Redox-Dependent Copper Ion Modulation of Amyloid-β (1-42) Aggregation In Vitro
Abstract
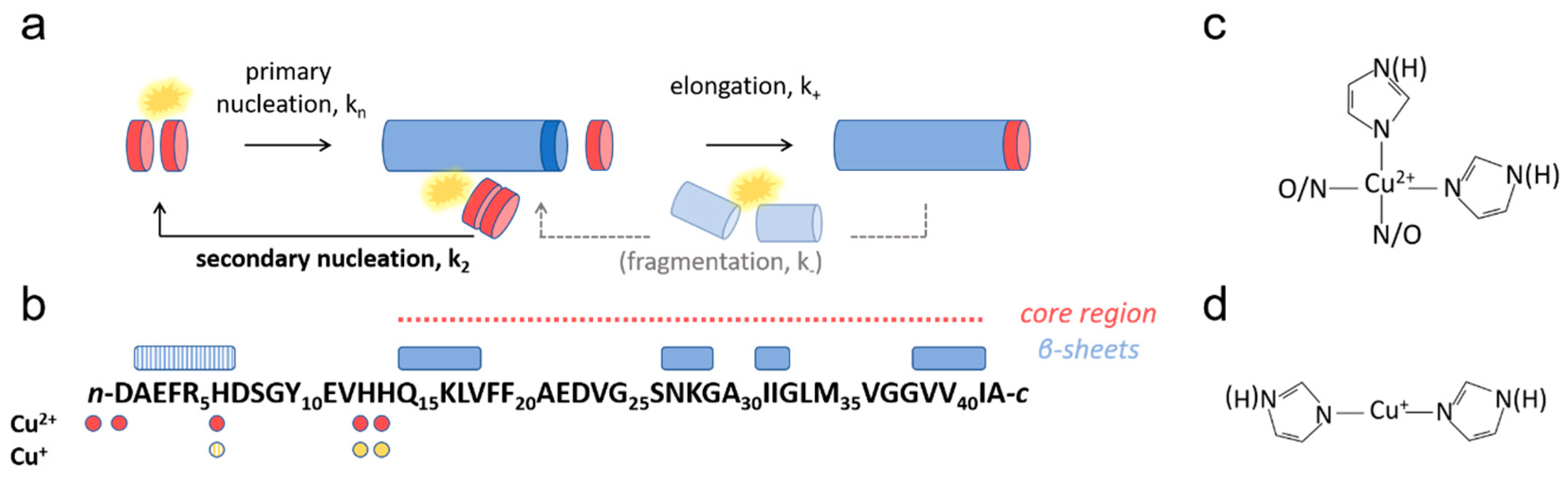
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Aβ(1-42) Aggregation Assays
2.3. Seeds Preparation
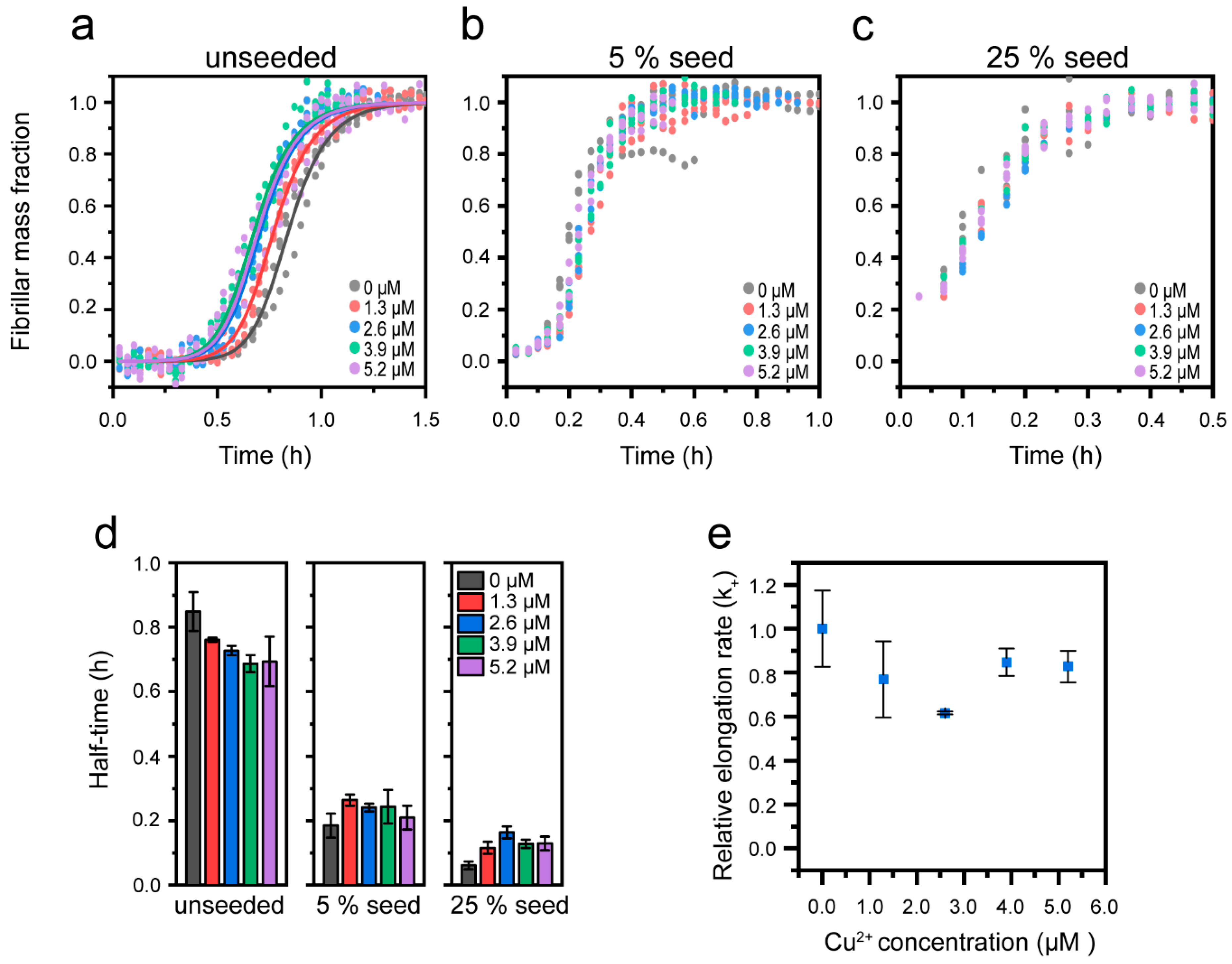
2.4. Analysis and Fitting of ThT Kinetic Curves
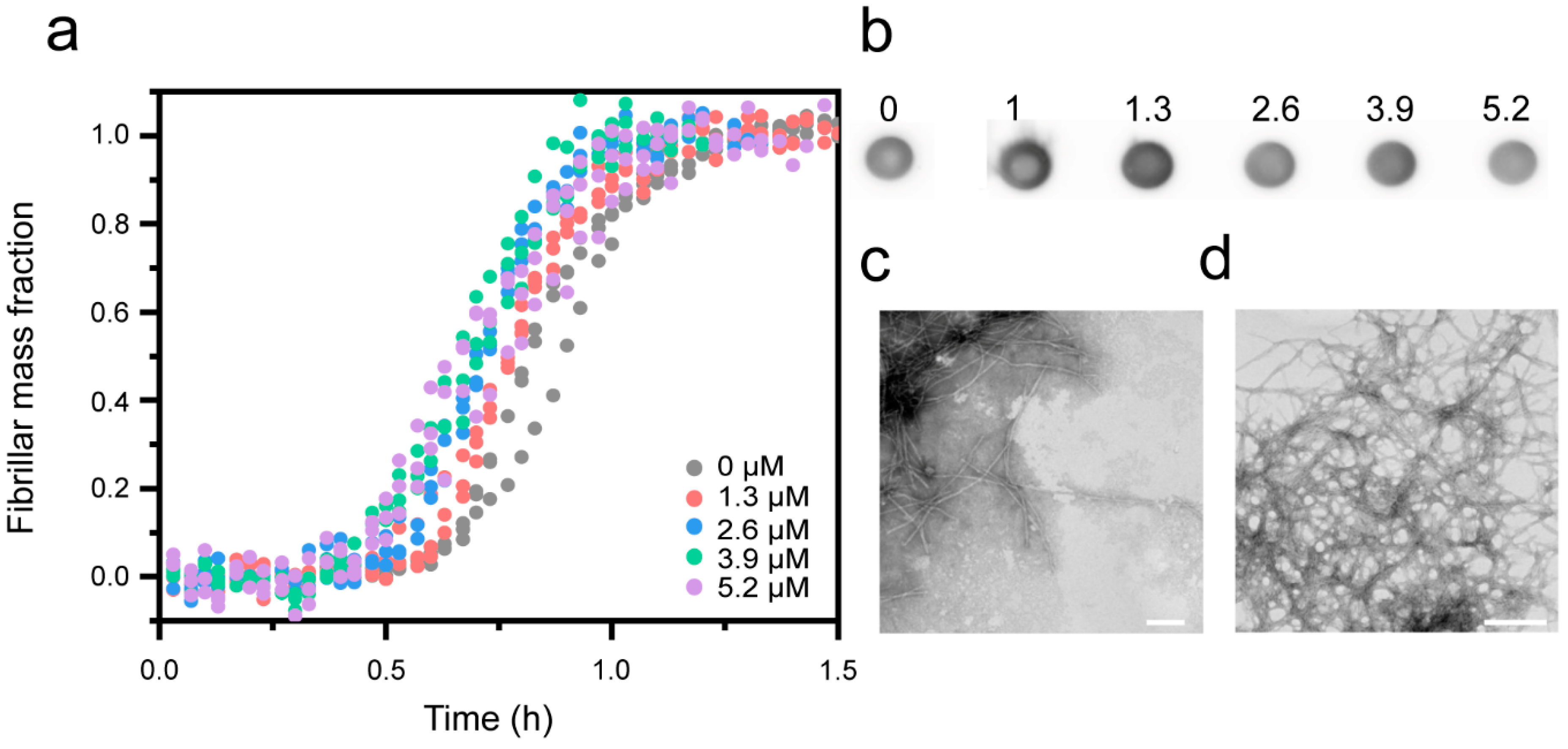
2.5. Dot-Blot Assay
2.6. Transmission Electron Microscopy (TEM)
3. Results and Discussion
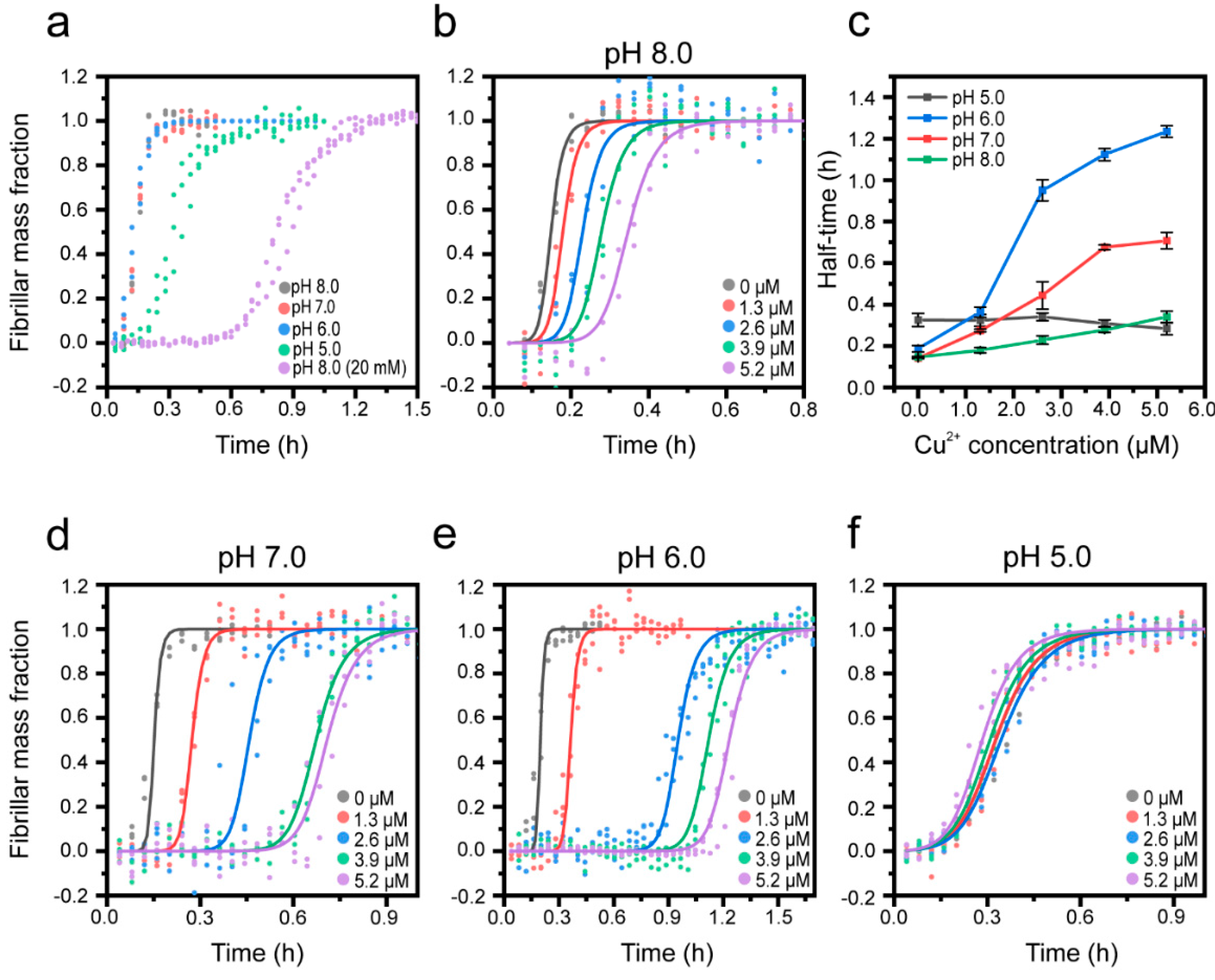
3.1. Cu2+ Inhibits Aβ(1-42) Amyloid Formation
3.2. pH and Salt Dependence of the Cu2+-Mediated Inhibition of Aβ(1-42) Amyloid Formation
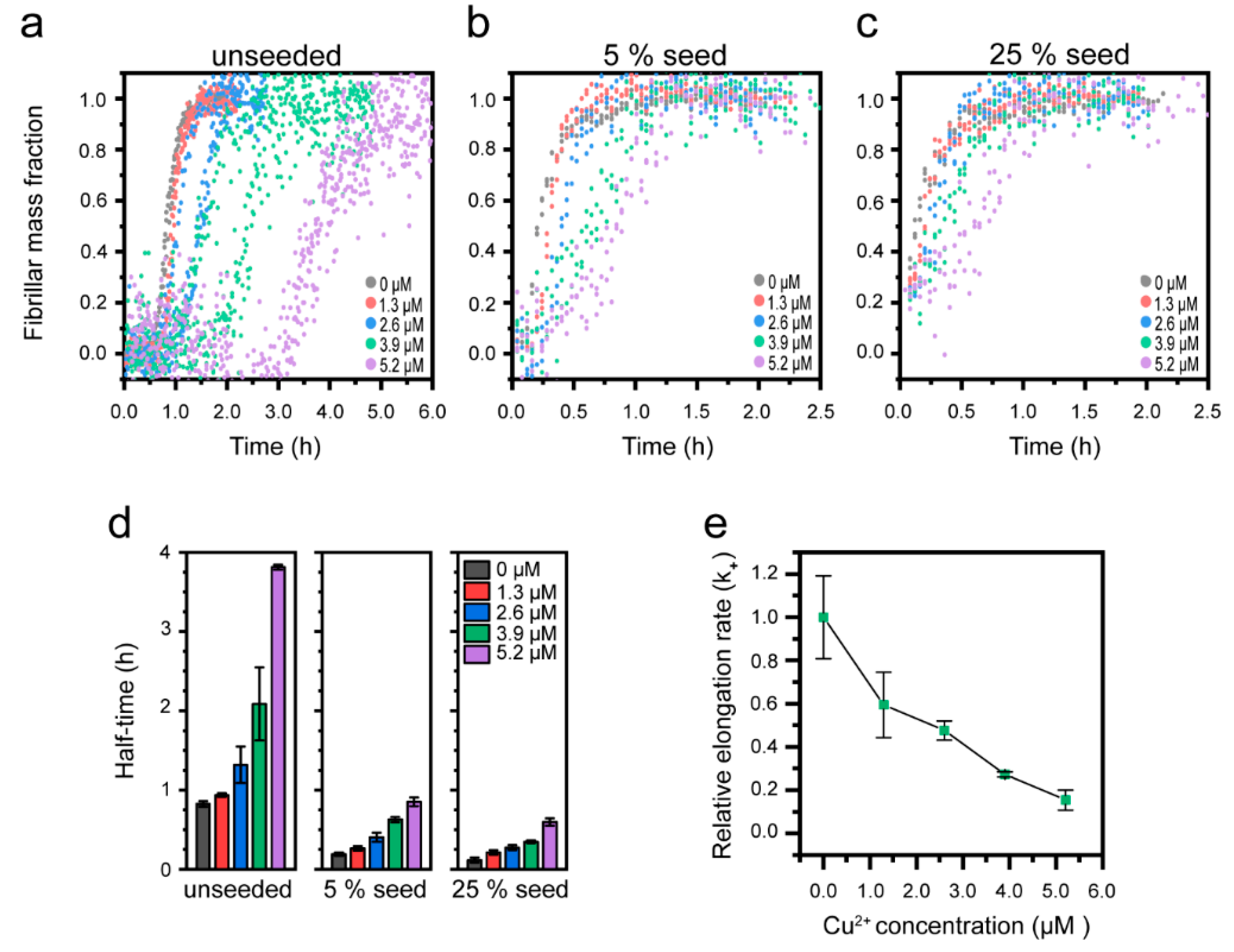
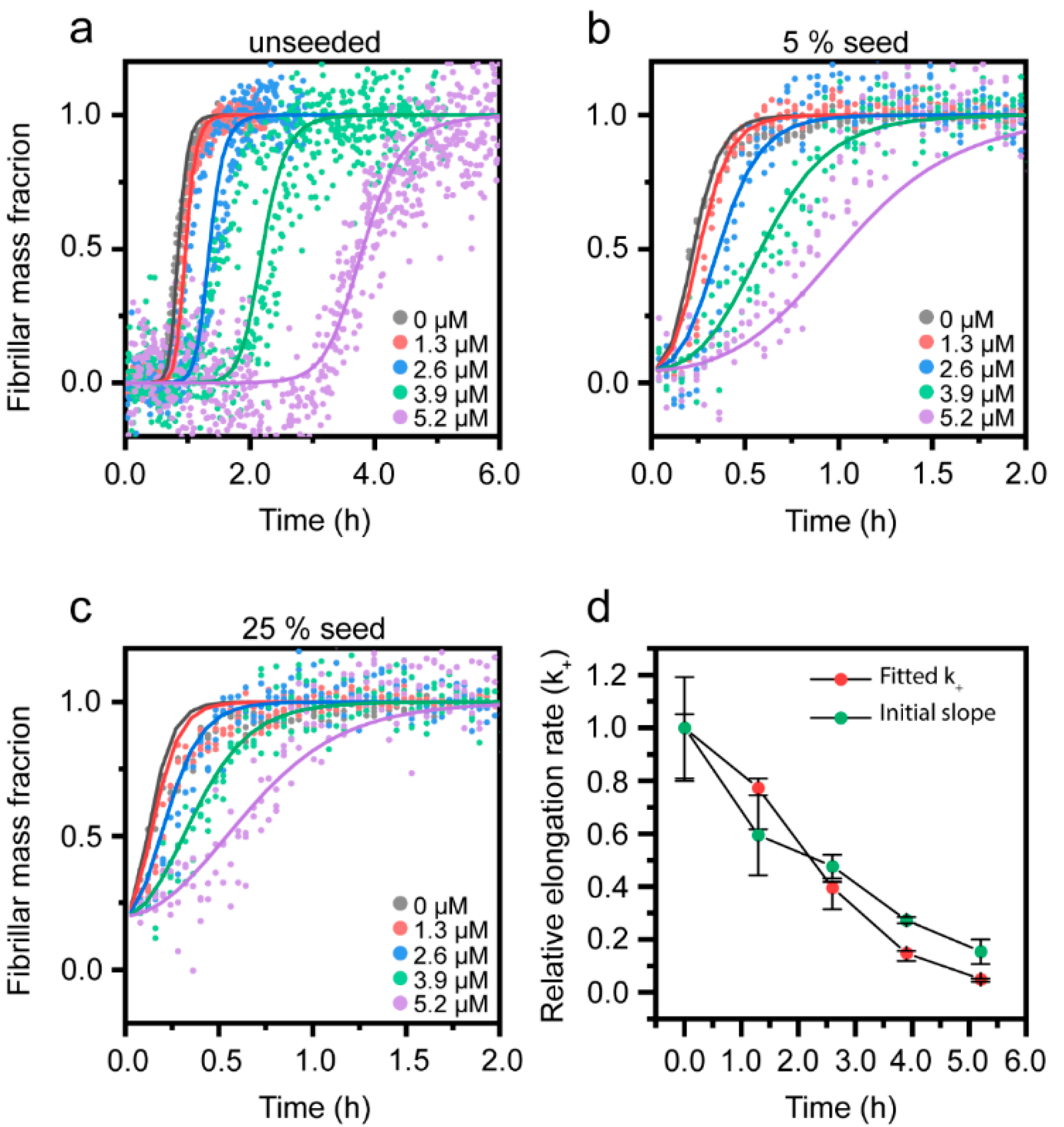
3.3. Cu2+-Mediated Inhibition of Aβ(1-42) Aggregation Affects the Fibril Elongation Step
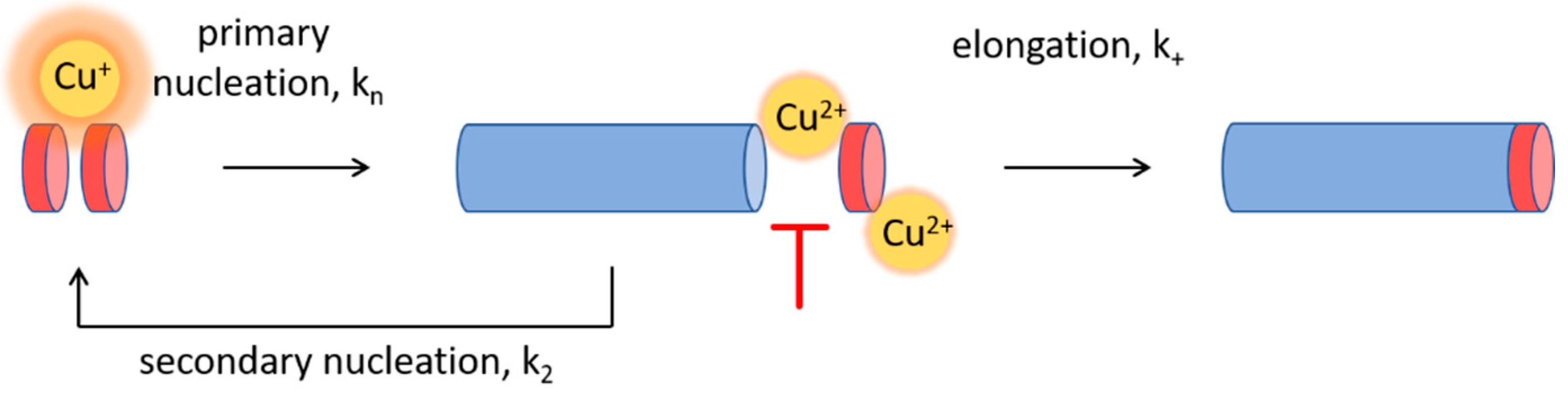
3.4. Effect of Cu+ on Aβ(1-42) Aggregation
3.5. Cu+ Enhances Aβ(1-42) Aggregation by Catalysis of Primary Nucleation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Masters, C.L.; Simms, G.; Weinman, N.A.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA 1985, 82, 4245–4249. [Google Scholar] [CrossRef] [PubMed]
- Hebert, L.E.; Weuve, J.; Scherr, P.A.; Evans, D.A. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 2013, 80, 1778–1783. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.J. World Alzheimer Report 2015: The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends; Alzheimer’s Disease International: London, UK, 2015. [Google Scholar]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Hardy, J.; Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef]
- LaFerla, F.M.; Green, K.N.; Oddo, S. Intracellular amyloid-β in Alzheimer’s disease. Nat. Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Sannerud, R.; Esselens, C.; Ejsmont, P.; Mattera, R.; Rochin, L.; Tharkeshwar, A.K.; De Baets, G.; De Wever, V.; Habets, R.; Baert, V. Restricted location of PSEN2/γ-secretase determines substrate specificity and generates an intracellular Aβ pool. Cell 2016, 166, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Esbjörner, E.K.; Chan, F.; Rees, E.; Erdelyi, M.; Luheshi, L.M.; Bertoncini, C.W.; Kaminski, C.F.; Dobson, C.M.; Schierle, G.S.K. Direct observations of amyloid β self-assembly in live cells provide insights into differences in the kinetics of Aβ (1–40) and Aβ (1–42) aggregation. Chem. Biol. 2014, 21, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Gouras, G.K.; Tsai, J.; Naslund, J.; Vincent, B.; Edgar, M.; Checler, F.; Greenfield, J.P.; Haroutunian, V.; Buxbaum, J.D.; Xu, H. Intraneuronal Aβ42 accumulation in human brain. Am. J. Pathol. 2000, 156, 15–20. [Google Scholar] [CrossRef]
- Jarrett, J.T.; Berger, E.P.; Lansbury, P.T., Jr. The carboxy terminus of the. beta. amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry 1993, 32, 4693–4697. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; LeVine, H., III. Alzheimer’s disease and the amyloid-β peptide. J. Alzheimer’s Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Linse, S. Monomer-dependent secondary nucleation in amyloid formation. Biophys. Rev. 2017, 9, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Meisl, G.; Yang, X.; Frohm, B.; Knowles, T.P.; Linse, S. Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated Aβ-peptide. Sci. Rep. 2016, 6, 18728. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.I.; Linse, S.; Luheshi, L.M.; Hellstrand, E.; White, D.A.; Rajah, L.; Otzen, D.E.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Michaels, T.C.; Linse, S.; Månsson, C.; Emanuelsson, C.; Presto, J.; Johansson, J.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P. Kinetic analysis reveals the diversity of microscopic mechanisms through which molecular chaperones suppress amyloid formation. Nat. Commun. 2016, 7, 10948. [Google Scholar] [CrossRef]
