Caudatin Isolated from Cynanchum auriculatum Inhibits Breast Cancer Stem Cell Formation via a GR/YAP Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical and Reagents
2.2. Plant Material
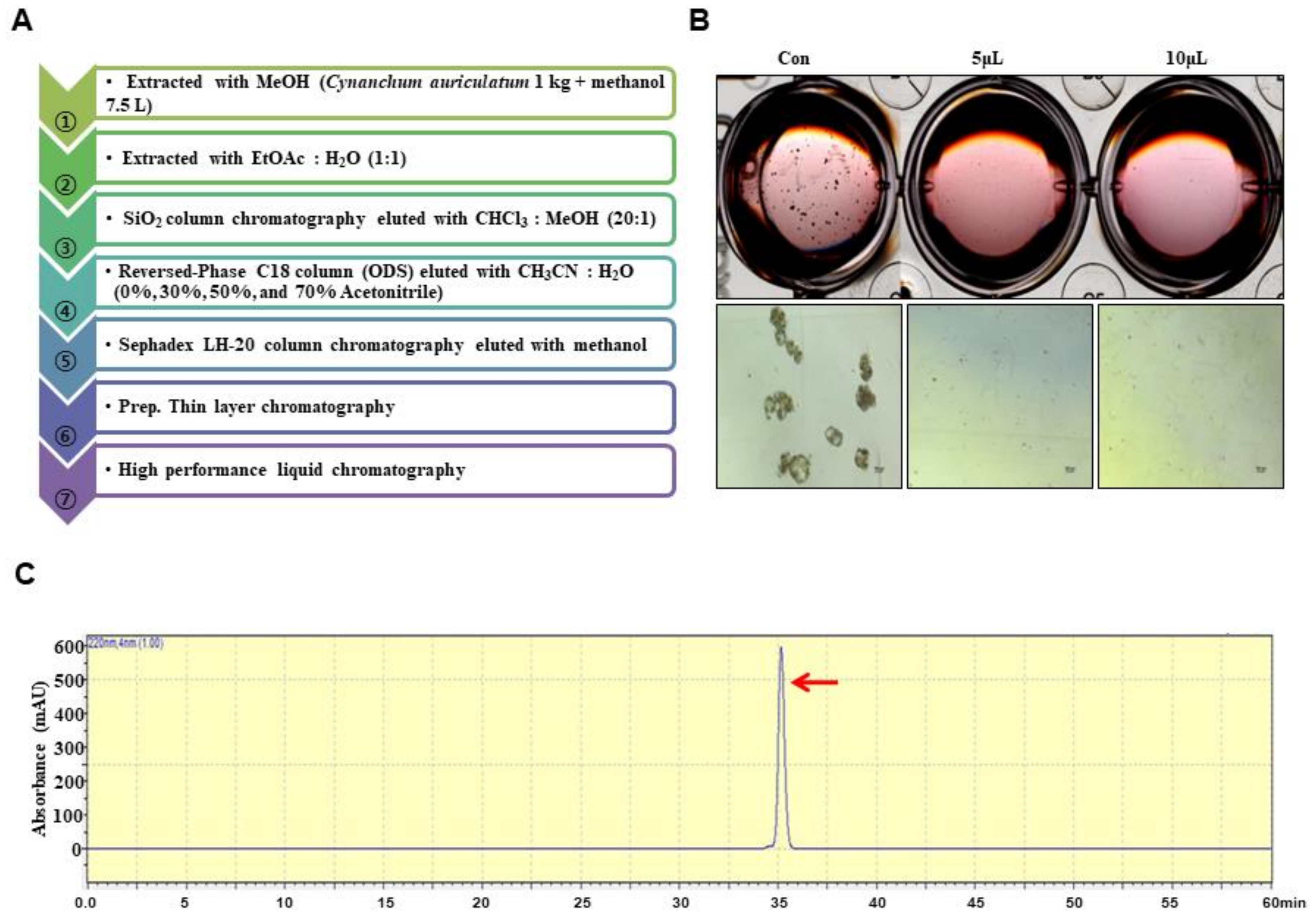
2.3. Extraction and Isolation
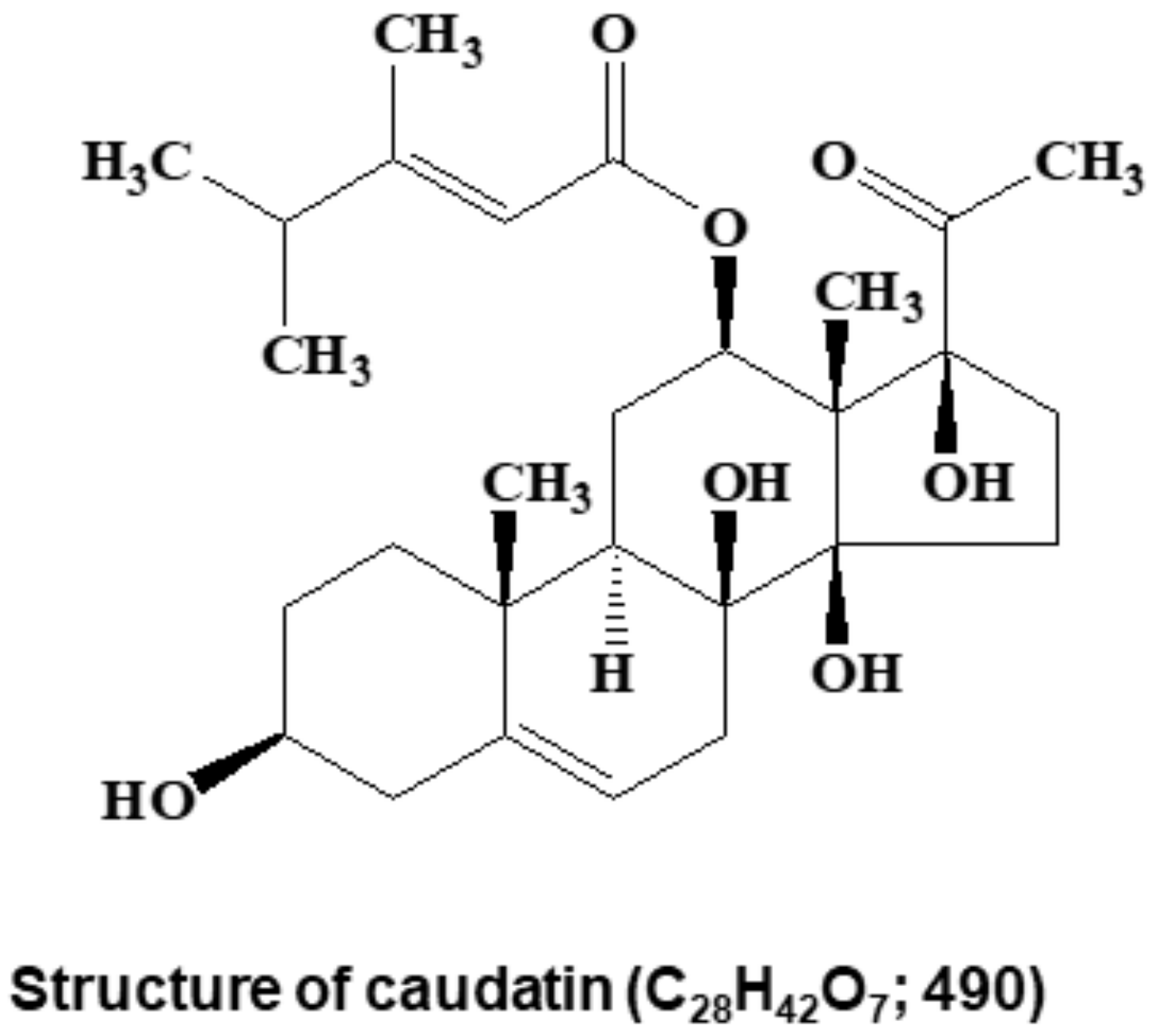
2.4. Structural Analysis
2.5. Cell Culture and Mammosphere Formation Assay
2.6. Cell Proliferation
2.7. Annexin V/Propidium Iodide (PI) Assay to Detect Cell Apoptosis and Hoechst 33,342 Staining of Apoptotic Nuclei
2.8. Scratch Assay
2.9. Transwell Assay
2.10. Colony Formation Assay
2.11. Flow Cytometric Analysis and ALDH1 Activity
2.12. Western Blot Analysis
2.13. Immunoprecipitation (IP)
2.14. Gene Expression Analysis
2.15. Immunofluorescence (IF) Assay
2.16. Small Interfering RNA (siRNA)
2.17. Xenograft Transplantation
2.18. Statistical Analysis
3. Results
3.1. BCSC Inhibitor from C. auriculatum
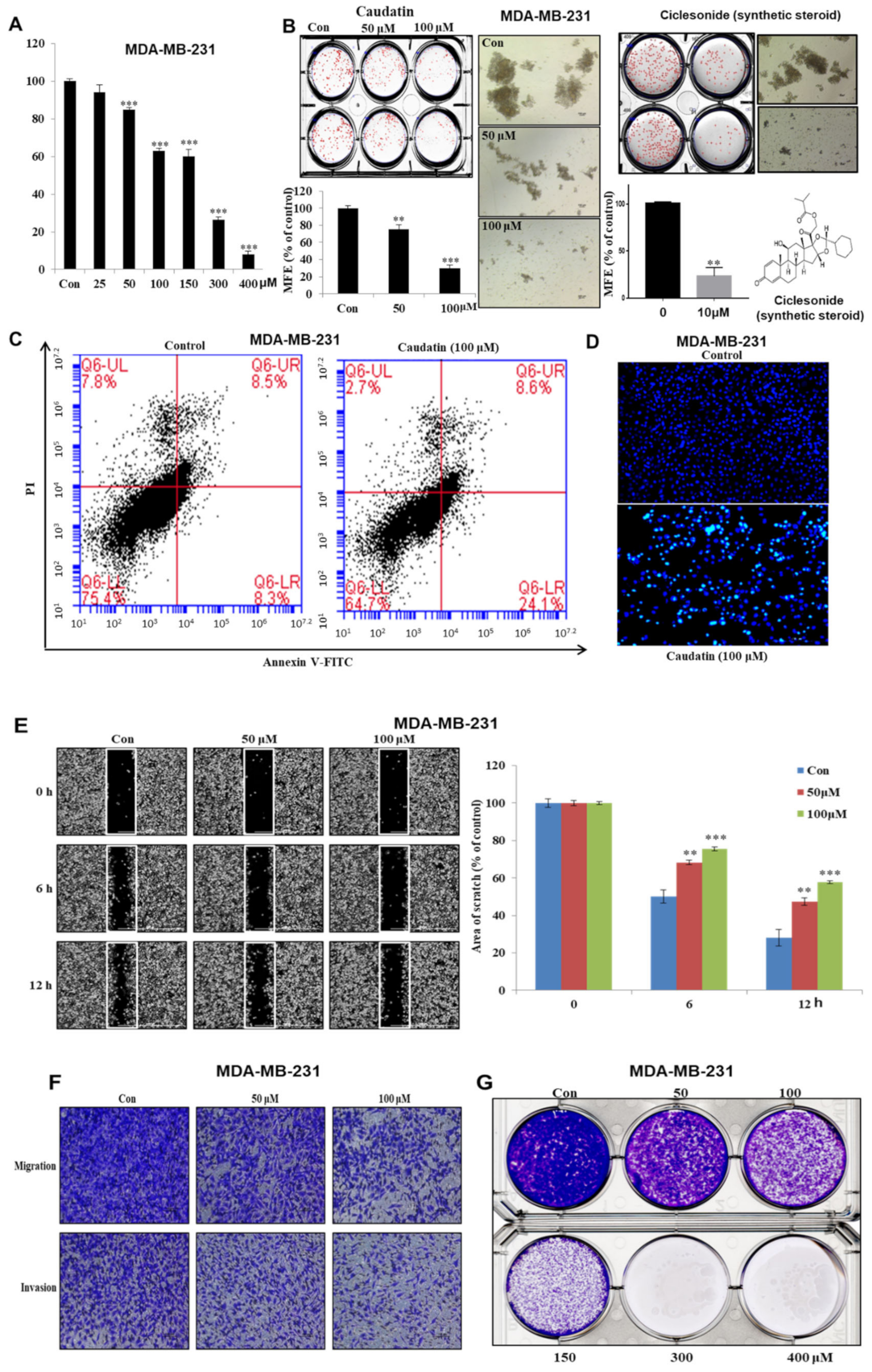
3.2. Caudatin Suppresses Cell Growth and Mammosphere Formation
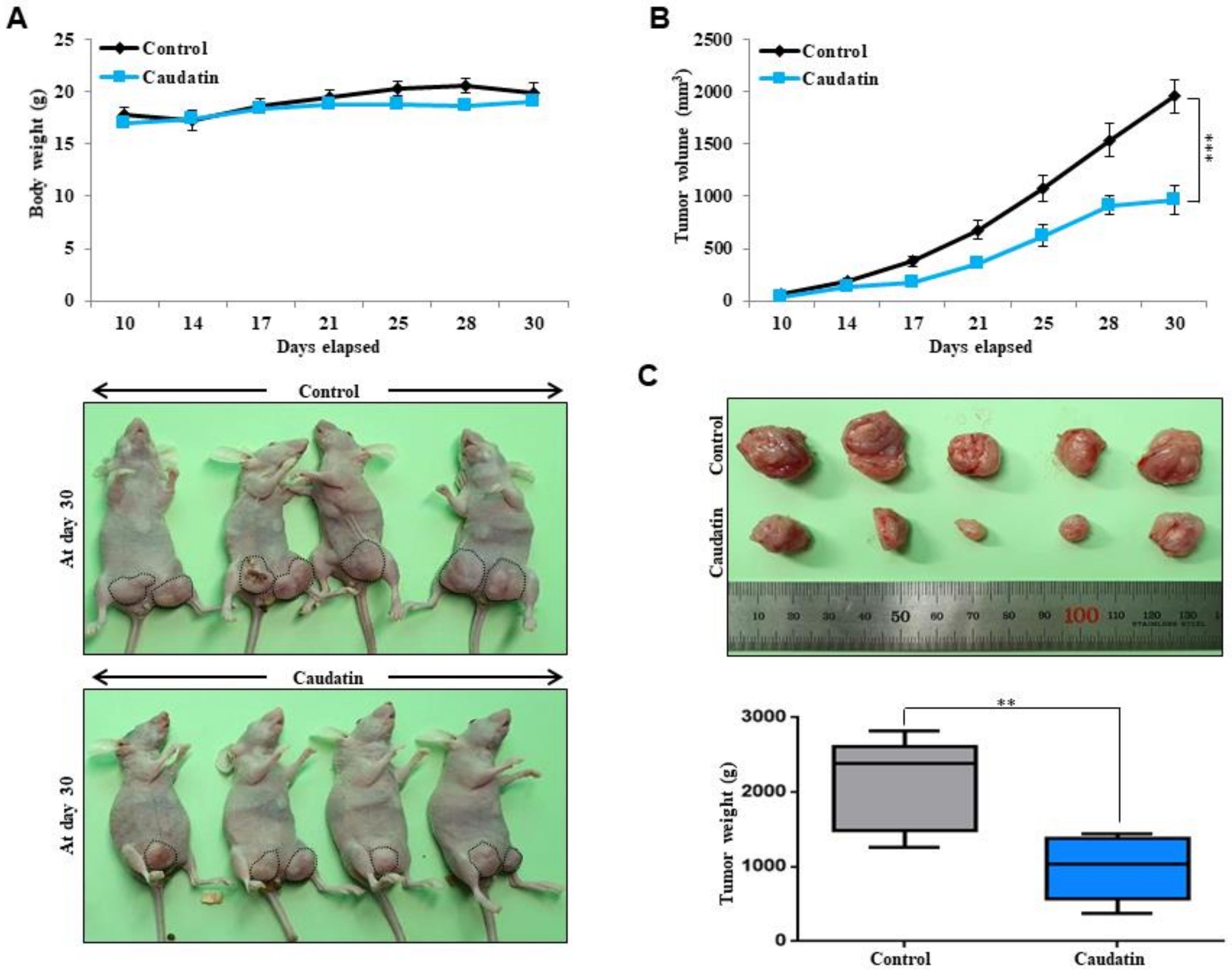
3.3. Caudatin Suppresses Xenograft Tumor Growth
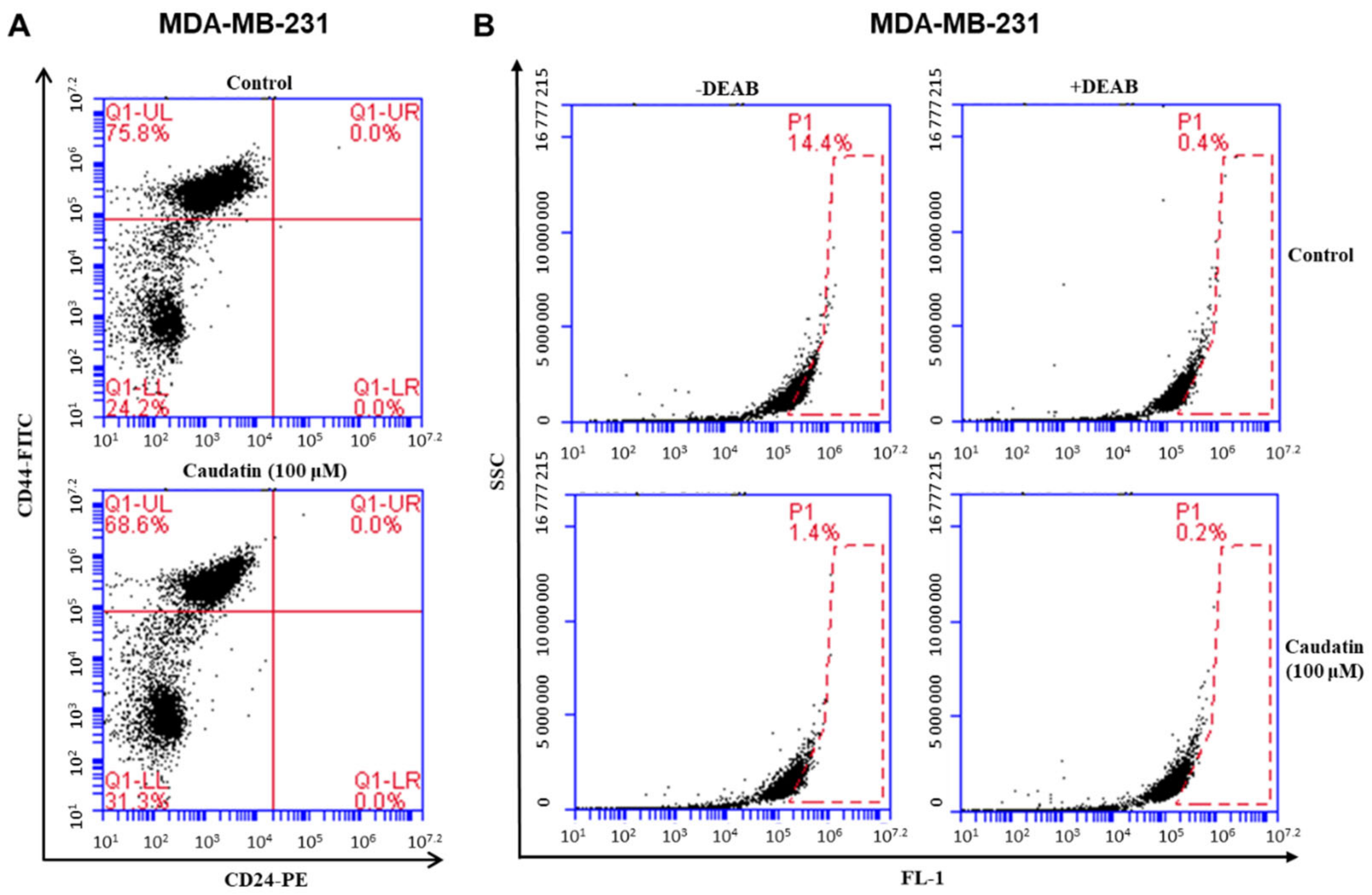
3.4. Caudatin Decreases the Populations of CD44+/CD24− and ALDH-expressing Cancer Cells
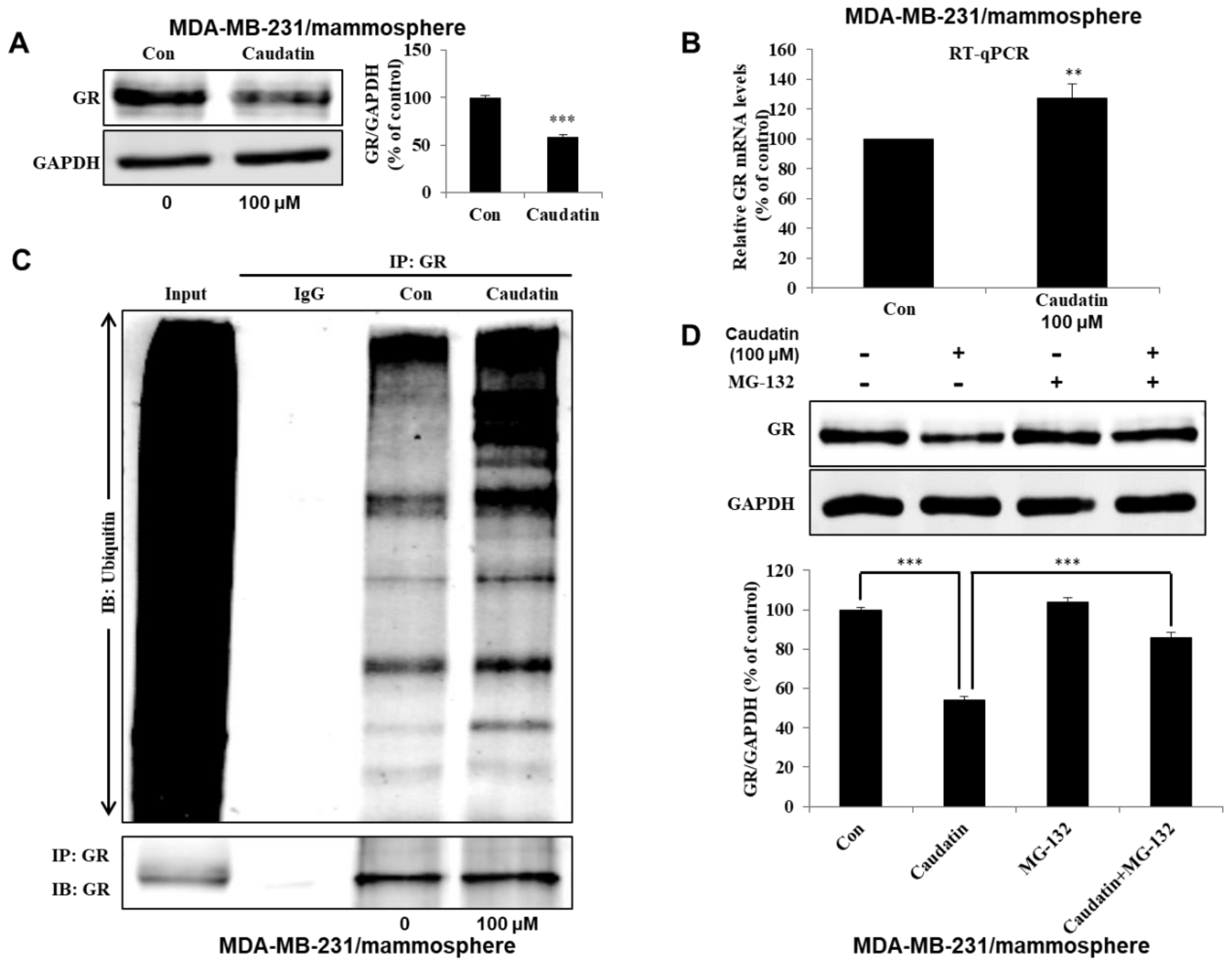
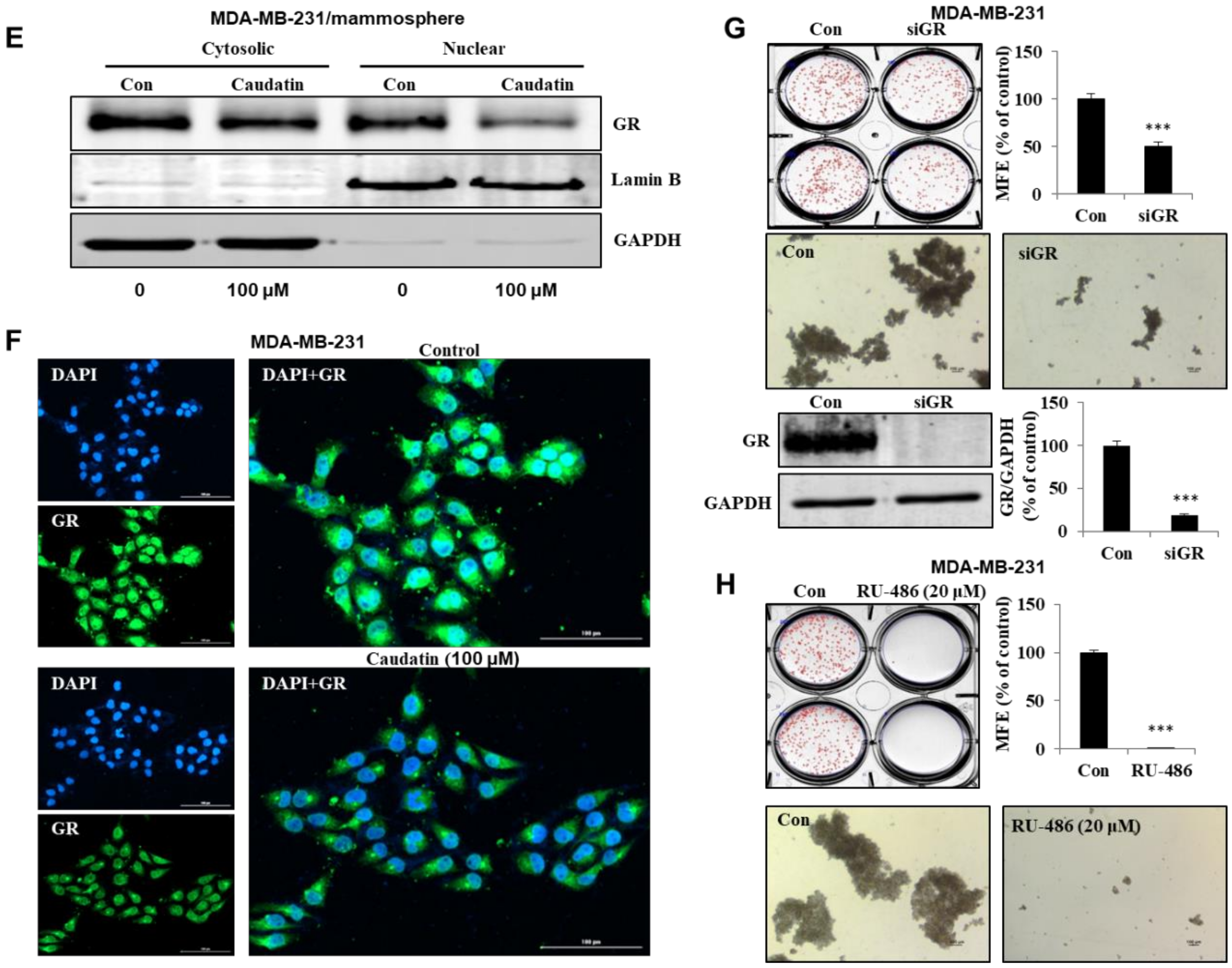
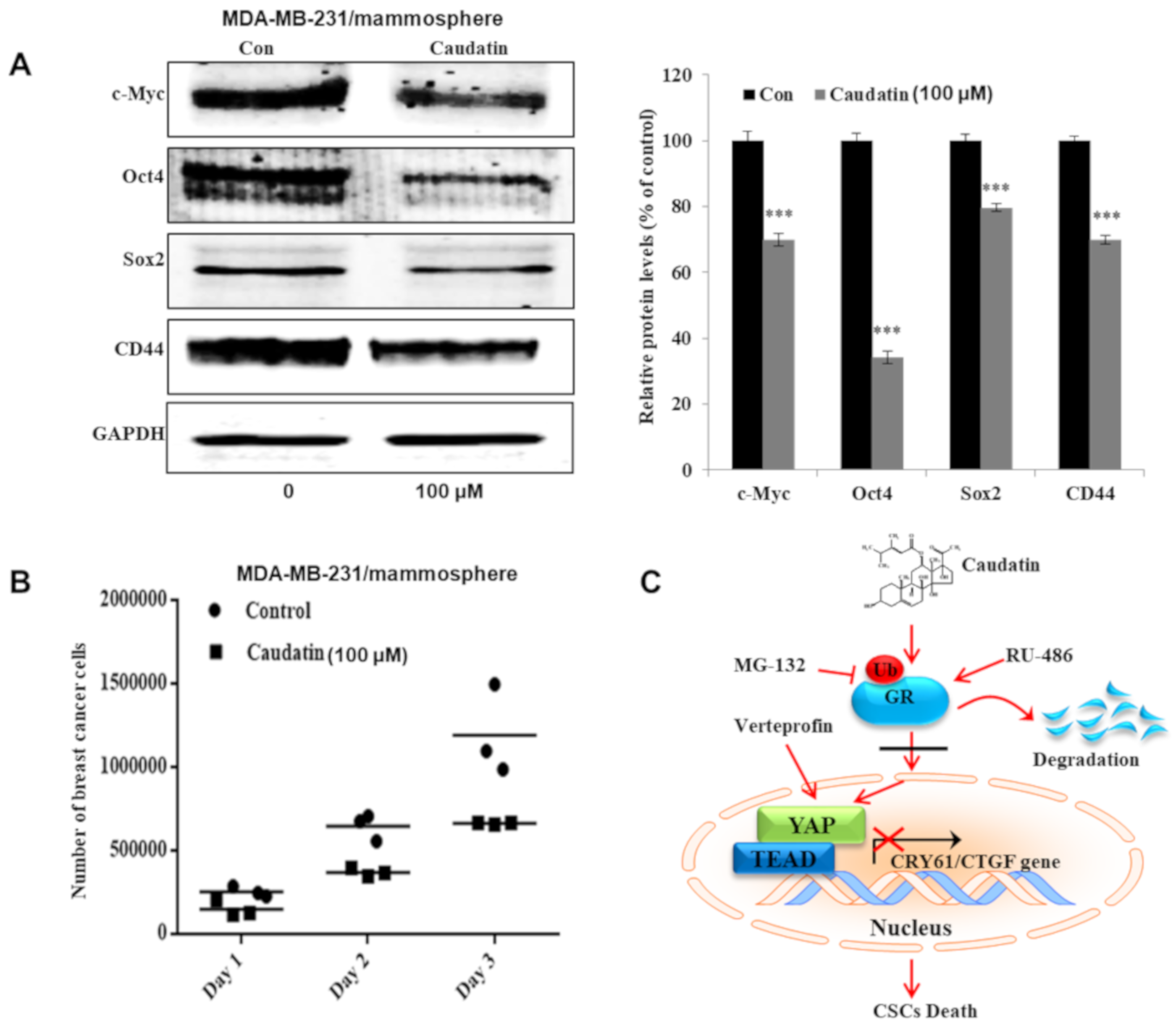
3.5. Caudatin Blocks the GR Signal through the Ubiquitin (Ub)-Dependent Degradation of GR in BCSCs
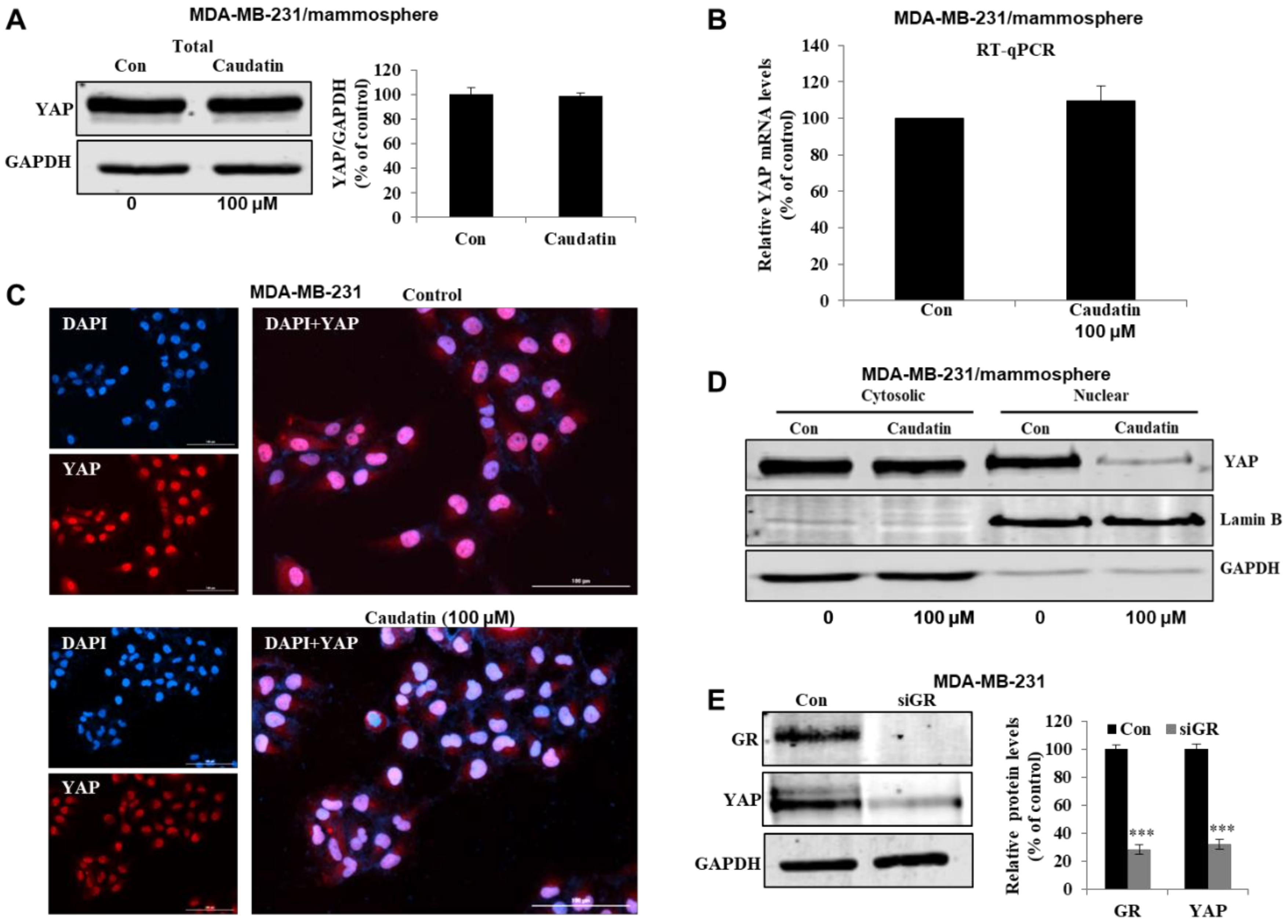
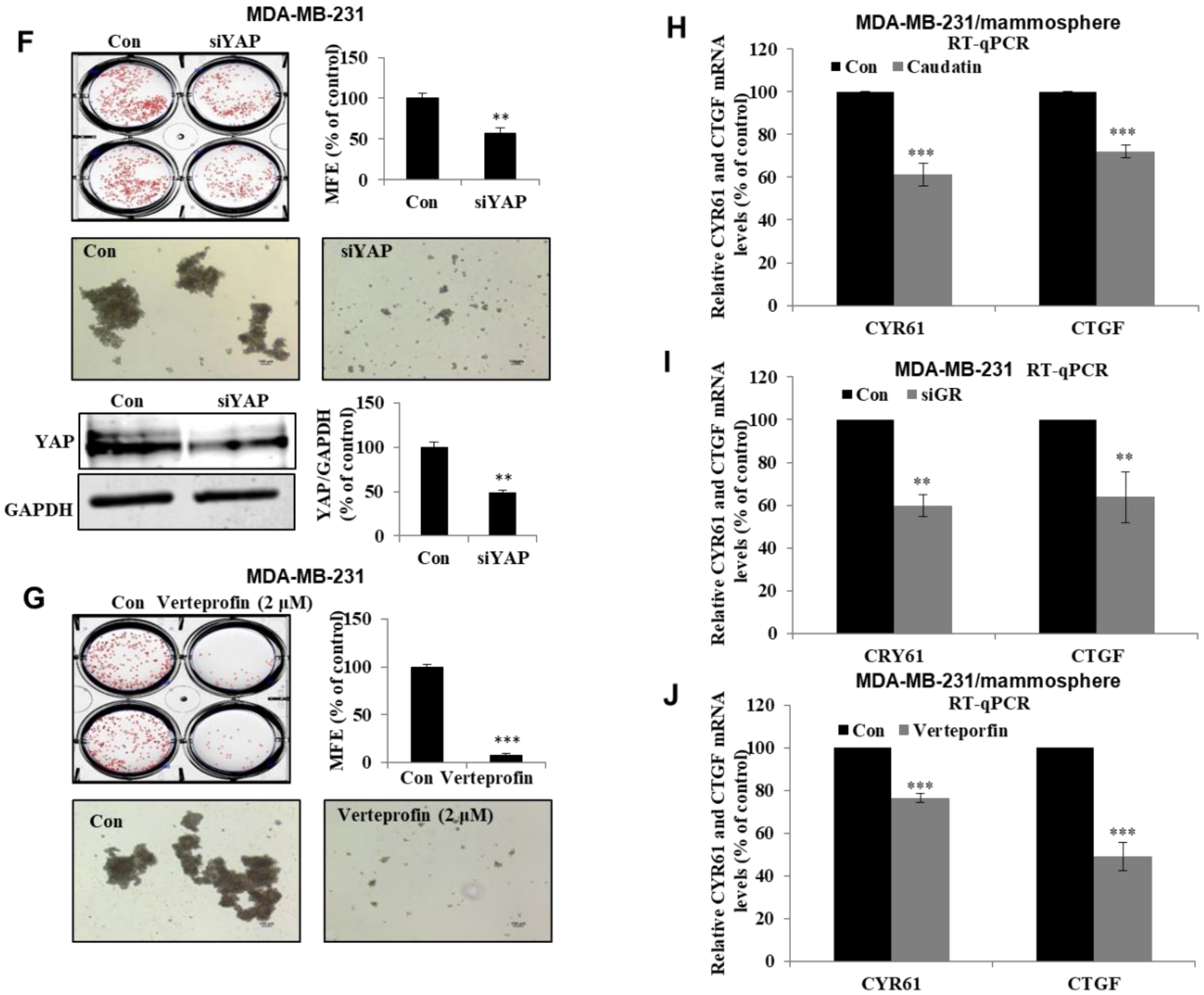
3.6. Caudatin Regulates the GR-YAP Signaling Pathway
3.7. Caudatin Regulates the Expression Levels of CSC Marker Proteins and Mammosphere Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chai, Z.; Huang, W.; Zhao, X.; Wu, H.; Zeng, X.; Li, C. Preparation, characterization, antioxidant activity and protective effect against cellular oxidative stress of polysaccharide from Cynanchum auriculatum Royle ex Wight. Int. J. Biol. Macromol. 