Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability
Abstract
1. Introduction—Modes of Mitochondrial Protein Quality Control
2. Mechanisms of Mitochondrial Precursor Protein Ubiquitination and Degradation in Cytosol
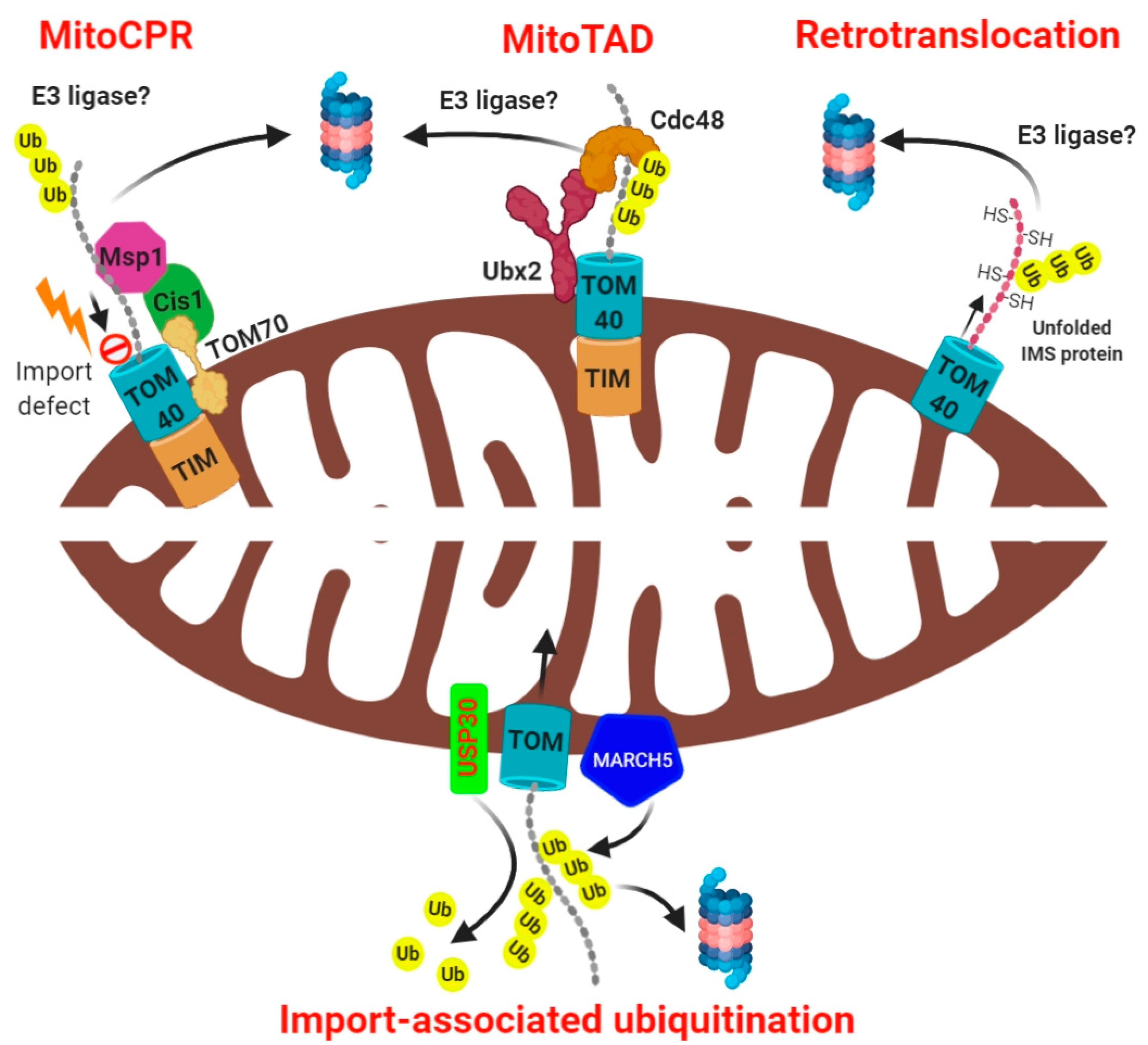
3. Mechanisms of Intramitochondrial Protein Ubiquitination at Mitochondria
4. Mitochondrial Import-Associated Ubiquitination
5. Mechanisms of Mitochondrial Export-Associated Ubiquitination
6. Implications of Intramitochondrial Protein Ubiquitination: Metabolic Regulation by UPS
7. Turnover of Respiratory Chain Subunits by UPS
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Burchell, V.S.; Gandhi, S.; Deas, E.; Wood, N.W.; Abramov, A.Y.; Plun-Favreau, H. Targeting mitochondrial dysfunction in neurodegenerative disease: Part I. Expert Opin. Ther. Targets 2010, 14, 369–385. [Google Scholar] [CrossRef]
- Yan, M.H.; Wang, X.; Zhu, X. Mitochondrial defects and oxidative stress in Alzheimer disease and Parkinson disease. Free Radic. Biol. Med. 2013, 62, 90–101. [Google Scholar] [CrossRef]
- Conley, K.E.; Marcinek, D.J.; Villarin, J. Mitochondrial dysfunction and age. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 688–692. [Google Scholar] [CrossRef]
- Sun, N.; Youle, R.J.; Finkel, T. The Mitochondrial Basis of Aging. Mol. Cell. 2016, 61, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Shkurat, T.P.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. The role of mitochondrial dysfunction in cardiovascular disease: A brief review. Ann. Med. 2018, 50, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.L.; Chourasia, A.H.; Macleod, K.F. Mitochondrial dysfunction in cancer. Front. Oncol. 2013, 3, 292. [Google Scholar] [CrossRef]
- Kudryavtseva, A.V.; Krasnov, G.S.; Dmitriev, A.A.; Alekseev, B.Y.; Kardymon, O.L.; Sadritdinova, A.F.; Fedorova, M.S.; Pokrovsky, A.V.; Melnikova, N.V.; Kaprin, A.D.; et al. Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget 2016, 7, 44879–44905. [Google Scholar] [CrossRef]
- Bulteau, A.L.; Szweda, L.I.; Friguet, B. Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp. Gerontol. 2006, 41, 653–657. [Google Scholar] [CrossRef]
- Cadenas, E.; Davies, K.J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Harbauer, A.B.; Zahedi, R.P.; Sickmann, A.; Pfanner, N.; Meisinger, C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metabolism 2014, 19, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef] [PubMed]
- von Heijne, G. Why mitochondria need a genome. FEBS Lett. 1986, 198, 1–4. [Google Scholar] [CrossRef]
- Taylor, E.B.; Rutter, J. Mitochondrial quality control by the ubiquitin–proteasome system. Biochem. Soc. Trans. 2011, 39, 1509–1513. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Tatsuta, T.; Langer, T. Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 2011, 3, a007559. [Google Scholar] [CrossRef]
- Rehling, P.; Pfanner, N.; Meisinger, C. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J. Mol. Biol. 2003, 326, 639–657. [Google Scholar] [CrossRef]
- Young, J.C.; Hoogenraad, N.J.; Hartl, F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003, 112, 41–50. [Google Scholar] [CrossRef]