- Munke, A.; Persson, J.; Weiffert, T.; De Genst, E.; Meisl, G.; Arosio, P.; Carnerup, A.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P. Phage display and kinetic selection of antibodies that specifically inhibit amyloid self-replication. Proc. Natl. Acad. Sci. USA 2017, 114, 6444–6449. [Google Scholar] [CrossRef] [PubMed]
- Limbocker, R.; Chia, S.; Ruggeri, F.S.; Perni, M.; Cascella, R.; Heller, G.T.; Meisl, G.; Mannini, B.; Habchi, J.; Michaels, T.C. Trodusquemine enhances Aβ 42 aggregation but suppresses its toxicity by displacing oligomers from cell membranes. Nat. Commun. 2019, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, D.J.; Wesén, E.; Björkeroth, J.; Rocha, S.; Esbjörner, E.K. Lipid membranes catalyse the fibril formation of the amyloid-β (1–42) peptide through lipid-fibril interactions that reinforce secondary pathways. Biochim. Biophys. Acta (BBA)-Biomembr. 2017, 1859, 1921–1929. [Google Scholar] [CrossRef]
- Bush, A.I. The metallobiology of Alzheimer’s disease. Trends Neurosci. 2003, 26, 207–214. [Google Scholar] [CrossRef]
- Li, F.; Calingasan, N.Y.; Yu, F.; Mauck, W.M.; Toidze, M.; Almeida, C.G.; Takahashi, R.H.; Carlson, G.A.; Flint Beal, M.; Lin, M.T. Increased plaque burden in brains of APP mutant MnSOD heterozygous knockout mice. J. Neurochem. 2004, 89, 1308–1312. [Google Scholar] [CrossRef]
- Smith, D.G.; Cappai, R.; Barnham, K.J. The redox chemistry of the Alzheimer’s disease amyloid β peptide. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 1976–1990. [Google Scholar] [CrossRef]
- Huang, X.; Atwood, C.S.; Hartshorn, M.A.; Multhaup, G.; Goldstein, L.E.; Scarpa, R.C.; Cuajungco, M.P.; Gray, D.N.; Lim, J.; Moir, R.D. The Aβ peptide of Alzheimer’s disease directly produces hydrogen peroxide through metal ion reduction. Biochemistry 1999, 38, 7609–7616. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Moir, R.D.; Tanzi, R.E.; Bush, A.I.; Rogers, J.T. Redox-active metals, oxidative stress, and Alzheimer’s disease pathology. Ann. N. Y. Acad. Sci. 2004, 1012, 153–163. [Google Scholar] [CrossRef]
- Atwood, C.S.; Moir, R.D.; Huang, X.; Scarpa, R.C.; Bacarra, N.M.E.; Romano, D.M.; Hartshorn, M.A.; Tanzi, R.E.; Bush, A.I. Dramatic aggregation of Alzheimer Aβ by Cu (II) is induced by conditions representing physiological acidosis. J. Biol. Chem. 1998, 273, 12817–12826. [Google Scholar] [CrossRef] [PubMed]
- Bush, A.I.; Pettingell, W.H.; Multhaup, G.; d Paradis, M.; Vonsattel, J.-P.; Gusella, J.F.; Beyreuther, K.; Masters, C.L.; Tanzi, R.E. Rapid induction of Alzheimer A beta amyloid formation by zinc. Science 1994, 265, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Esler, W.P.; Stimson, E.R.; Jennings, J.M.; Ghilardi, J.R.; Mantyh, P.W.; Maggio, J.E. Zinc-induced aggregation of human and rat β-amyloid peptides in vitro. J. Neurochem. 1996, 66, 723–732. [Google Scholar] [CrossRef]
