Effects of Protein Unfolding on Aggregation and Gelation in Lysozyme Solutions
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Apparent Monomer Size
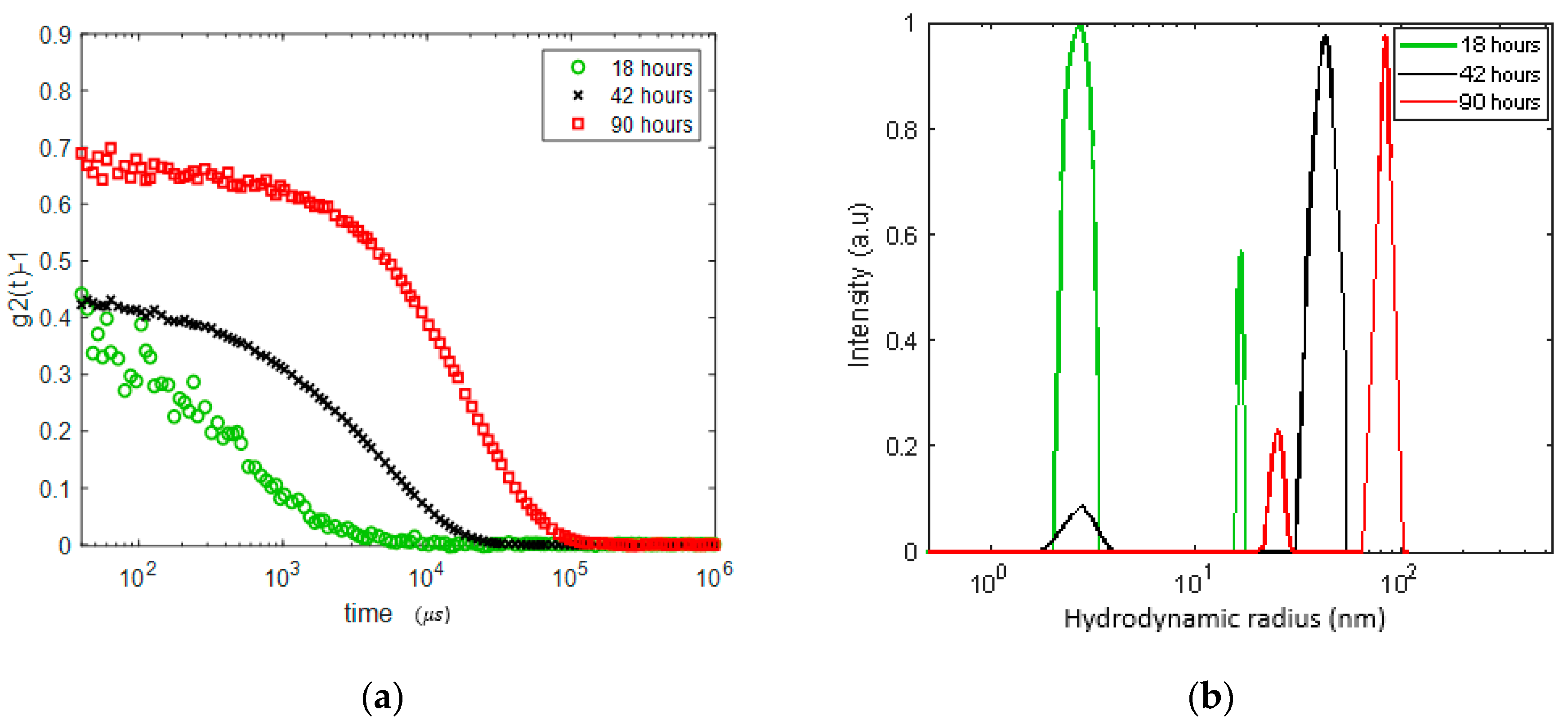
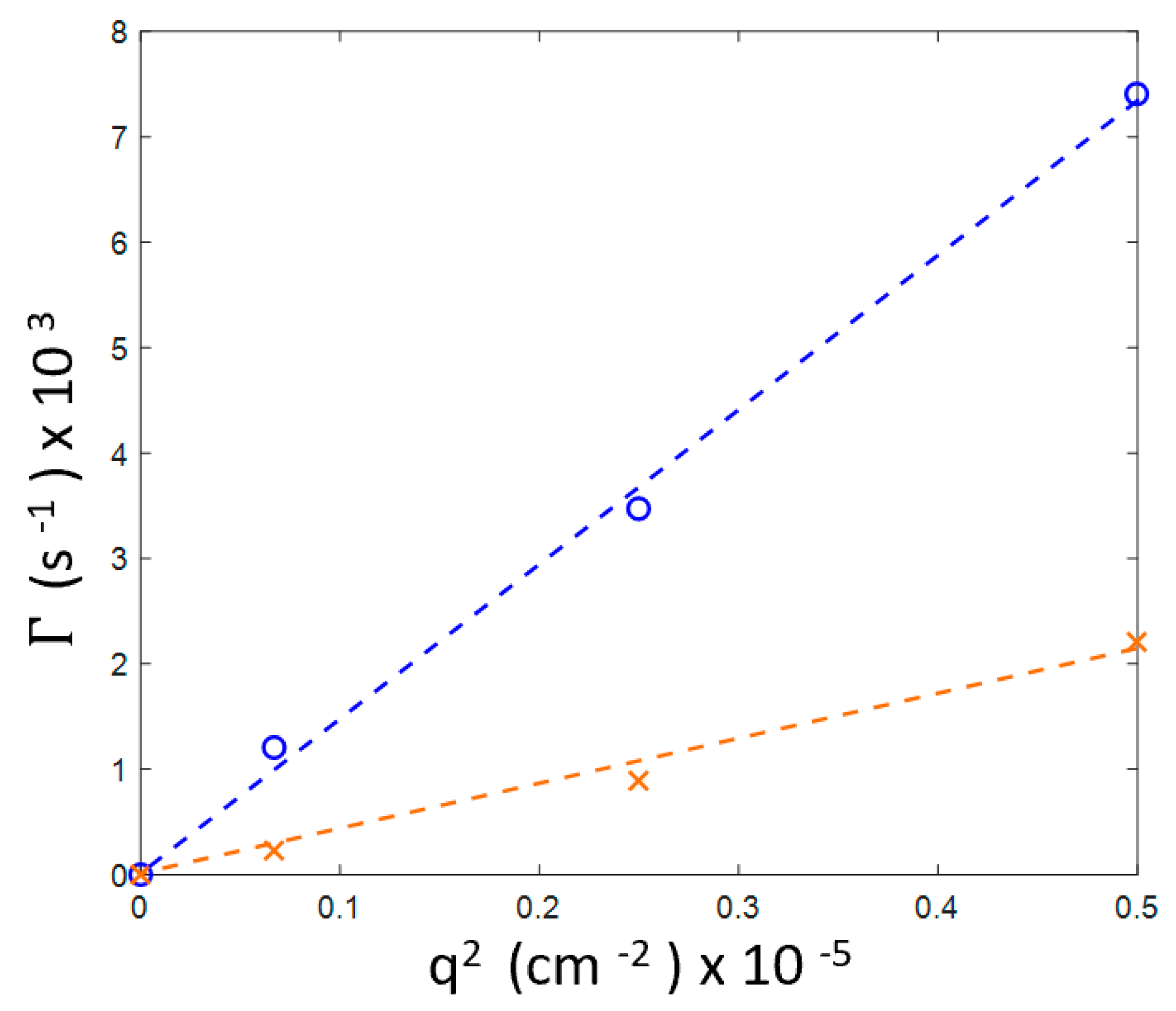
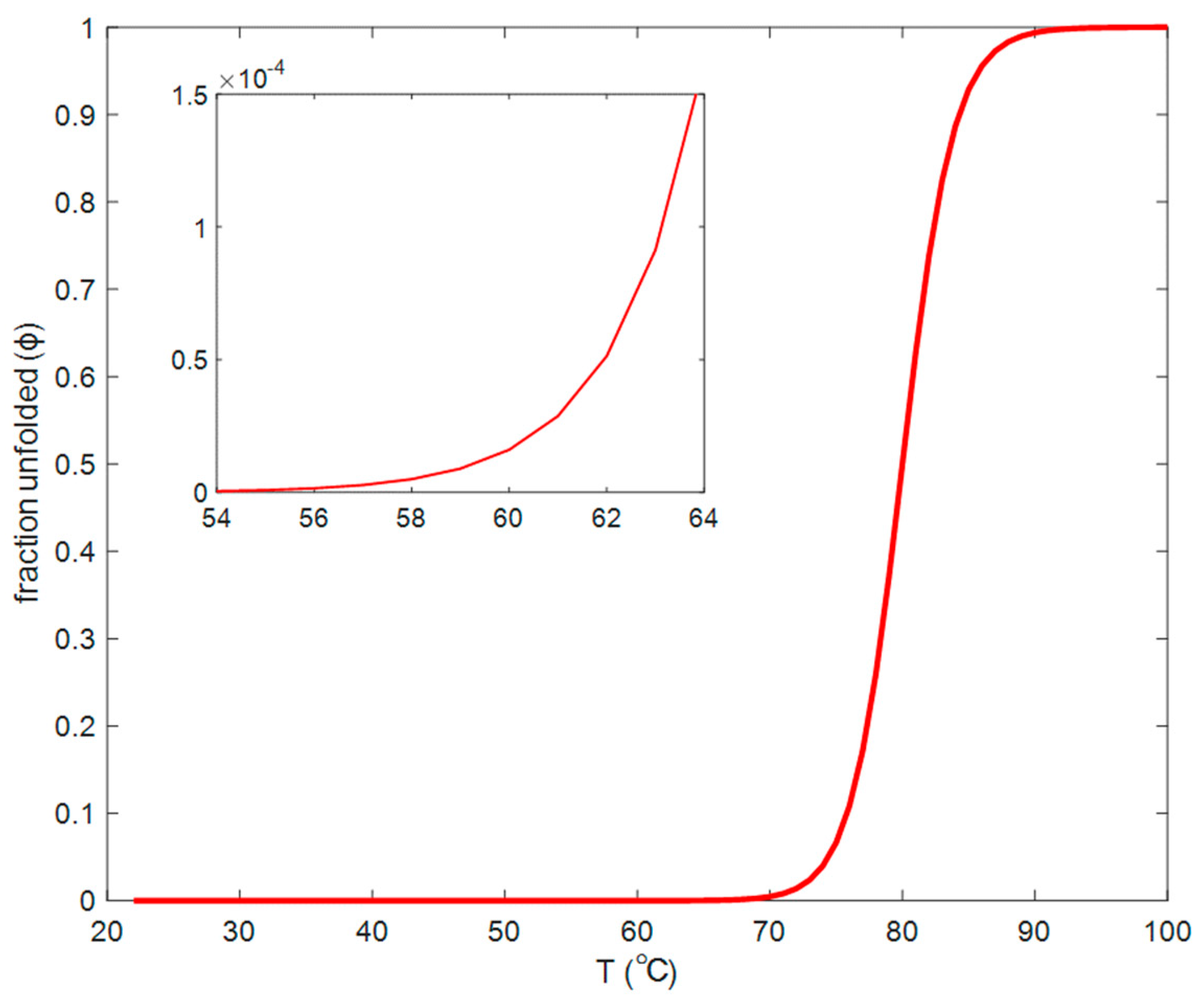
3.2. Continuous Heating
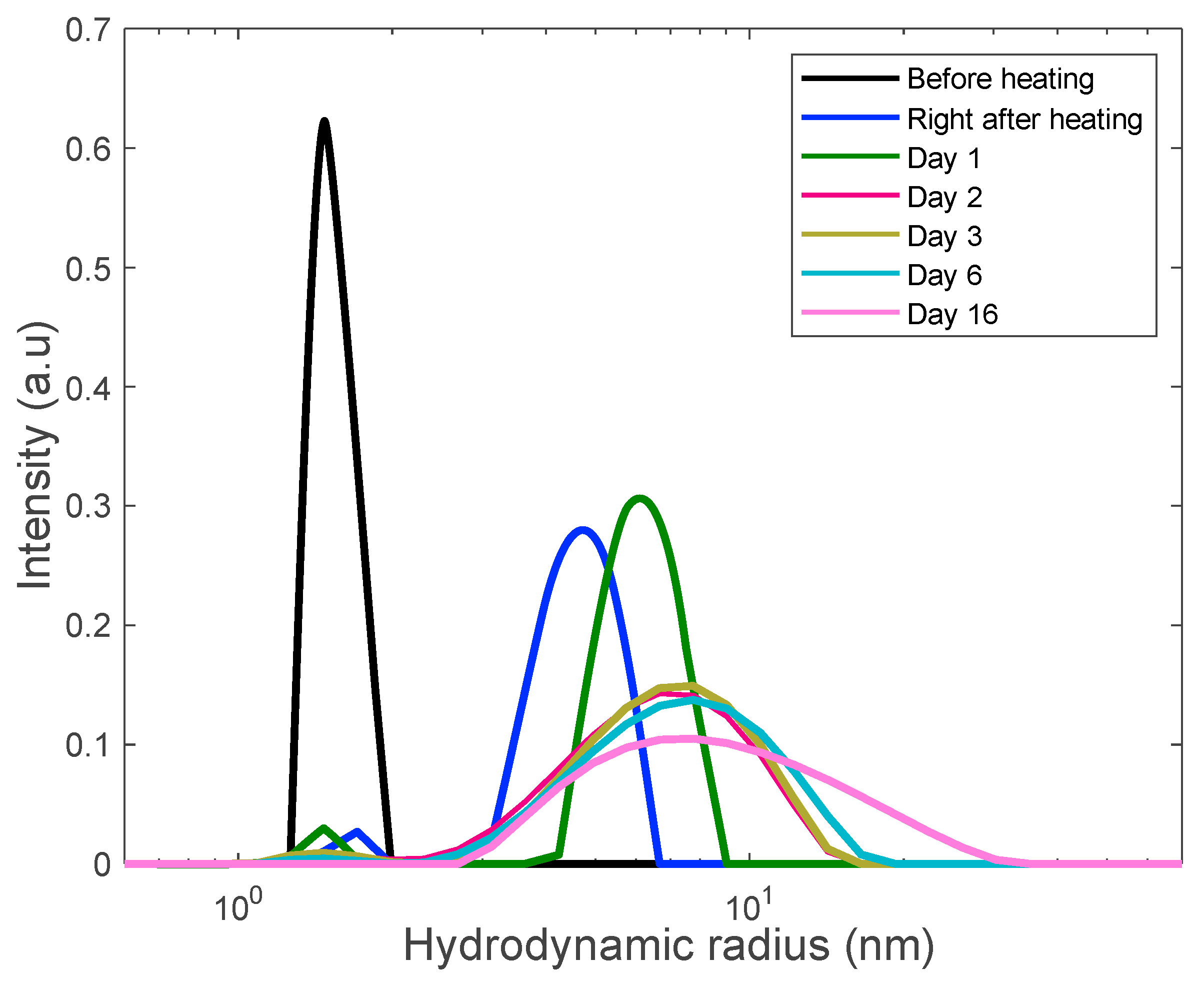
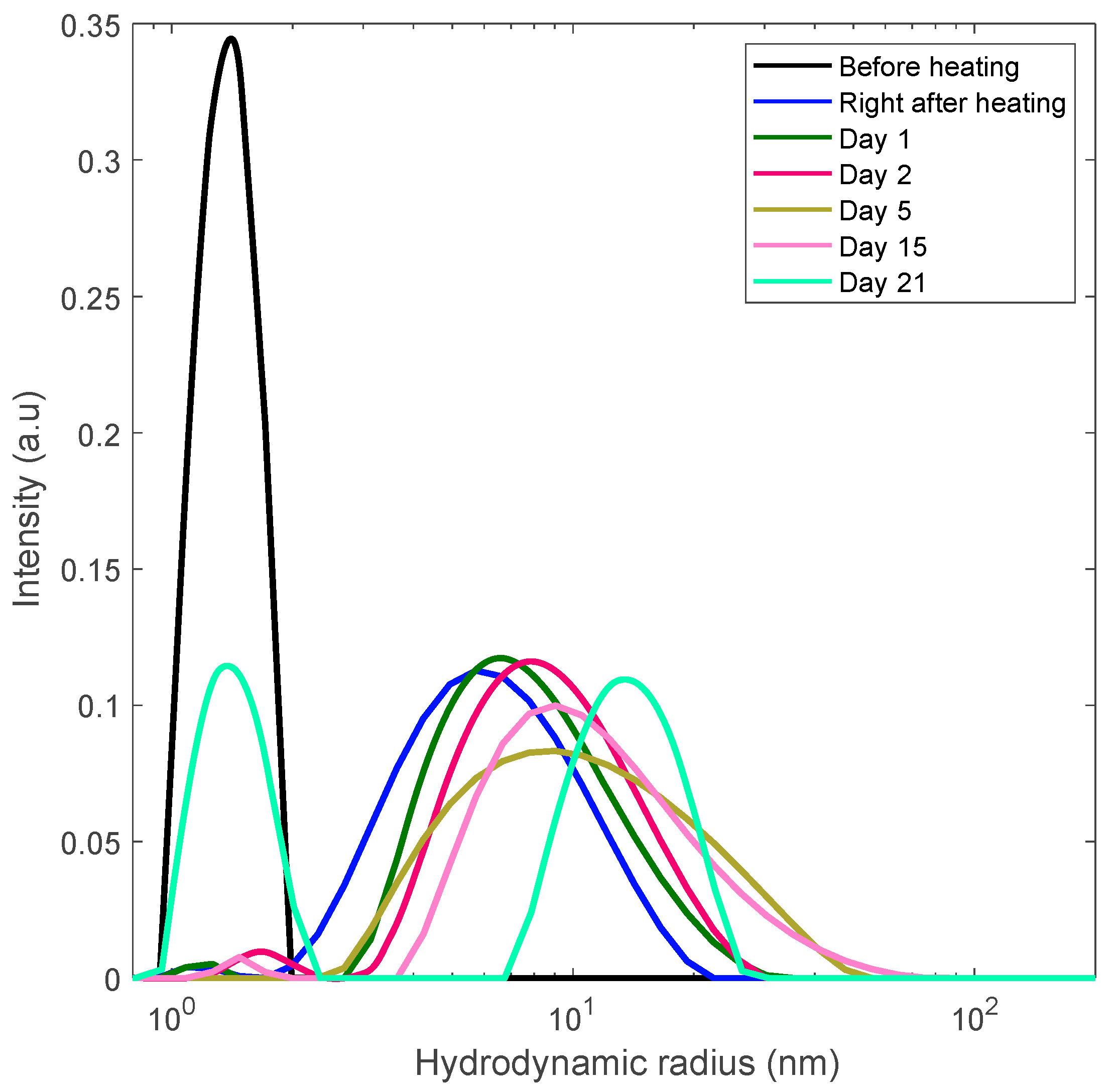
3.3. Preventing Gelation and Long-Time Monitoring
3.4. Formation of Polydisperse Aggregates
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Narhi, L.O.; Schmit, J.; Bechtold--Peters, K.; Sharma, D. Classification of protein aggregates. J. Pharm. Sci. 2012, 101, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Poirier, M.A. What is the role of protein aggregation in neurodegeneration? Nat. Rev. Mol. Cell Biol. 2005, 6, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Majhi, P.R.; Ganta, R.R.; Vanam, R.P.; Seyrek, E.; Giger, K.; Dubin, P.L. Electrostatically Driven Protein Aggregation: β-Lactoglobulin at Low Ionic Strength. Langmuir 2006, 22, 9150–9159. [Google Scholar] [CrossRef] [PubMed]
- Eisele, Y.S.; Monteiro, C.; Fearns, C.; Encalada, S.E.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 2015, 14, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Roberts, C.J. Protein aggregation—Mechanisms, detection, and control. Int. J. Pharm. 2018, 550, 251–268. [Google Scholar] [CrossRef] [PubMed]