2018, 119, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Choi, H.G.; Li, Y.; Park, Y.M.; Lee, J.H.; Kim, D.H.; Lee, J.H.; Son, J.K.; Na, M.; Lee, S.H. Chemical constituents of Cynanchum wilfordii and the chemotaxonomy of two species of the family Asclepiadacease, C. wilfordii and C. auriculatum. Arch. Pharm. Res. 2011, 34, 2021–2027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hong, L.Z.; Gu, M.F.; Wu, C.D.; Zhang, G. Transcriptome analyses revealed molecular responses of Cynanchum auriculatum leaves to saline stress. Sci. Rep. 2020, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Geng, C.A.; Ma, Y.B.; Huang, X.Y.; Luo, J.; Chen, H.; Guo, R.H.; Zhang, X.M.; Chen, J.J. Synthesis, structure-activity relationships and biological evaluation of caudatin derivatives as novel anti-hepatitis B virus agents. Bioorg. Med. Chem. 2012, 20, 2877–2888. [Google Scholar] [CrossRef]
- Shan, L.; Liu, R.H.; Shen, Y.H.; Zhang, W.D.; Zhang, C.; Wu, D.Z.; Min, L.; Su, J.; Xu, X.K. Gastroprotective effect of a traditional Chinese herbal drug “Baishouwu” on experimental gastric lesions in rats. J. Ethnopharmacol. 2006, 107, 389–394. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, Y.; Meng, X.; Wang, X.; Li, Z.; Qian, S.; Wei, Y.; Shu, L.; Ding, Y.; Wang, P.; et al. Total C-21 steroidal glycosides, isolated from the root tuber of Cynanchum auriculatum Royle ex Wight, attenuate hydrogen peroxide-induced oxidative injury and inflammation in L02 cells. Int. J. Mol. Med. 2018, 42, 3157–3170. [Google Scholar] [CrossRef]
- Ji, C.X.; Li, X.Y.; Jia, S.B.; Liu, L.L.; Ge, Y.C.; Yang, Q.X.; Zhang, J.J. The antidepressant effect of Cynanchum auriculatum in mice. Pharm. Biol. 2012, 50, 1067–1072. [Google Scholar] [CrossRef][Green Version]
- Ding, Y.F.; Peng, Z.X.; Ding, L.; Peng, Y.R. Baishouwu Extract Suppresses the Development of Hepatocellular Carcinoma via TLR4/MyD88/NF-kappaB Pathway. Front. Pharmacol. 2019, 10, 389. [Google Scholar] [CrossRef]
- Luo, Y.; Sun, Z.; Li, Y.; Liu, L.; Cai, X.; Li, Z. Caudatin inhibits human hepatoma cell growth and metastasis through modulation of the Wnt/beta-catenin pathway. Oncol. Rep. 2013, 30, 2923–2928. [Google Scholar] [CrossRef]