- Mantyh, P.W.; Ghilardi, J.R.; Rogers, S.; DeMaster, E.; Allen, C.J.; Stimson, E.R.; Maggio, J.E. Aluminum, iron, and zinc ions promote aggregation of physiological concentrations of β-amyloid peptide. J. Neurochem. 1993, 61, 1171–1174. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, D.; Kisilevsky, R. A method based on ICP-MS for the analysis of Alzheimer’s amyloid plaques. Anal. Chem. 1998, 70, 1026–1029. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Atwood, C.S.; Anderson, V.E.; Siedlak, S.L.; Smith, M.A.; Perry, G.; Carey, P.R. Metal binding and oxidation of amyloid-β within isolated senile plaque cores: Raman microscopic evidence. Biochemistry 2003, 42, 2768–2773. [Google Scholar] [CrossRef]
- Miller, L.M.; Wang, Q.; Telivala, T.P.; Smith, R.J.; Lanzirotti, A.; Miklossy, J. Synchrotron-based infrared and X-ray imaging shows focalized accumulation of Cu and Zn co-localized with β-amyloid deposits in Alzheimer’s disease. J. Struct. Biol. 2006, 155, 30–37. [Google Scholar] [CrossRef]
- Curtain, C.C.; Ali, F.; Volitakis, I.; Cherny, R.A.; Norton, R.S.; Beyreuther, K.; Barrow, C.J.; Masters, C.L.; Bush, A.I.; Barnham, K.J. Alzheimer’s disease amyloid-β binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J. Biol. Chem. 2001, 276, 20466–20473. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Cuajungco, M.P.; Atwood, C.S.; Hartshorn, M.A.; Tyndall, J.D.; Hanson, G.R.; Stokes, K.C.; Leopold, M.; Multhaup, G.; Goldstein, L.E. Cu (II) potentiation of Alzheimer Aβ neurotoxicity correlation with cell-free hydrogen peroxide production and metal reduction. J. Biol. Chem. 1999, 274, 37111–37116. [Google Scholar] [CrossRef] [PubMed]
- Opazo, C.; Huang, X.; Cherny, R.A.; Moir, R.D.; Roher, A.E.; White, A.R.; Cappai, R.; Masters, C.L.; Tanzi, R.E.; Inestrosa, N.C. Metalloenzyme-like activity of Alzheimer’s disease β-amyloid Cu-dependent catalytic conversion of dopamine, cholesterol, and biological reducing agents to neurotoxic H2O2. J. Biol. Chem. 2002, 277, 40302–40308. [Google Scholar] [CrossRef] [PubMed]
- Reybier, K.; Ayala, S.; Alies, B.; Rodrigues, J.V.; Bustos Rodriguez, S.; La Penna, G.; Collin, F.; Gomes, C.M.; Hureau, C.; Faller, P. Free Superoxide is an Intermediate in the Production of H2O2 by Copper (I)-Aβ Peptide and O2. Angew. Chem. Int. Ed. 2016, 55, 1085–1089. [Google Scholar] [CrossRef] [PubMed]
- Guilloreau, L.; Combalbert, S.; Sournia-Saquet, A.; Mazarguil, H.; Faller, P. Redox Chemistry of Copper–Amyloid-β: The Generation of Hydroxyl Radical in the Presence of Ascorbate is Linked to Redox-Potentials and Aggregation State. ChemBioChem 2007, 8, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Mathys, Z.K.; White, A.R. Copper and Alzheimer’s Disease. In Neurotoxicity of Metals; Springer: Berlin/Heidelberg, Germany, 2017; pp. 199–216. [Google Scholar]
- Santos, M.A.; Chand, K.; Chaves, S. Recent progress in multifunctional metal chelators as potential drugs for Alzheimer’s disease. Coord. Chem. Rev. 2016, 327, 287–303. [Google Scholar] [CrossRef]
- Matlack, K.E.; Tardiff, D.F.; Narayan, P.; Hamamichi, S.; Caldwell, K.A.; Caldwell, G.A.; Lindquist, S. Clioquinol promotes the degradation of metal-dependent amyloid-β (Aβ) oligomers to restore endocytosis and ameliorate Aβ toxicity. Proc. Natl. Acad. Sci. USA 2014, 111, 4013–4018. [Google Scholar] [CrossRef] [PubMed]