- Dudek, J.; Rehling, P.; van der Laan, M. Mitochondrial protein import: Common principles and physiological networks. Biochim. Biophys. Acta 2013, 1833, 274–285. [Google Scholar] [CrossRef]
- Neupert, W.; Pfanner, N. Roles of molecular chaperones in protein targeting to mitochondria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1993, 339, 355–361. [Google Scholar]
- Voos, W.; Pollecker, K. The Mitochondrial Lon Protease: Novel Functions off the Beaten Track? Biomolecules 2020, 10, 253. [Google Scholar] [CrossRef]
- Jackson, M.W.; Silva-Herzog, E.; Plano, G.V. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 2004, 54, 1364–1378. [Google Scholar] [CrossRef]
- Koppen, M.; Langer, T. Protein degradation within mitochondria: Versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol. Biol. 2007, 42, 221–242. [Google Scholar] [CrossRef]
- Moisoi, N.; Klupsch, K.; Fedele, V.; East, P.; Sharma, S.; Renton, A.; Plun-Favreau, H.; Edwards, R.E.; Teismann, P.; Esposti, M.D.; et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009, 16, 449–464. [Google Scholar] [CrossRef]
- Stiburek, L.; Cesnekova, J.; Kostkova, O.; Fornuskova, D.; Vinsova, K.; Wenchich, L.; Houstek, J.; Zeman, J. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 2012, 23, 1010–1023. [Google Scholar] [CrossRef]
- Pryde, K.R.; Taanman, J.W.; Schapira, A.H. A LON-ClpP proteolytic axis degrades complex I to extinguish ROS production in depolarized mitochondria. Cell Rep. 2016, 17, 2522–2531. [Google Scholar] [CrossRef]
- Bota, D.A.; Davies, K.J. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 2002, 4, 674–680. [Google Scholar] [CrossRef]
- König, T.; Tröder, S.E.; Bakka, K.; Korwitz, A.; Richter-Dennerlein, R.; Lampe, P.A.; Patron, M.; Mühlmeister, M.; Guerrero-Castillo, S.; Brandt, U.; et al. The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol. Cell 2016, 64, 148–162. [Google Scholar]
- Potting, C.; Wilmes, C.; Engmann, T.; Osman, C.; Langer, T. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 2010, 29, 2888–2898. [Google Scholar] [CrossRef]
- MacVicar, T.; Ohba, Y.; Nolte, H.; Mayer, F.C.; Tatsuta, T.; Sprenger, H.-G.; Lindner, B.; Zhao, Y.; Li, J.; Bruns, C.; et al. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 2019, 575, 361–365. [Google Scholar] [CrossRef]
- Deshwal, S.; Fiedler, K.U.; Langer, T. Mitochondrial proteases: Multifaceted regulators of mitochondrial plasticity. Annu. Rev. Biochem. 2020, 89, 501–528. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.; Kevei, E.; Hoppe, T. Double-edged alliance: Mitochondrial surveillance by the UPS and autophagy. Curr. Opin. Cell Biol. 2015, 37, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.M.; Rutter, J. Ubiquitin-dependent mitochondrial protein degradation. Int. J. Biochem. Cell Biol. 2011, 43, 1422–1426. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Ordureau, A.; Heo, J.M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 93–108. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Bence, N.F.; Sampat, R.M.; Kopito, R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001, 292, 1552–1555. [Google Scholar] [CrossRef]
- Leboucher, G.P.; Tsai, Y.C.; Yang, M.; Shaw, K.C.; Zhou, M.; Veenstra, T.D.; Glickman, M.H.; Weissman, A.M. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell 2012, 47, 547–557. [Google Scholar] [CrossRef]