- Philo, J.S.; Arakawa, T. Mechanisms of protein aggregation. Curr. Pharm. Biotechnol. 2009, 10, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.C.; Erdös, E.A.; Chiang, H.S.; Calderwood, T.; Tsai, K.; Visor, G.C.; Duffy, J.; Hsu, W.C.; Foster, L.C. Stability of interleukin 1 beta (IL-1 beta) in aqueous solution: Analytical methods, kinetics, products, and solution formulation implications. Pharm. Res. 1991, 8, 485–490. [Google Scholar] [CrossRef]
- Tsai, A.M.; van Zanten, J.H.; Betenbaugh, M.J., II. Electrostatic effect in the aggregation of heat-denatured RNase A and implications for protein additive design. Biotechnol. Bioeng. 1998, 59, 281–285. [Google Scholar] [CrossRef]
- Maa, Y.F.; Hsu, C.C. Protein denaturation by combined effect of shear and air-liquid interface. Biotechnol. Bioeng. 1997, 54, 503–512. [Google Scholar] [CrossRef]
- Stradner, A.; Sedgwick, H.; Cardinaux, F.; Poon, W.C.K.; Egelhaaf, S.U.; Schurtenberger, P. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 2004, 432, 492–495. [Google Scholar] [CrossRef]
- Šarić, A.; Chebaro, Y.C.; Knowles, T.P.J.; Frenkel, D. Crucial role of nonspecific interactions in amyloid nucleation. Proc. Natl. Acad. Sci. USA 2014, 111, 17869–17874. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 2005, 289, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.J. Therapeutic protein aggregation: Mechanisms, design, and control. Trends Biotechnol. 2014, 32, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Takata, S.; Norisuye, T.; Tanaka, N.; Shibayama, M. Heat-Induced Gelation of β-Lactoglobulin. 1. Time-Resolved Dynamic Light Scattering. Macromolecules 2000, 33, 5470–5475. [Google Scholar] [CrossRef]
- Mine, Y. Recent advances in the understanding of egg white protein functionality. Trends Food Sci. Technol. 1995, 6, 225–232. [Google Scholar] [CrossRef]
- Aymard, P.; Gimel, J.C.; Nicolai, T.; Durand, D. Experimental evidence for a two-step process in the aggregation of β-lactoglobulin at pH 7. J. Chim. Phys. 1996, 93, 987–997. [Google Scholar] [CrossRef]
- Nicolai, T.; Urban, C.; Schurtenberger, P. Light Scattering Study of Turbid Heat-Set Globular Protein Gels Using Cross-Correlation Dynamic Light Scattering. J. Colloid Interface Sci. 2001, 240, 419–424. [Google Scholar] [CrossRef]
- Mezzenga, R.; Fischer, P. The self-assembly, aggregation and phase transitions of food protein systems in one, two and three dimensions. Rep. Prog. Phys. 2013, 76, 046601. [Google Scholar] [CrossRef]