- Adlard, P.A.; Bush, A.I. Metals and Alzheimer’s disease. J. Alzheimer’s Dis. 2006, 10, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Deibel, M.; Ehmann, W.; Markesbery, W. Copper, iron, and zinc imbalances in severely degenerated brain regions in Alzheimer’s disease: Possible relation to oxidative stress. J. Neurol. Sci. 1996, 143, 137–142. [Google Scholar] [CrossRef]
- Squitti, R.; Ghidoni, R.; Simonelli, I.; Ivanova, I.D.; Colabufo, N.A.; Zuin, M.; Benussi, L.; Binetti, G.; Cassetta, E.; Rongioletti, M. Copper dyshomeostasis in Wilson disease and Alzheimer’s disease as shown by serum and urine copper indicators. J. Trace Elem. Med. Biol. 2018, 45, 181–188. [Google Scholar] [CrossRef]
- Xu, J.; Begley, P.; Church, S.J.; Patassini, S.; McHarg, S.; Kureishy, N.; Hollywood, K.A.; Waldvogel, H.J.; Liu, H.; Zhang, S. Elevation of brain glucose and polyol-pathway intermediates with accompanying brain-copper deficiency in patients with Alzheimer’s disease: Metabolic basis for dementia. Sci. Rep. 2016, 6, 27524. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.D.; Wen, X.; Gazzard, L.; Nelson, C.; Scheller, R.H.; Scales, S.J. Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody–drug conjugates. Proc. Natl. Acad. Sci. USA 2005, 102, 17987–17992. [Google Scholar] [CrossRef] [PubMed]
- Bolognin, S.; Messori, L.; Drago, D.; Gabbiani, C.; Cendron, L.; Zatta, P. Aluminum, copper, iron and zinc differentially alter amyloid-Aβ1–42 aggregation and toxicity. Int. J. Biochem. Cell Biol. 2011, 43, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Brzyska, M.; Trzesniewska, K.; Wieckowska, A.; Szczepankiewicz, A.; Elbaum, D. Electrochemical and Conformational Consequences of Copper (CuI and CuII) Binding to β-Amyloid (1–40). ChemBioChem 2009, 10, 1045–1055. [Google Scholar] [CrossRef]
- Dai, X.; Sun, Y.; Gao, Z.; Jiang, Z. Copper enhances amyloid-β peptide neurotoxicity and non β-aggregation: A series of experiments conducted upon copper-bound and copper-free amyloid-β peptide. J. Mol. Neurosci. 2010, 41, 66–73. [Google Scholar] [CrossRef] [PubMed]
- House, E.; Mold, M.; Collingwood, J.; Baldwin, A.; Goodwin, S.; Exley, C. Copper Abolishes the β-Sheet Secondary Structure of Preformed Amyloid Fibrils of Amyloid-β 42. J. Alzheimer’s Dis. 2009, 18, 811–817. [Google Scholar] [CrossRef]
- Mold, M.; Ouro-Gnao, L.; Wieckowski, B.M.; Exley, C. Copper prevents amyloid-β 1–42 from forming amyloid fibrils under near-physiological conditions in vitro. Sci. Rep. 2013, 3, 1256. [Google Scholar] [CrossRef]
- Yoshiike, Y.; Tanemura, K.; Murayama, O.; Akagi, T.; Murayama, M.; Sato, S.; Sun, X.; Tanaka, N.; Takashima, A. New insights on how metals disrupt amyloid β-aggregation and their effects on amyloid-β cytotoxicity. J. Biol. Chem. 2001, 276, 32293–32299. [Google Scholar] [CrossRef]
- Zou, J.; Kajita, K.; Sugimoto, N. Cu2+ Inhibits the Aggregation of Amyloid β-Peptide (1–42) in vitro. Angew. Chem. Int. Ed. 2001, 40, 2274–2277. [Google Scholar] [CrossRef]
- Abelein, A.; Gräslund, A.; Danielsson, J. Zinc as chaperone-mimicking agent for retardation of amyloid β peptide fibril formation. Proc. Natl. Acad. Sci. USA 2015, 112, 5407–5412. [Google Scholar] [CrossRef]
