Structural Features Mediating Zinc Binding and Transfer in the AztABCD Zinc Transporter System
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of Proteins
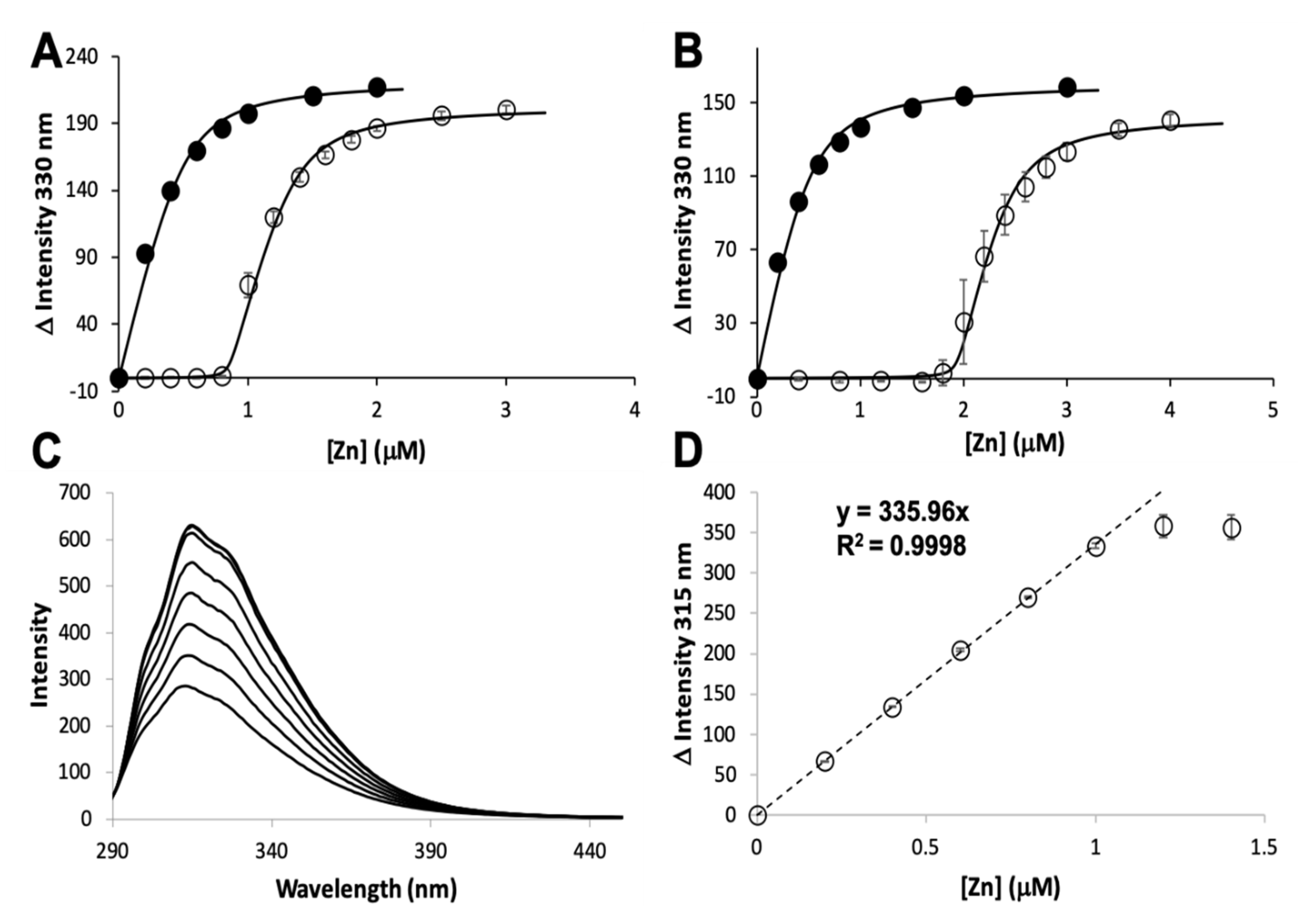
2.2. Mag-Fura 2 Competition Assay
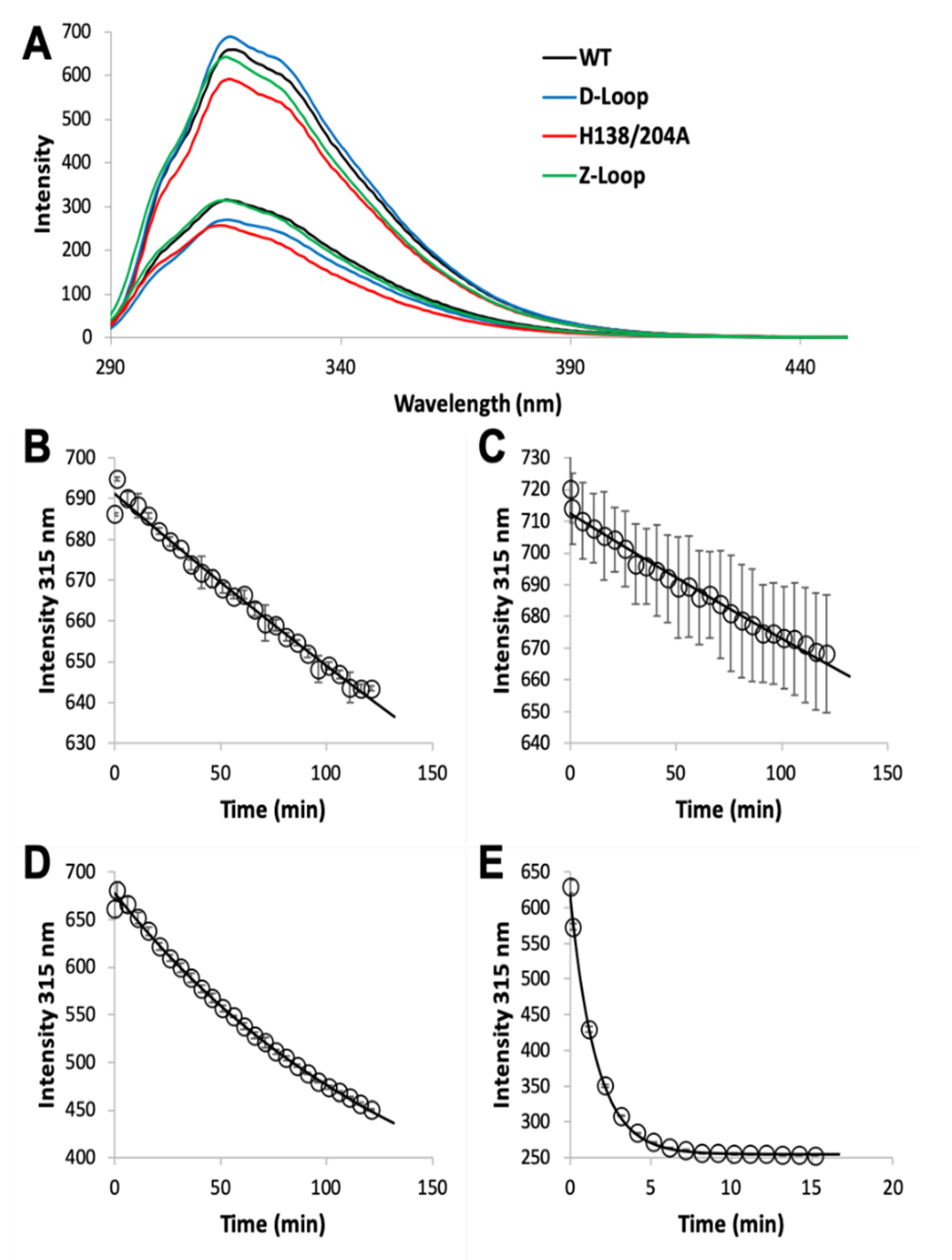
2.3. Zinc Dissociation
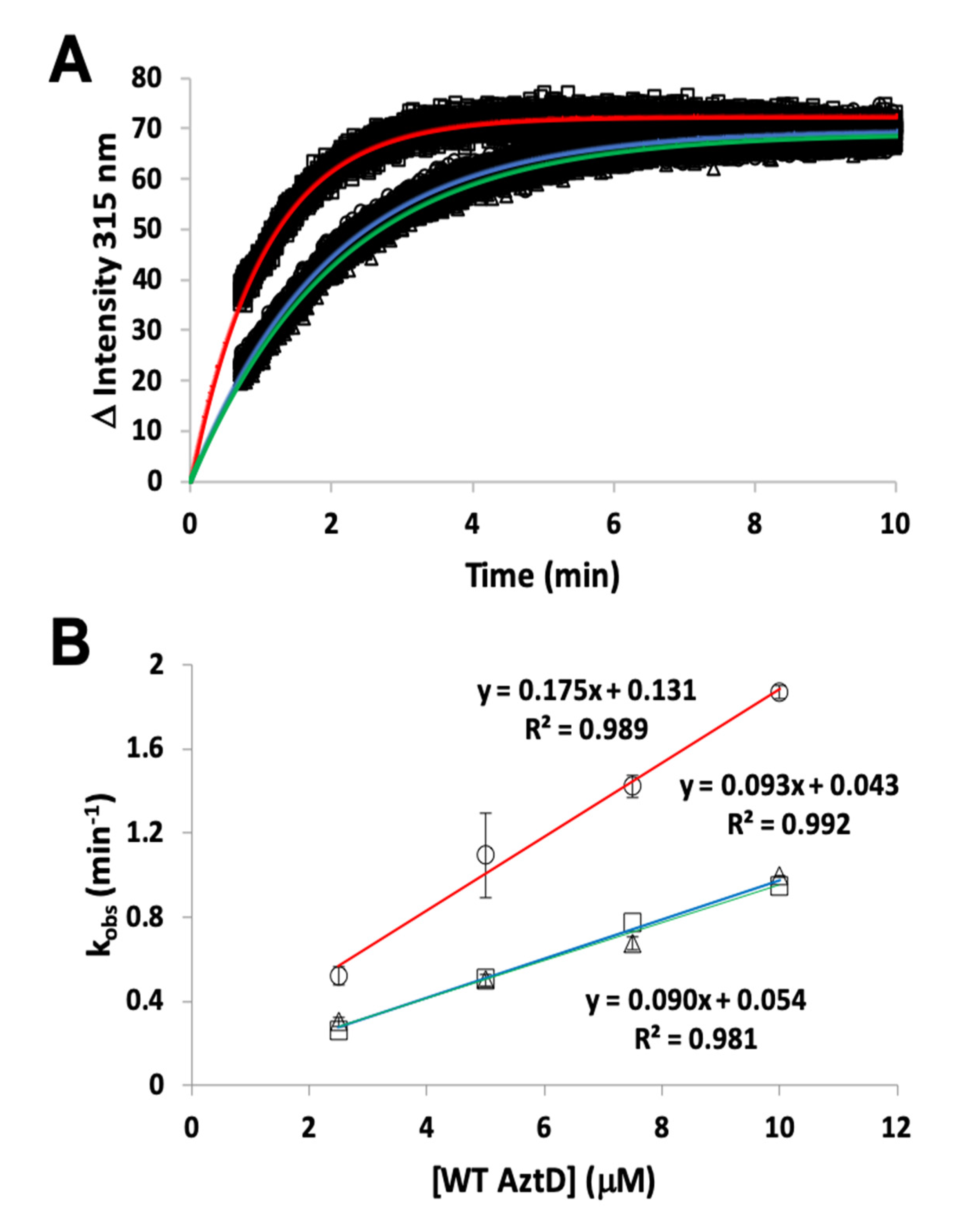
2.4. Zinc Binding and Transfer by Intrinsic Fluorescence
2.5. Crystallization and Structure Determination
3. Results
3.1. Zinc Binding and Dissociation
3.2. Zinc Transfer from AztD
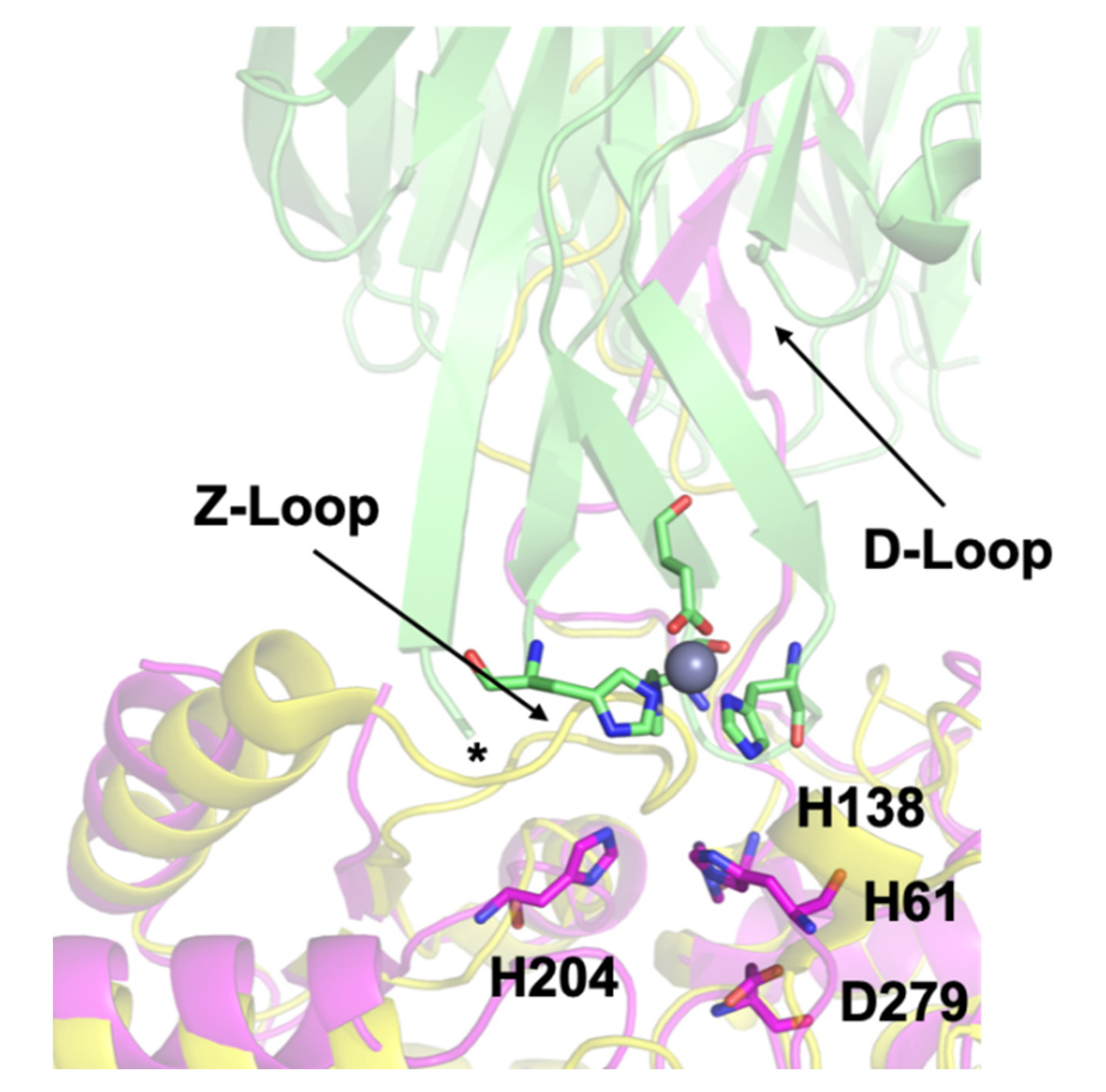
3.3. Crystal Structure of ΔZ-Loop
3.4. Competition Between AztC Mutants
4. Discussion
4.1. The AztD N-terminal Motif (NTM)
4.2. The AztC Z-Loop
4.3. H138/204A AztC
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Magistrato, A.; Pavlin, M.; Qasem, Z.; Ruthstein, S. Copper trafficking in eukaryotic systems: Current knowledge from experimental and computational efforts. Curr. Opin. Struct. Biol. 2019, 58, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Kambe, T.; Tsuji, T.; Hashimoto, A.; Itsumura, N. The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism. Physiol. Rev. 2015, 95, 749–784. [Google Scholar] [CrossRef] [PubMed]
- Blindauer, C.A. Bacterial metallothioneins: Past, present, and questions for the future. JBIC J. Biol. Inorg. Chem. 2011, 16, 1011–1024. [Google Scholar] [CrossRef]