- Li, Y.; Lubchenko, V.; Vorontsova, M.A.; Filobelo, L.; Vekilov, P.G. Ostwald-Like Ripening of the Anomalous Mesoscopic Clusters in Protein Solutions. J. Phys. Chem. B 2012, 116, 10657–10664. [Google Scholar] [CrossRef]
- Vorontsova, M.A.; Chan, H.Y.; Lubchenko, V.; Vekilov, P.G. Lack of Dependence of the Sizes of the Mesoscopic Protein Clusters on Electrostatics. Biophys. J. 2015, 109, 1959–1968. [Google Scholar] [CrossRef]
- Pan, W.; Vekilov, P.G.; Lubchenko, V. Origin of Anomalous Mesoscopic Phases in Protein Solutions. J. Phys. Chem. B 2010, 114, 7620–7630. [Google Scholar] [CrossRef] [PubMed]
- Safari, M.S.; Byington, M.C.; Conrad, J.C.; Vekilov, P.G. Polymorphism of Lysozyme Condensates. J. Phys. Chem. B 2017, 121, 9091–9101. [Google Scholar] [CrossRef] [PubMed]
- Vekilov, P.G.; McCabe, J.W.; Angel, L.A.; Hawke, D.H.; Byington, M.C.; Safari, M.S.; Lubchenko, V.; Conrad, J.C. Weakly-bound Dimers that Underlie the Crystal Nucleation Precursors in Lysozyme Solutions. bioRxiv 2018, 275222. [Google Scholar] [CrossRef]
- Nikfarjam, S.; Ghorbani, M.; Adhikari, S.; Karlsson, A.J.; Jouravleva, E.V.; Woehl, T.J.; Anisimov, M.A. Irreversible Nature of Mesoscopic Aggregates in Lysozyme Solutions. Colloid J. 2019, 81, 546–554. [Google Scholar] [CrossRef]