- Gu, M.; Bode, D.C.; Viles, J.H. Copper redox cycling inhibits Aβ fibre formation and promotes fibre fragmentation, while generating a dityrosine Aβ dimer. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sarell, C.J.; Wilkinson, S.R.; Viles, J.H. Substoichiometric levels of Cu2+ ions accelerate the kinetics of fiber formation and promote cell toxicity of amyloid-β from Alzheimer disease. J. Biol. Chem. 2010, 285, 41533–41540. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P.; Ciccotosto, G.D.; Tew, D.J.; Fodero-Tavoletti, M.T.; Johanssen, T.; Masters, C.L.; Barnham, K.J.; Cappai, R. Concentration dependent Cu2+ induced aggregation and dityrosine formation of the Alzheimer’s disease amyloid-β peptide. Biochemistry 2007, 46, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Pavlova, S.T.; Kim, J.; Kim, J.; Mirica, L.M. The effect of Cu 2+ and Zn 2+ on the Aβ 42 peptide aggregation and cellular toxicity. Metallomics 2013, 5, 1529–1536. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.; Zhu, Y.; Lenczowski, E.; Tian, Y.; Yang, J.; Zhang, C.; Hardt, M.; Qiao, C.; Tanzi, R.E. The double-edged role of copper in the fate of amyloid beta in the presence of anti-oxidants. Chem. Sci. 2017, 8, 6155–6164. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, Z.; Zheng, Y.; Li, H.; Ni, J.; Liu, Q. Inhibitory effect of selenoprotein P on Cu+/Cu2+-induced Aβ42 aggregation and toxicity. Inorg. Chem. 2014, 53, 1672–1678. [Google Scholar] [CrossRef]
- Faller, P.; Hureau, C. Bioinorganic chemistry of copper and zinc ions coordinated to amyloid-β peptide. Dalton Trans. 2009, 1080–1094. [Google Scholar] [CrossRef]
- Garzon-Rodriguez, W.; Yatsimirsky, A.K.; Glabe, C.G. Binding of Zn (II), Cu (II), and Fe (II) ions to alzheimer’s Aß peptide studied by fluorescence. Bioorganic Med. Chem. Lett. 1999, 9, 2243–2248. [Google Scholar] [CrossRef]
- Guilloreau, L.; Damian, L.; Coppel, Y.; Mazarguil, H.; Winterhalter, M.; Faller, P. Structural and thermodynamical properties of Cu II amyloid-β16/28 complexes associated with Alzheimer’s disease. JBIC J. Biol. Inorg. Chem. 2006, 11, 1024–1038. [Google Scholar] [CrossRef]
- Jiang, D.; Men, L.; Wang, J.; Zhang, Y.; Chickenyen, S.; Wang, Y.; Zhou, F. Redox reactions of copper complexes formed with different β-amyloid peptides and their neuropathalogical relevance. Biochemistry 2007, 46, 9270–9282. [Google Scholar] [CrossRef]
- Tõugu, V.; Tiiman, A.; Palumaa, P. Interactions of Zn (II) and Cu (II) ions with Alzheimer’s amyloid-beta peptide. Metal ion binding, contribution to fibrillization and toxicity. Metallomics 2011, 3, 250–261. [Google Scholar] [CrossRef]
- Karr, J.W.; Szalai, V.A. Cu (II) binding to monomeric, oligomeric, and fibrillar forms of the Alzheimer’s disease amyloid-β peptide. Biochemistry 2008, 47, 5006–5016. [Google Scholar] [CrossRef] [PubMed]
- Sarell, C.J.; Syme, C.D.; Rigby, S.E.; Viles, J.H. Copper (II) binding to amyloid-β fibrils of Alzheimer’s disease reveals a picomolar affinity: Stoichiometry and coordination geometry are independent of Aβ oligomeric form. Biochemistry 2009, 48, 4388–4402. [Google Scholar] [CrossRef] [PubMed]
- Shearer, J.; Szalai, V.A. The amyloid-β peptide of Alzheimer’s disease binds CuI in a linear bis-his coordination environment: Insight into a possible neuroprotective mechanism for the amyloid-β peptide. J. Am. Chem. Soc. 2008, 130, 17826–17835. [Google Scholar] [CrossRef] [PubMed]