- Nairn, B.L.; Lonergan, Z.; Wang, J.; Braymer, J.; Zhang, Y.; Calcutt, M.W.; Lisher, J.P.; Gilston, B.A.; Chazin, W.J.; De Crécy-Lagard, V.; et al. The Response of Acinetobacter baumannii to Zinc Starvation. Cell Host Microbe 2016, 19, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Chandrangsu, P.; Huang, X.; Gaballa, A.; Helmann, J.D. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 2019, 112, 751–765. [Google Scholar] [CrossRef]
- Neupane, D.P.; Jacquez, B.; Sundararajan, A.; Ramaraj, T.; Schilkey, F.D.; Yukl, E.T. Zinc-Dependent Transcriptional Regulation in Paracoccus denitrificans. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Biemans-Oldehinkel, E.; Doeven, M.K.; Poolman, B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2005, 580, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F. ABC Transporters: From Microorganisms to Man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, R.P.-A.; Smits, S.H.J.; Schmitt, L.; Slotboom, D.-J.; Poolman, B. A structural classification of substrate-binding proteins. FEBS Lett. 2010, 584, 2606–2617. [Google Scholar] [CrossRef]
- Loisel, E.; Jacquamet, L.; Serre, L.; Bauvois, C.; Ferrer, J.L.; Vernet, T.; Di Guilmi, A.M.; Durmort, C. AdcAII, A New Pneumococcal Zn-Binding Protein Homologous with ABC Transporters: Biochemical and Structural Analysis. J. Mol. Biol. 2008, 381, 594–606. [Google Scholar] [CrossRef]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Genet. 2012, 10, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Kehl-Fie, T.E.; Skaar, E.P. Nutritional immunity beyond iron: A role for manganese and zinc. Curr. Opin. Chem. Biol. 2010, 14, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Panina, E.M.; Mironov, A.A.; Gelfand, M.S. Comparative genomics of bacterial zinc regulons: Enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 9912–9917. [Google Scholar] [CrossRef] [PubMed]