- Livnat-Levanon, N.; Glickman, M.H. Ubiquitin–proteasome system and mitochondria—reciprocity. Biochim. Biophys. Acta 2011, 1809, 80–87. [Google Scholar] [CrossRef]
- Karbowski, M.; Youle, R.J. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 2011, 23, 476–482. [Google Scholar] [CrossRef]
- Bramasole, L.; Sinha, A.; Gurevich, S.; Radzinski, M.; Klein, Y.; Panat, N.; Gefen, E.; Rinaldi, T.; Jimenez-Morales, D.; Johnson, J.; et al. Proteasome lid bridges mitochondrial stress with Cdc53/Cullin1 NEDDylation status. Redox Biol. 2019, 20, 533–543. [Google Scholar] [CrossRef]
- Cohen, M.M.; Leboucher, G.P.; Livnat-Levanon, N.; Glickman, M.H.; Weissman, A.M. Ubiquitin–proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 2008, 19, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.-M.; Livnat-Levanon, N.; Taylor, E.B.; Jones, K.T.; Dephoure, N.; Ring, J.; Xie, J.; Brodsky, J.L.; Madeo, F.; Gygi, S.P.; et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell. 2010, 40, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Hoppins, S.; Lackner, L.; Nunnari, J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007, 76, 751–780. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.S.; Osellame, L.D.; Laine, D.; Koutsopoulos, O.S.; Frazier, A.E.; Ryan, M.T. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011, 12, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Henriques, M.; Altin, S.; Brave, F. Interplay between the ubiquitin proteasome system and mitochondria for protein homeostasis. Curr Issues Mol. Biol. 2019, 35, 35–58. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.-M.; Williams, J.A.; Ding, W.-X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015, 4, 6–13. [Google Scholar] [CrossRef]
- Pickles, S.; Vigie, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef]
- Bingol, B.; Sheng, M. Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond. Free Radic. Biol. Med. 2016, 100, 210–222. [Google Scholar] [CrossRef]
- Sugiura, A.; McLelland, G.L.; Fon, E.A.; McBride, H.M. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014, 33, 2142–2156. [Google Scholar] [CrossRef]
- Soubannier, V.; McLelland, G.L.; Zunino, R.; Braschi, E.; Rippstein, P.; Fon, E.A.; McBride, H.M. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 2012, 22, 135–141. [Google Scholar] [CrossRef]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef]
- Neupert, W.; Herrmann, J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007, 76, 723–749. [Google Scholar] [CrossRef]
- Azzu, V.; Brand, M.D. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2010, 123, 578–585. [Google Scholar] [CrossRef]
- Wrobel, L.; Topf, U.; Bragoszewski, P.; Wiese, S.; Sztolsztener, M.E.; Oeljeklaus, S.; Varabyova, A.; Lirski, M.; Chroscicki, P.; Mroczek, S.; et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 2015, 524, 485–488. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484. [Google Scholar] [CrossRef]
- Sulkshane, P.; Duek, I.; Ram, J.; Thakur, A.; Reis, N.; Ziv, T.; Glickman, M.H. Inhibition of proteasome reveals basal mitochondrial ubiquitination. J. Proteom. 2020, 229, 103949. [Google Scholar] [CrossRef]