- Syme, C.D.; Nadal, R.C.; Rigby, S.E.; Viles, J.H. Copper Binding to the Amyloid-β (Aβ) Peptide Associated with Alzheimer’s Disease Folding, Coordination Geometry, ph Dependence, Stoichiometry, and Affinity of aβ-(1–28): Insights from a Range of Complementary Spectroscopic Techniques. J. Biol. Chem. 2004, 279, 18169–18177. [Google Scholar] [CrossRef]
- Bin, Y.; Chen, S.; Xiang, J. pH-dependent kinetics of copper ions binding to amyloid-β peptide. J. Inorg. Biochem. 2013, 119, 21–27. [Google Scholar] [CrossRef]
- Ma, Q.F.; Hu, J.; Wu, W.H.; Liu, H.D.; Du, J.T.; Fu, Y.; Wu, Y.W.; Lei, P.; Zhao, Y.F.; Li, Y.M. Characterization of copper binding to the peptide amyloid-β (1–16) associated with Alzheimer’s disease. Biopolym. Orig. Res. Biomol. 2006, 83, 20–31. [Google Scholar] [CrossRef]
- Yako, N.; Young, T.R.; Jones, J.M.C.; Hutton, C.A.; Wedd, A.G.; Xiao, Z. Copper binding and redox chemistry of the Aβ16 peptide and its variants: Insights into determinants of copper-dependent reactivity. Metallomics 2017, 9, 278–291. [Google Scholar] [CrossRef]
- Atwood, C.S.; Scarpa, R.C.; Huang, X.; Moir, R.D.; Jones, W.D.; Fairlie, D.P.; Tanzi, R.E.; Bush, A.I. Characterization of Copper Interactions with Alzheimer Amyloid β Peptides: Identification of an Attomolar-Affinity Copper Binding Site on Amyloid β1-42. J. Neurochem. 2000, 75, 1219–1233. [Google Scholar] [CrossRef]
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Furlan, S.; Hureau, C.; Faller, P.; La Penna, G. Modeling the Cu+ Binding in the 1− 16 Region of the Amyloid-β Peptide Involved in Alzheimer’s Disease. J. Phys. Chem. B 2010, 114, 15119–15133. [Google Scholar] [CrossRef]
- Colvin, M.T.; Silvers, R.; Ni, Q.Z.; Can, T.V.; Sergeyev, I.; Rosay, M.; Donovan, K.J.; Michael, B.; Wall, J.; Linse, S. Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J. Am. Chem. Soc. 2016, 138, 9663–9674. [Google Scholar] [CrossRef] [PubMed]
- Wälti, M.A.; Ravotti, F.; Arai, H.; Glabe, C.G.; Wall, J.S.; Böckmann, A.; Güntert, P.; Meier, B.H.; Riek, R. Atomic-resolution structure of a disease-relevant Aβ (1–42) amyloid fibril. Proc. Natl. Acad. Sci. USA 2016, 113, E4976–E4984. [Google Scholar] [CrossRef] [PubMed]
- Hellstrand, E.; Boland, B.; Walsh, D.M.; Linse, S. Amyloid β-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem. Neurosci. 2010, 1, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Abelein, A.; Chen, G.; Kitoka, K.; Aleksis, R.; Oleskovs, F.; Sarr, M.; Landreh, M.; Pahnke, J.; Nordling, K.; Kronqvist, N. High-yield Production of Amyloid-β Peptide Enabled by a Customized Spider Silk Domain. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kronqvist, N.; Sarr, M.; Lindqvist, A.; Nordling, K.; Otikovs, M.; Venturi, L.; Pioselli, B.; Purhonen, P.; Landreh, M.; Biverstål, H. Efficient protein production inspired by how spiders make silk. Nat. Commun. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tropea, J.E.; Cherry, S.; Waugh, D.S. Expression and Purification of Soluble His 6-tagged TEV Protease. In High Throughput Protein Expression and Purification; Springer: Berlin/Heidelberg, Germany, 2009; pp. 297–307. [Google Scholar]
- Cohen, S.I.; Arosio, P.; Presto, J.; Kurudenkandy, F.R.; Biverstål, H.; Dolfe, L.; Dunning, C.; Yang, X.; Frohm, B.; Vendruscolo, M. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat. Struct. Mol. Biol. 2015, 22, 207. [Google Scholar] [CrossRef]
