1998, 391, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Hemesath, T.J.; Takemoto, C.M.; Horstmann, M.A.; Wells, A.G.; Price, E.R.; Fisher, D.Z.; Fisher, D.E. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes. Dev. 2000, 14, 301–312. [Google Scholar] [PubMed]
- Price, E.R.; Horstmann, M.A.; Wells, A.G.; Weilbaecher, K.N.; Takemoto, C.M.; Landis, M.W.; Fisher, D.E. alpha-Melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J. Biol. Chem. 1998, 273, 33042–33047. [Google Scholar] [CrossRef] [Green Version]
- Ploper, D.; Taelman, V.F.; Robert, L.; Perez, B.S.; Titz, B.; Chen, H.W.; Graeber, T.G.; von Euw, E.; Ribas, A.; De Robertis, E.M. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, E420–E429. [Google Scholar] [CrossRef] [Green Version]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef]
- Johnson, M.L.; Rajamannan, N. Diseases of Wnt signaling. Rev. Endocr. Metab. Disord. 2006, 7, 41–49. [Google Scholar] [CrossRef] [Green Version]
- De Ferrari, G.V.; Inestrosa, N.C. Wnt signaling function in Alzheimer’s disease. Brain Res. Brain Res. Rev. 2000, 33, 1–12. [Google Scholar] [CrossRef]
- Larue, L.; Beermann, F. Cutaneous melanoma in genetically modified animals. Pigment Cell Res. 2007, 20, 485–497. [Google Scholar] [CrossRef]
- Larue, L.; Luciani, F.; Kumasaka, M.; Champeval, D.; Demirkan, N.; Bonaventure, J.; Delmas, V. Bypassing melanocyte senescence by beta-catenin: A novel way to promote melanoma. Pathol. Biol. 2009, 57, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Delmas, V.; Beermann, F.; Martinozzi, S.; Carreira, S.; Ackermann, J.; Kumasaka, M.; Denat, L.; Goodall, J.; Luciani, F.; Viros, A.; et al. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes. Dev. 2007, 21, 2923–2935. [Google Scholar] [CrossRef] [Green Version]
- Pafumi, I.; Festa, M.; Papacci, F.; Lagostena, L.; Giunta, C.; Gutla, V.; Cornara, L.; Favia, A.; Palombi, F.; Gambale, F.; et al. Naringenin Impairs Two-Pore Channel 2 Activity And Inhibits VEGF-Induced Angiogenesis. Sci. Rep. 2017, 7, 5121. [Google Scholar] [CrossRef] [PubMed]
- Sterea, A.M.; Almasi, S.; El Hiani, Y. The hidden potential of lysosomal ion channels: A new era of oncogenes. Cell Calcium 2018, 72, 91–103. [Google Scholar] [CrossRef]
- Santoni, G.; Santoni, M.; Maggi, F.; Marinelli, O.; Morelli, M.B. Emerging Role of Mucolipins TRPML Channels in Cancer. Front. Oncol. 2020, 10, 659. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Dong, X.P. Endolysosomal TRPMLs in Cancer. Biomolecules 2021, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Kasitinon, S.Y.; Eskiocak, U.; Martin, M.; Bezwada, D.; Khivansara, V.; Tasdogan, A.; Zhao, Z.; Mathews, T.; Aurora, A.B.; Morrison, S.J. TRPML1 Promotes Protein Homeostasis in Melanoma Cells by Negatively Regulating MAPK and mTORC1 Signaling. Cell Rep. 2019, 28, 2293–2305.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.J.; Xu, J.; Fu, C.; Zhang, J.; Zheng, Y.G.; Jia, H.; Liu, J.O. Regulation of mTORC1 by lysosomal calcium and calmodulin. eLife 2016, 5, e19360. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rydzewski, N.; Hider, A.; Zhang, X.; Yang, J.; Wang, W.; Gao, Q.; Cheng, X.; Xu, H. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 2016, 18, 404–417. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Gao, Q.; Yang, M.; Zhang, X.; Yu, L.; Lawas, M.; Li, X.; Bryant-Genevier, M.; Southall, N.T.; Marugan, J.; et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl. Acad. Sci. USA 2015, 112, E1373–E1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rühl, P.; Rosato, A.S.; Urban, N.; Gerndt, S.; Tang, R.; Abrahamian, C.; Leser, C.; Sheng, J.; Jha, A.; Vollmer, G.; et al. Estradiol analogs attenuate autophagy, cell migration and invasion by direct and selective inhibition of TRPML1, independent of estrogen receptors. Sci. Rep. 2021, 11, 8313. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abrahamian, C.; Grimm, C. Endolysosomal Cation Channels and MITF in Melanocytes and Melanoma. Biomolecules 2021, 11, 1021. https://doi.org/10.3390/biom11071021
Abrahamian C, Grimm C. Endolysosomal Cation Channels and MITF in Melanocytes and Melanoma. Biomolecules. 2021; 11(7):1021. https://doi.org/10.3390/biom11071021
Chicago/Turabian StyleAbrahamian, Carla, and Christian Grimm. 2021. "Endolysosomal Cation Channels and MITF in Melanocytes and Melanoma" Biomolecules 11, no. 7: 1021. https://doi.org/10.3390/biom11071021
APA StyleAbrahamian, C., & Grimm, C. (2021). Endolysosomal Cation Channels and MITF in Melanocytes and Melanoma. Biomolecules, 11(7), 1021. https://doi.org/10.3390/biom11071021