- Fu, X.Y.; Zhang, S.; Wang, K.; Yang, M.F.; Fan, C.D.; Sun, B.L. Caudatin Inhibits Human Glioma Cells Growth Through Triggering DNA Damage-Mediated Cell Cycle Arrest. Cell. Mol. Neurobiol. 2015, 35, 953–959. [Google Scholar] [CrossRef]
- Fei, H.R.; Yuan, C.; Wang, G.L.; Zhao, Y.; Li, Z.J.; Du, X.; Wang, F.Z. Caudatin potentiates the anti-tumor effects of TRAIL against human breast cancer by upregulating DR5. Phytomedicine 2019, 62, 152950. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.W.; Xie, S.; Hu, S.Y.; Liao, T.; Liu, P.; Peng, K.H.; Yang, X.Z.; He, Z.L.; Tang, H.Y.; Cui, Y.; et al. Caudatin targets TNFAIP1/NF-kappaB and cytochrome c/caspase signaling to suppress tumor progression in human uterine cancer. Int. J. Oncol. 2016, 49, 1638–1650. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Siegel, R.L.; Jemal, A.; Wender, R.C.; Gansler, T.; Ma, J.; Brawley, O.W. An assessment of progress in cancer control. CA Cancer J. Clin. 2018, 68, 329–339. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef] [PubMed]
- La Belle, A.; Khatib, J.; Schiemann, W.P.; Vinayak, S. Role of Platinum in Early-Stage Triple-Negative Breast Cancer. Curr. Treat. Options Oncol. 2017, 18, 68. [Google Scholar] [CrossRef]
- Day, C.M.; Hickey, S.M.; Song, Y.; Plush, S.E.; Garg, S. Novel Tamoxifen Nanoformulations for Improving Breast Cancer Treatment: Old Wine in New Bottles. Molecules 2020, 25, 1182. [Google Scholar] [CrossRef]
- Wu, H.J.; Chu, P.Y. Role of Cancer Stem Cells in Cholangiocarcinoma and Therapeutic Implications. Int. J. Mol. Sci. 2019, 20, 4154. [Google Scholar] [CrossRef]
- Sridharan, S.; Howard, C.M.; Tilley, A.M.C.; Subramaniyan, B.; Tiwari, A.K.; Ruch, R.J.; Raman, D. Novel and Alternative Targets Against Breast Cancer Stemness to Combat Chemoresistance. Front. Oncol. 2019, 9, 1003. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers (Basel) 2019, 11, 862. [Google Scholar] [CrossRef]