- Plumptre, C.D.; Eijkelkamp, B.A.; Morey, J.R.; Behr, F.; Couñago, R.M.; Ogunniyi, A.D.; Kobe, B.; O’Mara, M.L.; Paton, J.C.; McDevitt, C.A. AdcA and AdcAII employ distinct zinc acquisition mechanisms and contribute additively to zinc homeostasis in Streptococcus pneumoniae. Mol. Microbiol. 2014, 91, 834–851. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Li, N.; Wang, H.; Cao, X.; He, J.; Zhang, B.; He, Q.-Y.; Zhang, G.; Sun, X. Two zinc-binding domains in the transporter AdcA from Streptococcus pyogenes facilitate high-affinity binding and fast transport of zinc. J. Biol. Chem. 2018, 293, 6075–6089. [Google Scholar] [CrossRef] [PubMed]
- Ilari, A.; Alaleona, F.; Tria, G.; Petrarca, P.; Battistoni, A.; Zamparelli, C.; Verzili, D.; Falconi, M.; Chiancone, E. The Salmonella enterica ZinT structure, zinc affinity and interaction with the high-affinity uptake protein ZnuA provide insight into the management of periplasmic zinc. Biochim. Biophys. Acta 2014, 1840, 535–544. [Google Scholar] [CrossRef][Green Version]
- Petrarca, P.; Ammendola, S.; Pasquali, P.; Battistoni, A. The Zur-Regulated ZinT Protein Is an Auxiliary Component of the High-Affinity ZnuABC Zinc Transporter That Facilitates Metal Recruitment during Severe Zinc Shortage. J. Bacteriol. 2010, 192, 1553–1564. [Google Scholar] [CrossRef]