- Pecora, R. Dynamic Light Scattering Measurement of Nanometer Particles in Liquids. J. Nanoparticle Res. 2000, 2, 123–131. [Google Scholar] [CrossRef]
- Zheng, X.; Anisimov, M.A.; Sengers, J.V.; He, M. Mesoscopic Diffusion of Poly(ethylene oxide) in Pure and Mixed Solvents. J. Phys. Chem. B 2018, 122, 3454–3464. [Google Scholar] [CrossRef]
- Weitz, D.A.; Huang, J.S.; Lin, M.Y.; Sung, J. Limits of the Fractal Dimension for Irreversible Kinetic Aggregation of Gold Colloids. Phys. Rev. Lett. 1985, 54, 1416–1419. [Google Scholar] [CrossRef]
- Breure, B.; Subramanian, D.; Leys, J.; Peters, C.J.; Anisimov, M.A. Modeling Asphaltene Aggregation with a Single Compound. Energy Fuels 2013, 27, 172–176. [Google Scholar] [CrossRef]
- Kryven, I.; Lazzari, S.; Storti, G. Population Balance Modeling of Aggregation and Coalescence in Colloidal Systems. Macromol. Theory Simul. 2014, 23, 170–181. [Google Scholar] [CrossRef]
- Avanti, C.; Saluja, V.; van Streun, E.L.P.; Frijlink, H.W.; Hinrichs, W.L.J. Stability of Lysozyme in Aqueous Extremolyte Solutions during Heat Shock and Accelerated Thermal Conditions. PLoS ONE 2014, 9, e86244. [Google Scholar] [CrossRef]