- Scheidt, T.; Łapińska, U.; Kumita, J.R.; Whiten, D.R.; Klenerman, D.; Wilson, M.R.; Cohen, S.I.; Linse, S.; Vendruscolo, M.; Dobson, C.M. Secondary nucleation and elongation occur at different sites on Alzheimer’s amyloid-β aggregates. Sci. Adv. 2019, 5, eaau3112. [Google Scholar] [CrossRef] [PubMed]
- Niemiec, M.S.; Dingeldein, A.P.; Wittung-Stafshede, P. T versus D in the MTCXXC motif of copper transport proteins plays a role in directional metal transport. JBIC J. Biol. Inorg. Chem. 2014, 19, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Niemiec, M.S.; Dingeldein, A.P.; Wittung-Stafshede, P. Enthalpy-entropy compensation at play in human copper ion transfer. Sci. Rep. 2015, 5, 10518. [Google Scholar] [CrossRef] [PubMed]
- Niemiec, M.S.; Weise, C.F.; Wittung-Stafshede, P. In vitro thermodynamic dissection of human copper transfer from chaperone to target protein. PLoS ONE 2012, 7, e36102. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Waudby, C.A.; Devlin, G.L.; Cohen, S.I.; Aguzzi, A.; Vendruscolo, M.; Terentjev, E.M.; Welland, M.E.; Dobson, C.M. An analytical solution to the kinetics of breakable filament assembly. Science 2009, 326, 1533–1537. [Google Scholar] [CrossRef] [PubMed]
- Meisl, G.; Kirkegaard, J.B.; Arosio, P.; Michaels, T.C.; Vendruscolo, M.; Dobson, C.M.; Linse, S.; Knowles, T.P. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 2016, 11, 252. [Google Scholar] [CrossRef]
- Kayed, R.; Head, E.; Sarsoza, F.; Saing, T.; Cotman, C.W.; Necula, M.; Margol, L.; Wu, J.; Breydo, L.; Thompson, J.L. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2007, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Indi, S.; Rao, K. Copper-and iron-induced differential fibril formation in α-synuclein: TEM study. Neurosci. Lett. 2007, 424, 78–82. [Google Scholar]
- Levine, H., III. Thioflavine T interaction with synthetic Alzheimer’s disease β-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, D.J.; Wranne, M.S.; Gatty, M.G.; Westerlund, F.; Esbjörner, E.K. Steady-state and time-resolved Thioflavin-T fluorescence can report on morphological differences in amyloid fibrils formed by Aβ (1-40) and Aβ (1-42). Biochem. Biophys. Res. Commun. 2015, 458, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Saxena, S. The Aggregated State of Amyloid-β Peptide In Vitro Depends on Cu2+ Ion Concentration. Angew. Chem. Int. Ed. 2007, 46, 3959–3961. [Google Scholar] [CrossRef]
- Klement, K.; Wieligmann, K.; Meinhardt, J.; Hortschansky, P.; Richter, W.; Fändrich, M. Effect of different salt ions on the propensity of aggregation and on the structure of Alzheimer’s Aβ (1-40) amyloid fibrils. J. Mol. Biol. 2007, 373, 1321–1333. [Google Scholar] [CrossRef]
- Lindberg, D.J.; Esbjörner, E.K. Detection of amyloid-β fibrils using the DNA-intercalating dye YOYO-1: Binding mode and fibril formation kinetics. Biochem. Biophys. Res. Commun. 2016, 469, 313–318. [Google Scholar] [CrossRef]
- Meisl, G.; Yang, X.; Dobson, C.M.; Linse, S.; Knowles, T.P. Modulation of electrostatic interactions to reveal a reaction network unifying the aggregation behaviour of the Aβ42 peptide and its variants. Chem. Sci. 2017, 8, 4352–4362. [Google Scholar] [CrossRef]