- Gabbianelli, R.; Scotti, R.; Ammendola, S.; Petrarca, P.; Nicolini, L.; Battistoni, A. Role of ZnuABC and ZinT in Escherichia coli O157:H7 zinc acquisition and interaction with epithelial cells. BMC Microbiol. 2011, 11, 36. [Google Scholar] [CrossRef]
- Graham, A.I.; Hunt, S.; Stokes, S.L.; Bramall, N.; Bunch, J.; Cox, A.G.; McLeod, C.W.; Poole, R.K.; Oka, M.; Sumita, N.; et al. Severe Zinc Depletion of Escherichia coli. J. Biol. Chem. 2009, 284, 18377–18389. [Google Scholar] [CrossRef]
- Eijkelkamp, B.A.; Pederick, V.G.; Plumptre, C.D.; Harvey, R.M.; Hughes, C.E.; Paton, J.C.; McDevitt, C.A. The First Histidine Triad Motif of PhtD Is Critical for Zinc Homeostasis in Streptococcus pneumoniae. Infect. Immun. 2015, 84, 407–415. [Google Scholar] [CrossRef]
- Ogunniyi, A.D.; Grabowicz, M.; Mahdi, L.K.; Cook, J.; Gordon, D.L.; Sadlon, T.A.; Paton, J.C. Pneumococcal histidine triad proteins are regulated by the Zn 2+ -dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2008, 23, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Bersch, B.; Bougault, C.; Roux, L.; Favier, A.; Vernet, T.; Durmort, C. New Insights into Histidine Triad Proteins: Solution Structure of a Streptococcus pneumoniae PhtD Domain and Zinc Transfer to AdcAII. PLOS ONE 2013, 8, e81168. [Google Scholar] [CrossRef] [PubMed]
- Handali, M.; Roychowdhury, H.; Neupane, D.P.; Yukl, E.T. AztD, a Periplasmic Zinc Metallochaperone to an ATP-binding Cassette (ABC) Transporter System in Paracoccus denitrificans. J. Biol. Chem. 2015, 290, 29984–29992. [Google Scholar] [CrossRef] [PubMed]