- Ogara, M.F.; Rodriguez-Segui, S.A.; Marini, M.; Nacht, A.S.; Stortz, M.; Levi, V.; Presman, D.M.; Vicent, G.P.; Pecci, A. The glucocorticoid receptor interferes with progesterone receptor-dependent genomic regulation in breast cancer cells. Nucleic Acids Res. 2019, 47, 10645–10661. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, M.M.S.; Hamelin, B.; Manevski, N.; Couto, J.P.; Sethi, A.; Coissieux, M.M.; Munst, S.; Okamoto, R.; Kohler, H.; Schmidt, A.; et al. Glucocorticoids promote breast cancer metastasis. Nature 2019, 567, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, J.S.; Baldassarre, G.; Thorat, M.A.; Massarut, S. Role of glucocorticoids in breast cancer. Curr. Pharm. Des. 2010, 16, 3593–3600. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Gonzalez, R.; Kirchner, B.; Mazzone, C.; Pfaffl, M.W.; Schelling, G.; Steinlein, O.; Reithmair, M. Glucocorticoid receptor overexpression slightly shifts microRNA expression patterns in triple-negative breast cancer. Int. J. Oncol. 2018, 52, 1765–1776. [Google Scholar] [CrossRef]
- Sorrentino, G.; Ruggeri, N.; Zannini, A.; Ingallina, E.; Bertolio, R.; Marotta, C.; Neri, C.; Cappuzzello, E.; Forcato, M.; Rosato, A.; et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat. Commun. 2017, 8, 14073. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yuan, L.; Sun, Y.; Wang, P.; Zhang, H.; Feng, X.; Wang, Z.; Zhang, W.; Yang, C.; Zeng, Y.A.; et al. Glucocorticoid Receptor Signaling Activates TEAD4 to Promote Breast Cancer Progression. Cancer Res. 2019, 79, 4399–4411. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, J.E.; Park, H.W. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol. Cells 2018, 41, 83–92. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef]