- Antzutkin, O.N. Amyloidosis of Alzheimer’s Aβ peptides: Solid-state nuclear magnetic resonance, electron paramagnetic resonance, transmission electron microscopy, scanning transmission electron microscopy and atomic force microscopy studies. Magn. Reson. Chem. 2004, 42, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Wesén, E.; Jeffries, G.D.; Dzebo, M.M.; Esbjörner, E.K. Endocytic uptake of monomeric amyloid-β peptides is clathrin-and dynamin-independent and results in selective accumulation of Aβ (1–42) compared to Aβ (1–40). Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Törnquist, M.; Michaels, T.C.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.; Knowles, T.P.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.I.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 2012, 421, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Bacci, M.; Vymětal, J.í.; Mihajlovic, M.; Caflisch, A.; Vitalis, A. Amyloid β fibril elongation by monomers involves disorder at the tip. J. Chem. Theory Comput. 2017, 13, 5117–5130. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.J.; Williams, A.D.; Wetzel, R.; Myszka, D.G. Kinetic analysis of beta-amyloid fibril elongation. Anal. Biochem. 2004, 328, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Vettore, N.; Buell, A.K. Thermodynamics of amyloid fibril formation from chemical depolymerization. Phys. Chem. Chem. Phys. 2019, 21, 26184–26194. [Google Scholar] [CrossRef] [PubMed]
- Gurry, T.; Stultz, C.M. Mechanism of amyloid-β fibril elongation. Biochemistry 2014, 53, 6981–6991. [Google Scholar] [CrossRef] [PubMed]
- Feaga, H.A.; Maduka, R.C.; Foster, M.N.; Szalai, V.A. Affinity of Cu+ for the copper-binding domain of the amyloid-β peptide of Alzheimer’s disease. Inorg. Chem. 2011, 50, 1614–1618. [Google Scholar] [CrossRef] [PubMed]

| [Cu2+] | k+ | Upper/Lower Error |
|---|---|---|
| (µM) | (M−1h−1) | (M−1h−1) |
| 0 | 2.1 × 108 | +1.1 × 107 |
| −4.2 × 107 | ||
| 1.3 | 1.6 × 106 | +7.5 × 106 |
| −3.2 × 107 | ||
| 2.6 | 8.2 × 107 | +4.5 × 106 |
| −1.7 × 107 | ||
| 3.9 | 3.1 × 107 | +1.6 × 106 |
| −6.2 × 106 | ||
| 5.2 | 1.0 × 107 | +5.3 × 105 |
| −2.1 × 106 |
| [Cu+] | k+kn | Upper/Lower Error |
|---|---|---|
| (µM) | (M−2h−2) | (M−2h−2) |
| 0 | 5.5 × 108 | +5.6 × 107 |
| −3.9 × 107 | ||
| 1.3 | 1.4 × 109 | +1.3 × 108 |
| −1.0 × 108 | ||
| 2.6 | 3.2 × 109 | +2.6 × 108 |
| −2.1 × 108 | ||
| 3.9 | 4.7 × 109 | +4.3 × 108 |
| −2.8 × 108 | ||
| 5.2 | 3.7 × 109 | +3.0 × 108 |
| −2.4 × 108 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasanian, N.; Bernson, D.; Horvath, I.; Wittung-Stafshede, P.; Esbjörner, E.K. Redox-Dependent Copper Ion Modulation of Amyloid-β (1-42) Aggregation In Vitro. Biomolecules 2020, 10, 924. https://doi.org/10.3390/biom10060924
Sasanian N, Bernson D, Horvath I, Wittung-Stafshede P, Esbjörner EK. Redox-Dependent Copper Ion Modulation of Amyloid-β (1-42) Aggregation In Vitro. Biomolecules. 2020; 10(6):924. https://doi.org/10.3390/biom10060924
Chicago/Turabian StyleSasanian, Nima, David Bernson, Istvan Horvath, Pernilla Wittung-Stafshede, and Elin K. Esbjörner. 2020. "Redox-Dependent Copper Ion Modulation of Amyloid-β (1-42) Aggregation In Vitro" Biomolecules 10, no. 6: 924. https://doi.org/10.3390/biom10060924
APA StyleSasanian, N., Bernson, D., Horvath, I., Wittung-Stafshede, P., & Esbjörner, E. K. (2020). Redox-Dependent Copper Ion Modulation of Amyloid-β (1-42) Aggregation In Vitro. Biomolecules, 10(6), 924. https://doi.org/10.3390/biom10060924