- Lesturgeon, V.; Nicolai, T.; Durand, D. Dynamic and static light scattering study of the formation of cross-linked PMMA gels. Eur. Phys. J. B 1999, 9, 71–82. [Google Scholar] [CrossRef]
- Martin, J.E.; Wilcoxon, J.; Odinek, J. Decay of density fluctuations in gels. Phys. Rev. A 1991, 43, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Tsurusawa, H.; Leocmach, M.; Russo, J.; Tanaka, H. Direct link between mechanical stability in gels and percolation of isostatic particles. Sci. Adv. 2019, 5, eaav6090. [Google Scholar] [CrossRef] [PubMed]
- Haynie, D.T. Biological Thermodynamics; Cambridge University Press: Cambridge, UK, 2001; ISBN 978-0-521-79549-4. [Google Scholar]
- Li-Blatter, X.; Seelig, J. Thermal and Chemical Unfolding of Lysozyme. Multistate Zimm–Bragg Theory Versus Two-State Model. J. Phys. Chem. B 2019, 123, 10181–10191. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Shaw, D.E. How Fast-Folding Proteins Fold. Science 2011, 334, 517–520. [Google Scholar] [CrossRef]
- Matagne, A.; Radford, S.E.; Dobson, C.M. Fast and slow tracks in lysozyme folding: Insight into the role of domains in the folding process. J. Mol. Biol. 1997, 267, 1068–1074. [Google Scholar] [CrossRef]
- Chung, H.S.; Eaton, W.A. Protein folding transition path times from single molecule FRET. Curr. Opin. Struct. Biol. 2018, 48, 30–39. [Google Scholar] [CrossRef]