- Clarke, M.L.; Burton, R.L.; Hill, A.N.; Litorja, M.; Nahm, M.H.; Hwang, J. Low-cost, high-throughput, automated counting of bacterial colonies. Cytom. A 2010, 77, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Kim, D.A.; Chung, H.; Park, I.H.; Kim, B.H.; Oh, E.S.; Kang, D.H. Screening of breast cancer stem cell inhibitors using a protein kinase inhibitor library. Cancer Cell. Int. 2017, 17, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Sun, H.N.; Liu, R.; Choi, H.S.; Lee, D.S. Non-thermal Plasma-activated Medium Induces Apoptosis of Aspc1 Cells Through the ROS-dependent Autophagy Pathway. In Vivo 2020, 34, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Kim, J.H.; Kim, S.L.; Deng, H.Y.; Lee, D.; Kim, C.S.; Yun, B.S.; Lee, D.S. Catechol derived from aronia juice through lactic acid bacteria fermentation inhibits breast cancer stem cell formation via modulation Stat3/IL-6 signaling pathway. Mol. Carcinog. 2018, 57, 1467–1479. [Google Scholar] [CrossRef]
- Choi, H.S.; Kim, S.L.; Kim, J.H.; Deng, H.Y.; Yun, B.S.; Lee, D.S. Triterpene Acid (3-O-p-Coumaroyltormentic Acid) Isolated from Aronia Extracts Inhibits Breast Cancer Stem Cell Formation through Downregulation of c-Myc Protein. Int. J. Mol. Sci. 2018, 19, 2528. [Google Scholar] [CrossRef]
- Zhen, X.; Choi, H.S.; Kim, J.H.; Kim, S.L.; Liu, R.; Yun, B.S.; Lee, D.S. Machilin D, a Lignin Derived from Saururus chinensis, Suppresses Breast Cancer Stem Cells and Inhibits NF-kappaB Signaling. Biomolecules 2020, 10, 245. [Google Scholar] [CrossRef]
- Bai, J.; Chen, W.B.; Zhang, X.Y.; Kang, X.N.; Jin, L.J.; Zhang, H.; Wang, Z.Y. HIF-2alpha regulates CD44 to promote cancer stem cell activation in triple-negative breast cancer via PI3K/AKT/mTOR signaling. World J. Stem Cells 2020, 12, 87–99. [Google Scholar] [CrossRef]
- Mao, Y.; Zhu, L.; Huang, Z.; Luo, C.; Zhou, T.; Li, L.; Wang, G.; Yang, Z.; Qi, W.; Yang, X.; et al. Stem-like tumor cells involved in heterogeneous vasculogenesis in breast cancer. Endocr. Relat. Cancer 2020, 27, 23–39. [Google Scholar] [CrossRef]
- Liu, H.; Ma, L.; Lin, J.; Cao, B.; Qu, D.; Luo, C.; Huang, W.; Han, L.; Xu, H.; Wu, Z.; et al. Advances in molecular mechanisms of drugs affecting abnormal glycosylation and metastasis of breast cancer. Pharmacol. Res. 2020, 10, 104738. [Google Scholar] [CrossRef]