- Handali, M.; Neupane, D.P.; Roychowdhury, H.; Yukl, E.T. Transcriptional Regulation, Metal Binding Properties and Structure of Pden1597, an Unusual Zinc Transport Protein from Paracoccus denitrificans. J. Biol. Chem. 2015, 290, 11878–11889. [Google Scholar] [CrossRef]
- Neupane, D.P.; Avalos, D.; Fullam, S.; Roychowdhury, H.; Yukl, E.T. Mechanisms of zinc binding to the solute-binding protein AztC and transfer from the metallochaperone AztD. J. Biol. Chem. 2017, 292, 17496–17505. [Google Scholar] [CrossRef]
- Neupane, D.P.; Fullam, S.H.; Chacón, K.N.; Yukl, E.T. Crystal structures of AztD provide mechanistic insights into direct zinc transfer between proteins. Commun. Biol. 2019, 2, 308–312. [Google Scholar] [CrossRef]
- Edelhoch, H. Spectroscopic Determination of Tryptophan and Tyrosine in Proteins*. Biochemistry 1967, 6, 1948–1954. [Google Scholar] [CrossRef]
- Golynskiy, M.V.; Gunderson, W.A.; Hendrich, M.P.; Cohen, S.M. Metal Binding Studies and EPR Spectroscopy of the Manganese Transport Regulator MntR. Biochemistry 2006, 45, 15359–15372. [Google Scholar] [CrossRef]
- Kuzmic, P. Program DYNAFIT for the Analysis of Enzyme Kinetic Data: Application to HIV Proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef]
- Kuzmic, P. DynaFit—A Software Package for Enzymology. Methods Enzymol. 2009, 467, 247–280. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Cryst. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Cryst. 2010, 66, 133–144. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Cryst. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Zheng, B.; Zhang, Q.; Gao, J.; Han, H.; Li, M.; Zhang, J.; Qi, J.; Yan, J.; Gao, G.F. Insight into the Interaction of Metal Ions with TroA from Streptococcus suis. PLOS ONE 2011, 6, e19510. [Google Scholar] [CrossRef]