- Weijers, M.; Barneveld, P.A.; Cohen Stuart, M.A.; Visschers, R.W. Heat-induced denaturation and aggregation of ovalbumin at neutral pH described by irreversible first-order kinetics. Protein Sci. 2003, 12, 2693–2703. [Google Scholar] [CrossRef]
- Woodard, D.; Bell, D.; Tipton, D.; Durrance, S.; Cole, L.; Li, B.; Xu, S. Gel Formation in Protein Amyloid Aggregation: A Physical Mechanism for Cytotoxicity. PLoS ONE 2014, 9, e94789. [Google Scholar] [CrossRef]
- Renkema, J.M.S.; van Vliet, T. Heat-Induced Gel Formation by Soy Proteins at Neutral pH. J. Agric. Food Chem. 2002, 50, 1569–1573. [Google Scholar] [CrossRef]
- Ako, K.; Nicolai, T.; Durand, D. Salt-induced gelation of globular protein aggregates: Structure and kinetics. Biomacromolecules 2010, 11, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Šarić, A.; Buell, A.K.; Meisl, G.; Michaels, T.C.T.; Dobson, C.M.; Linse, S.; Knowles, T.P.J.; Frenkel, D. Physical determinants of the self-replication of protein fibrils. Nat. Phys. 2016, 12, 874–880. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Beairsto, S.; Hartt, C.; Zavalov, O.; Saika-Voivod, I.; Bowles, R.K.; Poole, P.H. Phase transitions in fluctuations and their role in two-step nucleation. J. Chem. Phys. 2019, EDCH2019, 074501. [Google Scholar] [CrossRef] [PubMed]