- Albeshan, S.M.; Hossain, S.Z.; Mackey, M.G.; Brennan, P.C. Can Breast Self-examination and Clinical Breast Examination Along with Increasing Breast Awareness Facilitate Earlier Detection of Breast Cancer in Populations with Advanced Stages at Diagnosis? Clin. Breast Cancer 2020. [Google Scholar] [CrossRef]
- Hercules, S.M.; Hercules, J.C.; Ansari, A.; Date, S.A.J.; Skeete, D.H.A.; Smith Connell, S.P.; Pond, G.R.; Daniel, J.M. High triple-negative breast cancer prevalence and aggressive prognostic factors in Barbadian women with breast cancer. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell. Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Kim, J.H.; Kim, S.L.; Lee, D.S. Disruption of the NF-kappaB/IL-8 Signaling Axis by Sulconazole Inhibits Human Breast Cancer Stem Cell Formation. Cells 2019, 8, 1007. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Mukherjee, S.; Bhattacharya, A.; Basak, U.; Chakraborty, S.; Paul, S.; Khan, P.; Jana, K.; Hazra, T.K.; Das, T. Pyridoxine enhances chemo-responsiveness of breast cancer stem cells via redox reconditioning. Free Radic. Biol. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.F.; Wang, Y.Q.; Yang, B.; Zhang, R.S. Cytotoxic and apoptosis-inducing properties of a C21-steroidal glycoside isolated from the roots of Cynanchum auriculatum. Oncol. Lett. 2013, 5, 1407–1411. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Zhang, S.J.; Lu, H.; Yang, B.; Ye, L.F.; Zhang, R.S. A C 21 -Steroidal Glycoside Isolated from the Roots of Cynanchum auriculatum Induces Cell Cycle Arrest and Apoptosis in Human Gastric Cancer SGC-7901 Cells. Evid. Based. Complement. Alternat. Med. 2013, 2013, 180839. [Google Scholar] [CrossRef][Green Version]
- Zhu, L.Z.; Hou, Y.J.; Zhao, M.; Yang, M.F.; Fu, X.T.; Sun, J.Y.; Fu, X.Y.; Shao, L.R.; Zhang, H.F.; Fan, C.D.; et al. Caudatin induces caspase-dependent apoptosis in human glioma cells with involvement of mitochondrial dysfunction and reactive oxygen species generation. Cell Biol. Toxicol. 2016, 32, 333–345. [Google Scholar] [CrossRef]