- Lee, Y.H.; Deka, R.K.; Norgard, M.V.; Radolf, J.D.; A Hasemann, C. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Genet. 1999, 6, 628–633. [Google Scholar] [CrossRef]
- Ragunathan, P.; Spellerberg, B.; Ponnuraj, K. Structure of laminin-binding adhesin (Lmb) from Streptococcus agalactiae. Acta Cryst. 2009, 65, 1262–1269. [Google Scholar] [CrossRef]
- Linke, C.; Caradoc-Davies, T.T.; Young, P.G.; Proft, T.; Baker, E.N. The Laminin-Binding Protein Lbp from Streptococcus pyogenes Is a Zinc Receptor. J. Bacteriol. 2009, 191, 5814–5823. [Google Scholar] [CrossRef][Green Version]
- Lee, Y.-H.; Dorwart, M.R.; Hazlett, K.R.O.; Deka, R.K.; Norgard, M.V.; Radolf, J.D.; Hasemann, C.A. The Crystal Structure of Zn(II)-Free Treponema pallidum TroA, a Periplasmic Metal-Binding Protein, Reveals a Closed Conformation. J. Bacteriol. 2002, 184, 2300–2304. [Google Scholar] [CrossRef]
- Sridharan, U.; Ragunathan, P.; Spellerberg, B.; Ponnuraj, K. Molecular dynamics simulation of metal free structure of Lmb, a laminin-binding adhesin of Streptococcus agalactiae: Metal removal and its structural implications. J. Biomol. Struct. Dyn. 2018, 37, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Yatsunyk, L.A.; Easton, J.A.; Kim, L.R.; Sugarbaker, S.A.; Bennett, B.; Breece, R.M.; Vorontsov, I.I.; Tierney, D.L.; Crowder, M.W.; Rosenzweig, A.C. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 2007, 13, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Randich, A.M.; Bhattacharyya-Pakrasi, M.; Pakrasi, H.B.; Smith, T.J. Possible Regulatory Role for the Histidine-Rich Loop in the Zinc Transport Protein, ZnuA. Biochemistry 2007, 46, 8734–8743. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, D.C.; Sun, Y.C.; Zaidi, A.A.; Eggers, C.H.; Cox, D.L.; Radolf, J.D. The general transition metal (Tro) and Zn2+(Znu) transporters in Treponema pallidum: Analysis of metal specificities and expression profiles. Mol. Microbiol. 2007, 65, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Abate, F.; Malito, E.; Cozzi, R.; Surdo, P.L.; Maione, M.; Bottomley, M.J. Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius. Biosci. Rep. 2014, 34, 743–758. [Google Scholar] [CrossRef]
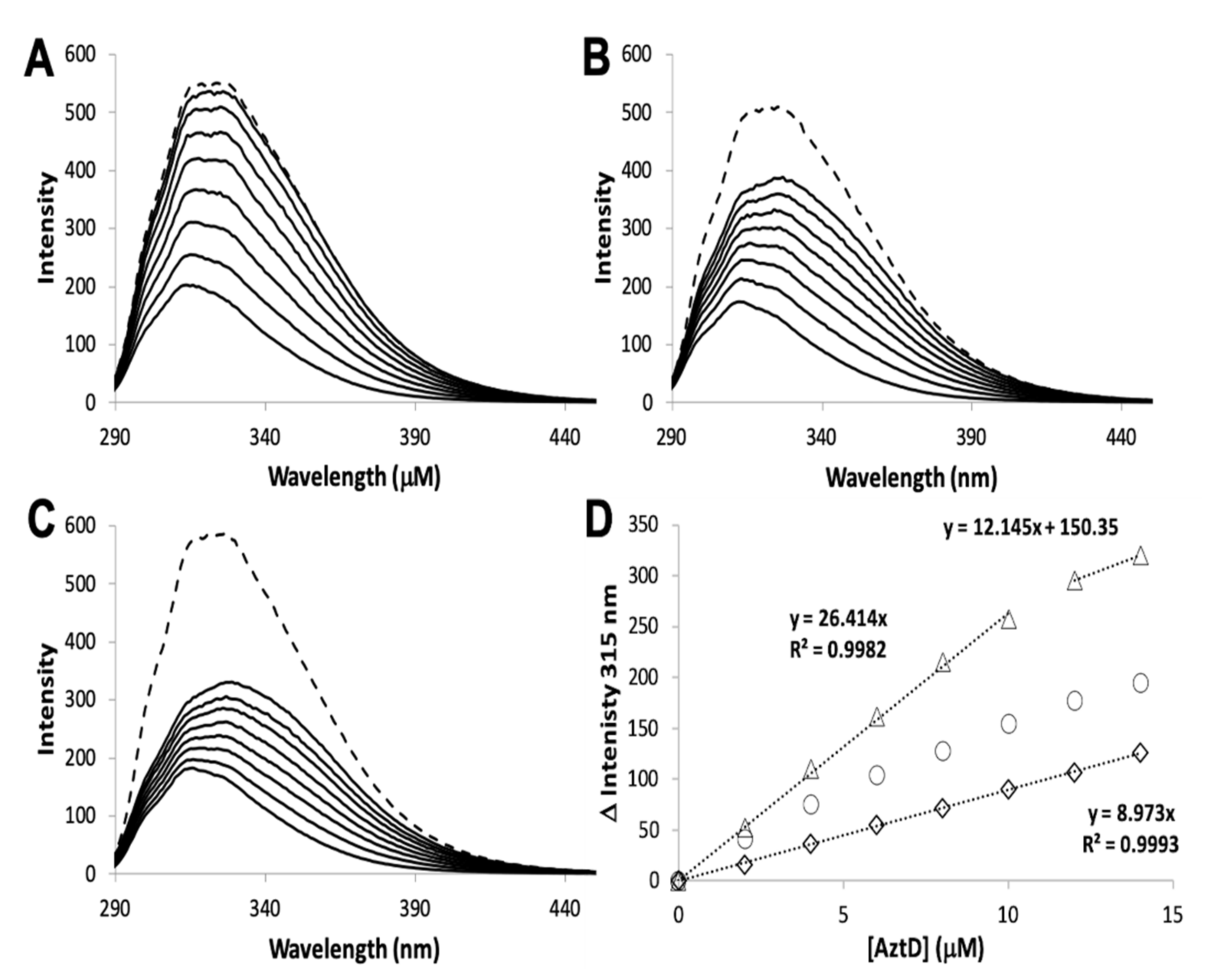
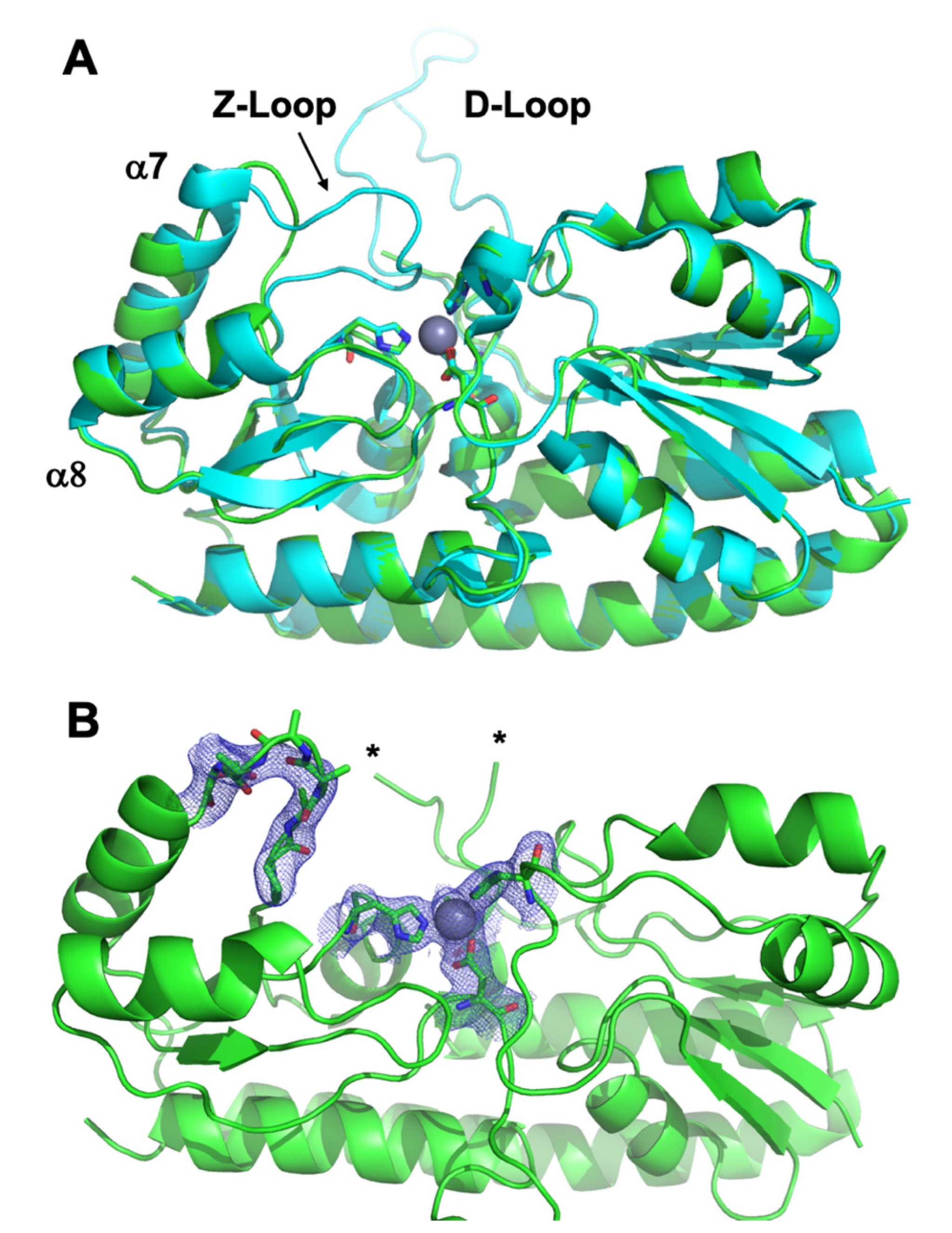
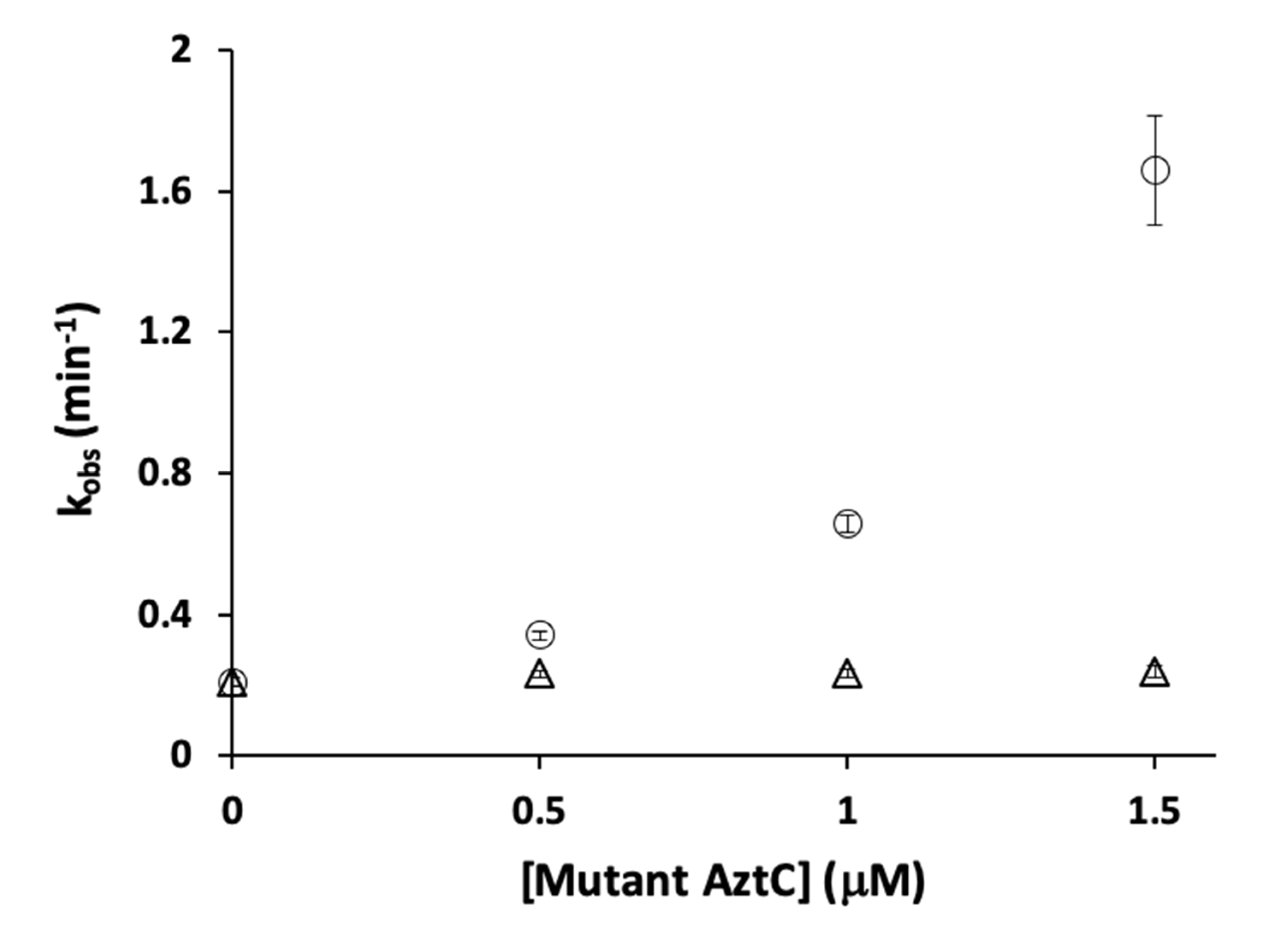
| Protein | Site | Kd ± S.D. (nM) | koff ± S.D. (min−1) | kT (M−1 × s−1) |
|---|---|---|---|---|
| WT AztD [23] | 1 | 0.7 ± 0.3 | ||
| 2 | 54 ± 8 | |||
| 3 | 340 ± 110 | |||
| ΔS1 AztD [26] | 2 | 1.3 ± 0.7 | ||
| 3 | 248 ± 81 | |||
| ΔS2 AztD [26] | 1 | 0.1 * | ||
| 3 | 158 ± 35 | |||
| ΔNTM AztD | 1 | 0.1 * | 1.50 × 10−3 | |
| 2 | 0.2 ± 0.1 | |||
| WT AztC [24] | 1 | 0.3 ± 0.1 | 1.2 ± 0.1 × 10−3 | 1.33 × 10−3 [26]; 1.55 × 10−3 (this work) |
| ΔD-Loop AztC [25] | 1 | 0.2 ± 0.1 | 0.9 ± 0.1 × 10−3 | nd |
| ΔZ-Loop AztC [25] | 1 | 0.2 * | 7.0 ± 0.1 × 10−3 | 2.92 × 10−3 |
| H138/204A AztC [24] | 1 | 0.1 * | 623.4 ± 0.5 × 10−3 |