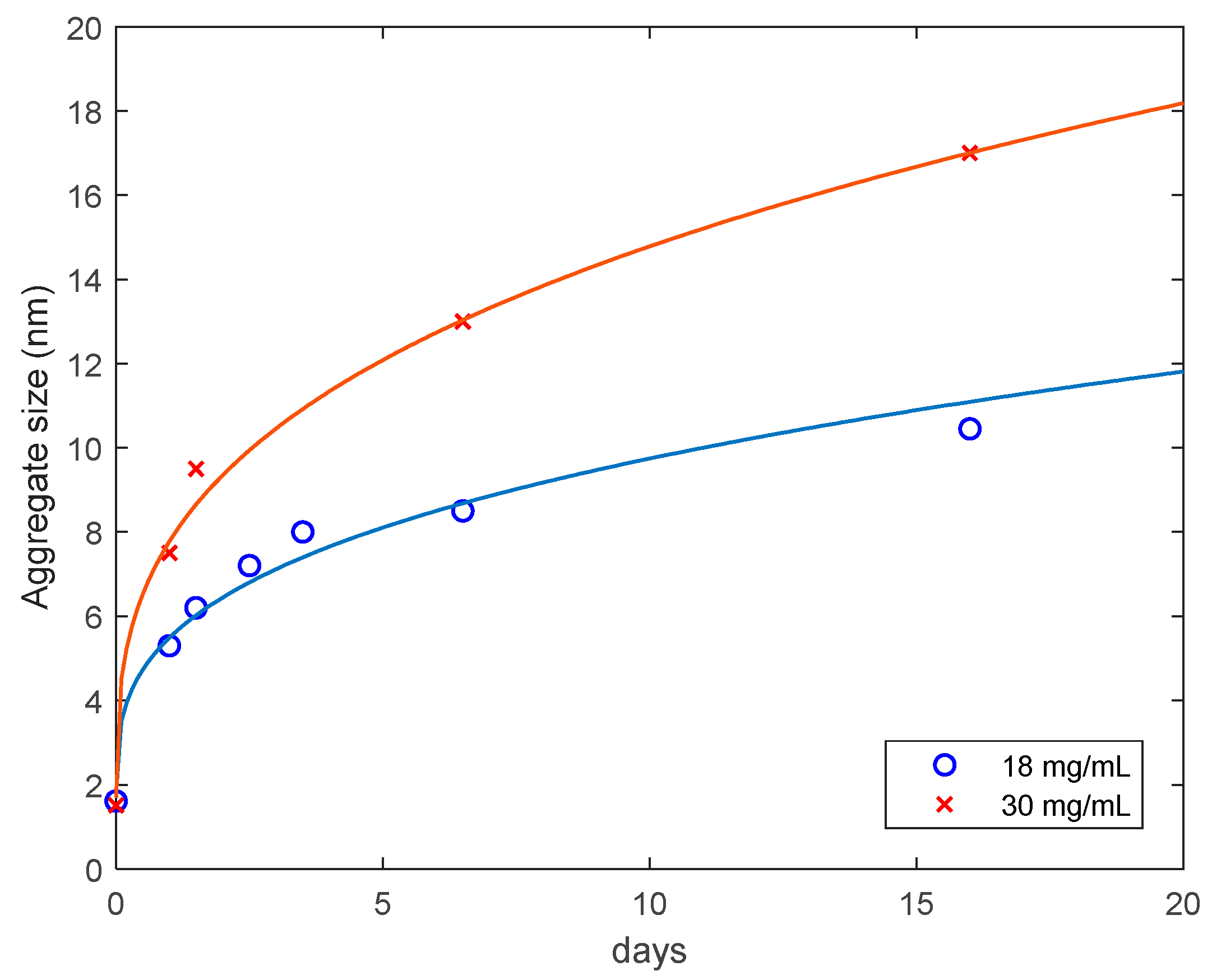
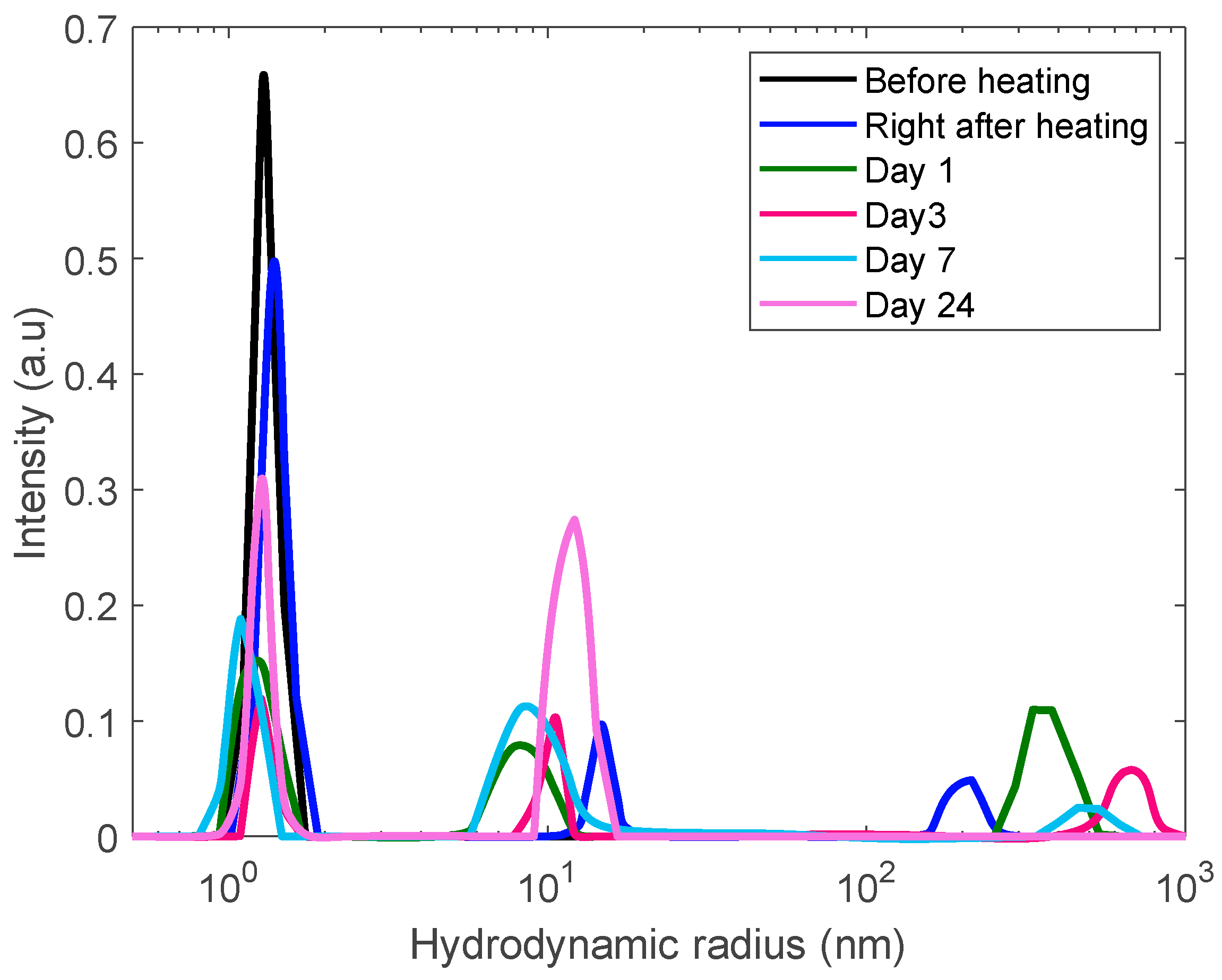
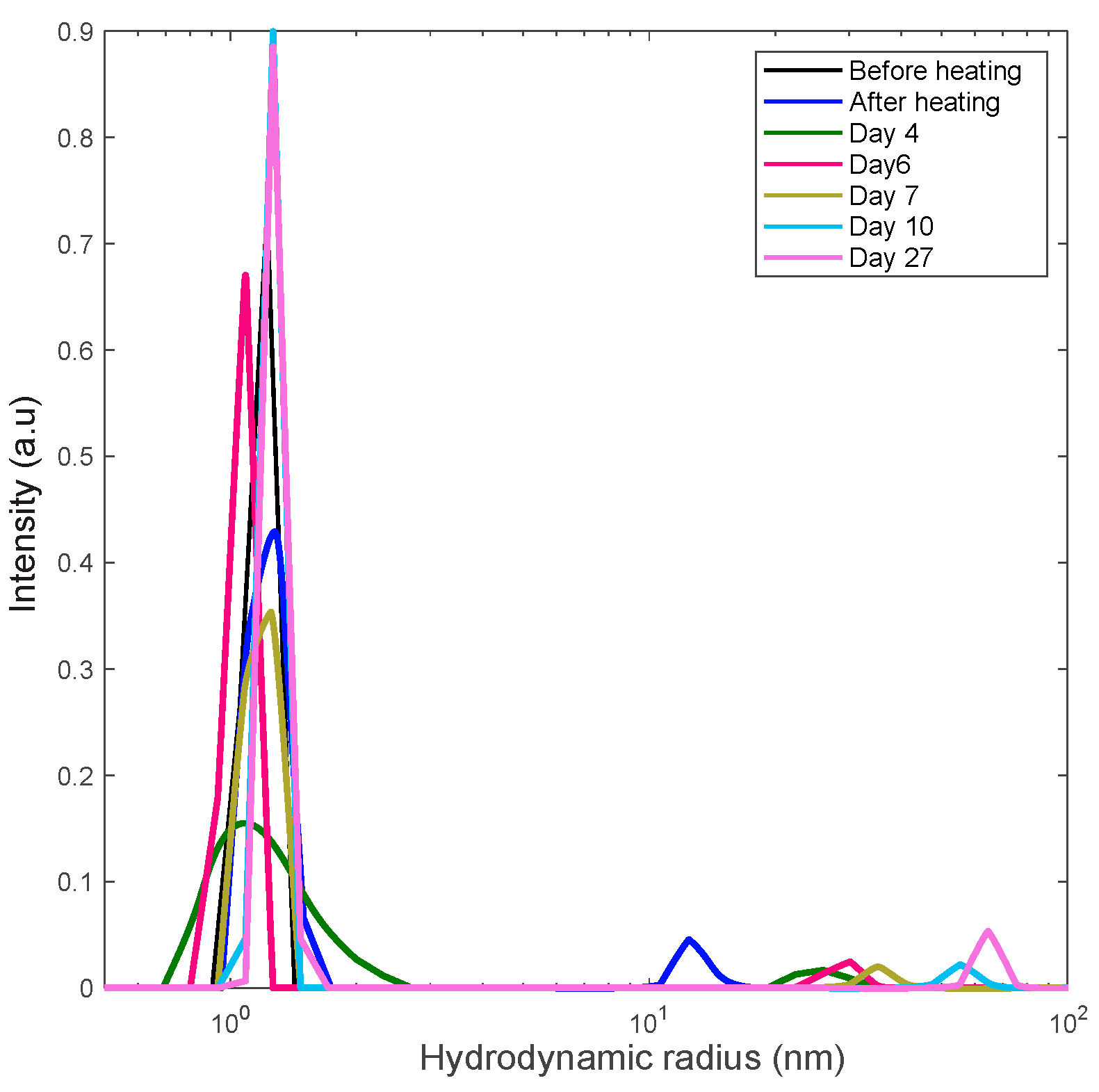
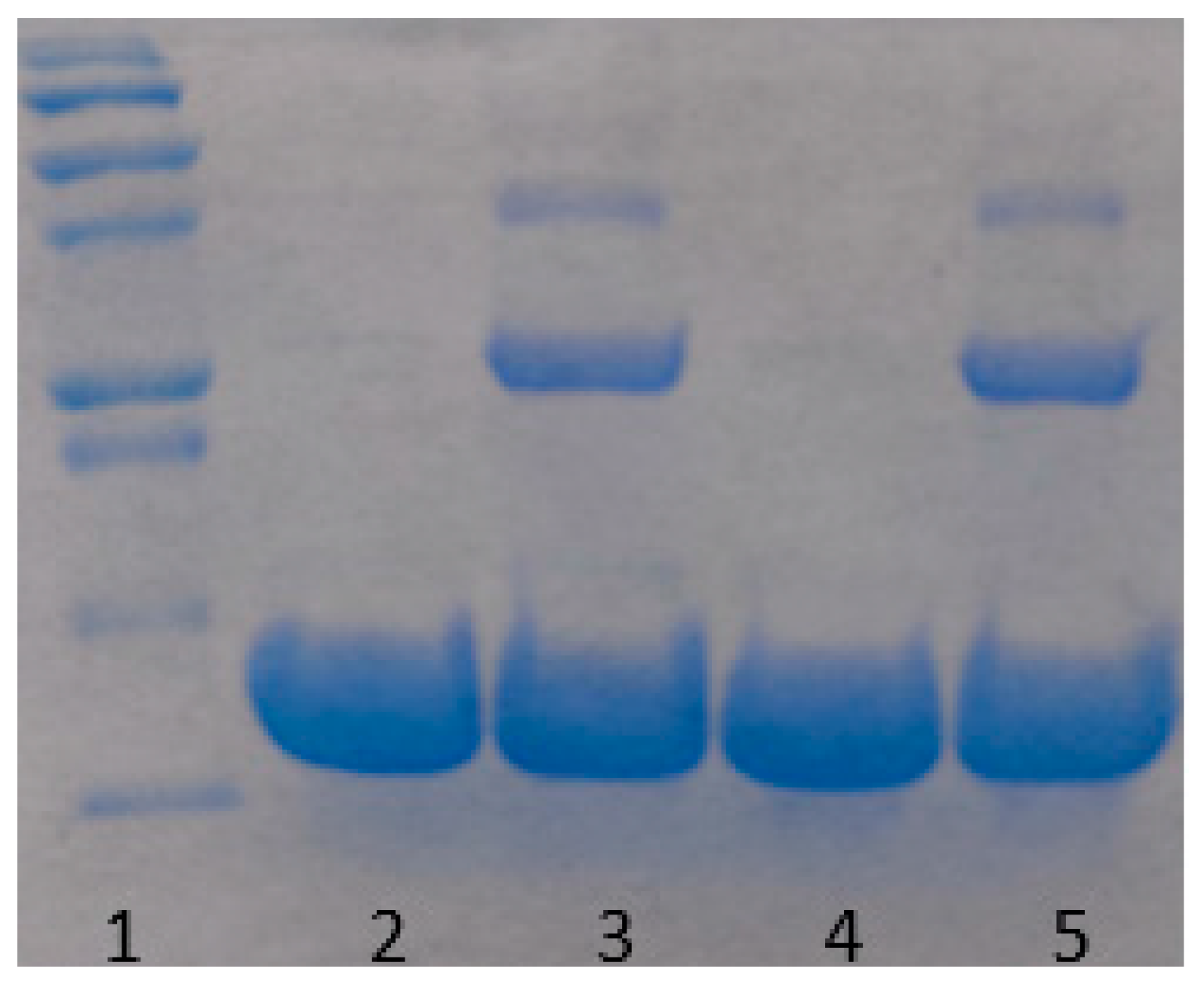
| Concentration (mg/mL) | Amplitude (A) | Standard Deviation |
|---|---|---|
| 18 | 3.68 | 0.37 |
| 30 | 6.07 | 0.36 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikfarjam, S.; Jouravleva, E.V.; Anisimov, M.A.; Woehl, T.J. Effects of Protein Unfolding on Aggregation and Gelation in Lysozyme Solutions. Biomolecules 2020, 10, 1262. https://doi.org/10.3390/biom10091262
Nikfarjam S, Jouravleva EV, Anisimov MA, Woehl TJ. Effects of Protein Unfolding on Aggregation and Gelation in Lysozyme Solutions. Biomolecules. 2020; 10(9):1262. https://doi.org/10.3390/biom10091262
Chicago/Turabian StyleNikfarjam, Shakiba, Elena V. Jouravleva, Mikhail A. Anisimov, and Taylor J. Woehl. 2020. "Effects of Protein Unfolding on Aggregation and Gelation in Lysozyme Solutions" Biomolecules 10, no. 9: 1262. https://doi.org/10.3390/biom10091262
APA StyleNikfarjam, S., Jouravleva, E. V., Anisimov, M. A., & Woehl, T. J. (2020). Effects of Protein Unfolding on Aggregation and Gelation in Lysozyme Solutions. Biomolecules, 10(9), 1262. https://doi.org/10.3390/biom10091262