- Quaglino, E.; Conti, L.; Cavallo, F. Breast cancer stem cell antigens as targets for immunotherapy. Semin. Immunol. 2020, 101386. [Google Scholar] [CrossRef]
- Sulaiman, A.; McGarry, S.; Han, X.; Liu, S.; Wang, L. CSCs in Breast Cancer-One Size Does Not Fit All: Therapeutic Advances in Targeting Heterogeneous Epithelial and Mesenchymal CSCs. Cancers (Basel) 2019, 11, 1128. [Google Scholar] [CrossRef]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 13856. [Google Scholar] [CrossRef]
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011, 10, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Vegiopoulos, A.; Herzig, S. Glucocorticoids, metabolism and metabolic diseases. Mol. Cell. Endocrinol. 2007, 275, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Topete, D.; Cidlowski, J.A. One hormone, two actions: Anti- and pro-inflammatory effects of glucocorticoids. Neuroimmunomodulation 2015, 22, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Nussinovitch, U.; de Carvalho, J.F.; Pereira, R.M.; Shoenfeld, Y. Glucocorticoids and the cardiovascular system: State of the art. Curr. Pharm. Des. 2010, 16, 3574–3585. [Google Scholar] [CrossRef] [PubMed]
- Lucafo, M.; Franzin, M.; Decorti, G.; Stocco, G. A patent review of anticancer glucocorticoid receptor modulators (2014-present). Expert. Opin. Ther. Pat. 2020. [Google Scholar] [CrossRef]
- Kumar, R. Emerging role of glucocorticoid receptor in castration resistant prostate cancer: A potential therapeutic target. J. Cancer. 2020, 11, 696–701. [Google Scholar] [CrossRef]
- Pan, D.; Kocherginsky, M.; Conzen, S.D. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011, 71, 6360–6370. [Google Scholar] [CrossRef]
- Sundahl, N.; Clarisse, D.; Bracke, M.; Offner, F.; Berghe, W.V.; Beck, I.M. Selective glucocorticoid receptor-activating adjuvant therapy in cancer treatments. Oncoscience 2016, 3, 188–202. [Google Scholar] [CrossRef]