| Holo ΔZ-Loop AztC | |
|---|---|
| Data collection | |
| Space group | P21 |
| Unit cell parameters | |
| a, b, c (Å) α, β, γ (°) | 62.3, 105.0, 64.3 90.0, 110.8, 90.0 |
| Resolution range (Å) | 46.6–2.26 |
| Number of reflections (measured/unique) | 133,590/36,075 |
| Rmerge | 0.04 (0.59) |
| I/σI | 18.5 (2.3) |
| Completeness (%) | 99.5 (99.9) |
| Redundancy | 3.7 (3.7) |
| Refinement Statistics | |
| Resolution (Å) | 39.5–2.26 |
| Rwork/Rfree | 17.4/20.8 |
| Number of atoms | |
| Protein | 3,840 |
| Zinc | 2 |
| Water | 57 |
| Other | 0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.20 |
| Ramachandran Statistics | |
| Allowed | 99.5% |
| Outliers | 0.5% |
| Average B-factor (Å2) | 58.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meni, A.; Yukl, E.T. Structural Features Mediating Zinc Binding and Transfer in the AztABCD Zinc Transporter System. Biomolecules 2020, 10, 1156. https://doi.org/10.3390/biom10081156
Meni A, Yukl ET. Structural Features Mediating Zinc Binding and Transfer in the AztABCD Zinc Transporter System. Biomolecules. 2020; 10(8):1156. https://doi.org/10.3390/biom10081156
Chicago/Turabian StyleMeni, Anusha, and Erik T. Yukl. 2020. "Structural Features Mediating Zinc Binding and Transfer in the AztABCD Zinc Transporter System" Biomolecules 10, no. 8: 1156. https://doi.org/10.3390/biom10081156
APA StyleMeni, A., & Yukl, E. T. (2020). Structural Features Mediating Zinc Binding and Transfer in the AztABCD Zinc Transporter System. Biomolecules, 10(8), 1156. https://doi.org/10.3390/biom10081156