- Shome, D.; von Woedtke, T.; Riedel, K.; Masur, K. The HIPPO Transducer YAP and Its Targets CTGF and Cyr61 Drive a Paracrine Signalling in Cold Atmospheric Plasma-Mediated Wound Healing. Oxid. Med. Cell. Longev. 2020, 2020, 4910280. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, X.; Feng, W.; Yu, Y.; Jeong, K.; Guo, W.; Lu, Y.; Mills, G.B. Verteporfin inhibits YAP function through up-regulating 14-3-3sigma sequestering YAP in the cytoplasm. Am. J. Cancer Res. 2016, 6, 27–37. [Google Scholar]
| Genes | Primers |
|---|---|
| GR | Forward: 5′-GAAGGAAACTCCAGCCAGAA-3′ Reverse: 5′-CAGCTAACATCTCGGGGAAT-3′ |
| YAP | Forward: 5′- GAACCCCAGATGACTTCCTG-3′ Reverse: 5′-CTCCTTCCAGTGTTCCAAGG-3′ |
| CTGF | Forward: 5′-CCAATGACAACGCCTCCTG-3′ Reverse: 5′-TGGTGCAGCCAGAAAGCTC-3′ |
| CYR61 | Forward: 5′-AGCCTCGCATCCTATACAACC-3′ Reverse: 5′-TTCTTTCACAAGGCGGCACTC-3′ |
| β-actin | Forward: 5′-TGTTACCAACTGGGACGACA-3′ Reverse: 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhen, X.; Choi, H.S.; Kim, J.-H.; Kim, S.-L.; Liu, R.; Ko, Y.-C.; Yun, B.-S.; Lee, D.-S. Caudatin Isolated from Cynanchum auriculatum Inhibits Breast Cancer Stem Cell Formation via a GR/YAP Signaling. Biomolecules 2020, 10, 925. https://doi.org/10.3390/biom10060925
Zhen X, Choi HS, Kim J-H, Kim S-L, Liu R, Ko Y-C, Yun B-S, Lee D-S. Caudatin Isolated from Cynanchum auriculatum Inhibits Breast Cancer Stem Cell Formation via a GR/YAP Signaling. Biomolecules. 2020; 10(6):925. https://doi.org/10.3390/biom10060925
Chicago/Turabian StyleZhen, Xing, Hack Sun Choi, Ji-Hyang Kim, Su-Lim Kim, Ren Liu, Yu-Chan Ko, Bong-Sik Yun, and Dong-Sun Lee. 2020. "Caudatin Isolated from Cynanchum auriculatum Inhibits Breast Cancer Stem Cell Formation via a GR/YAP Signaling" Biomolecules 10, no. 6: 925. https://doi.org/10.3390/biom10060925
APA StyleZhen, X., Choi, H. S., Kim, J.-H., Kim, S.-L., Liu, R., Ko, Y.-C., Yun, B.-S., & Lee, D.-S. (2020). Caudatin Isolated from Cynanchum auriculatum Inhibits Breast Cancer Stem Cell Formation via a GR/YAP Signaling. Biomolecules, 10(6), 925. https://doi.org/10.3390/biom10060925