- Stojanovski, D.; Bragoszewski, P.; Chacinska, A. The MIA pathway: A tight bond between protein transport and oxidative folding in mitochondria. Biochim. Biophys. Acta 2012, 1823, 1142–1150. [Google Scholar] [CrossRef]
- Bragoszewski, P.; Gornicka, A.; Sztolsztener, M.E.; Chacinska, A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol. Cell Biol. 2013, 33, 2136–2148. [Google Scholar] [CrossRef]
- Bragoszewski, P.; Wasilewski, M.; Sakowska, P.; Gornicka, A.; Bottinger, L.; Qiu, J.; Wiedemann, N.; Chacinska, A. Retro-translocation of mitochondrial intermembrane space proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 7713–7718. [Google Scholar] [CrossRef]
- Finger, Y.; Habich, M.; Gerlich, S.; Urbanczyk, S.; van de Logt, E.; Koch, J.; Schu, L.; Lapacz, K.J.; Ali, M.; Petrungaro, C.; et al. Proteasomal degradation induced by DPP9-mediated processing competes with mitochondrial protein import. EMBO J. 2020, 39, e103889. [Google Scholar] [CrossRef]
- Itakura, E.; Zavodszky, E.; Shao, S.; Wohlever, M.L.; Keenan, R.J.; Hegde, R.S. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell 2016, 63, 21–33. [Google Scholar] [CrossRef]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef]
- Margineantu, D.H.; Emerson, C.B.; Diaz, D.; Hockenbery, D.M. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS ONE 2007, 2, e1066. [Google Scholar] [CrossRef]
- Lehmann, G.; Ziv, T.; Braten, O.; Admon, A.; Udasin, R.G.; Ciechanover, A. Ubiquitination of specific mitochondrial matrix proteins. Biochem. Biophys. Res. Commun. 2016, 475, 13–18. [Google Scholar] [CrossRef]
- Baker, M.J.; Frazier, A.E.; Gulbis, J.M.; Ryan, M.T. Mitochondrial protein-import machinery: Correlating structure with function. Trends Cell Biol. 2007, 17, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Pfanner, N.; Meisinger, C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 655–667. [Google Scholar] [CrossRef]
- Martensson, C.U.; Priesnitz, C.; Song, J.; Ellenrieder, L.; Doan, K.N.; Boos, F.; Floerchinger, A.; Zufall, N.; Oeljeklaus, S.; Warscheid, B.; et al. Mitochondrial protein translocation-associated degradation. Nature 2019, 569, 679–683. [Google Scholar] [CrossRef]
- Weidberg, H.; Amon, A. MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science 2018, 360, eaan4146. [Google Scholar] [CrossRef]
- Phu, L.; Rose, C.M.; Tea, J.S.; Wall, C.E.; Verschueren, E.; Cheung, T.K.; Kirkpatrick, D.S.; Bingol, B. Dynamic Regulation of Mitochondrial Import by the Ubiquitin System. Mol. Cell 2020, 77, 1107–1123.e10. [Google Scholar] [CrossRef]
- Ordureau, A.; Paulo, J.A.; Zhang, J.; An, H.; Swatek, K.N.; Cannon, J.R.; Wan, Q.; Komander, D.; Harper, J.W. Global landscape and dynamics of parkin and USP30-dependent ubiquitylomes in ineurons during mitophagic signaling. Mol. Cell 2020, 77, 1124–1142.e10. [Google Scholar] [CrossRef]
- Kowalski, L.; Bragoszewski, P.; Khmelinskii, A.; Glow, E.; Knop, M.; Chacinska, A. Determinants of the cytosolic turnover of mitochondrial intermembrane space proteins. BMC Biol. 2018, 16, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Bragoszewski, P.; Turek, M.; Chacinska, A. Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system. Open Biol. 2017, 7, 170007. [Google Scholar] [CrossRef] [PubMed]
- Munn, E.A. The Structure of Mitochondria; Academic Press: London, UK, 2014. [Google Scholar]
- Mannella, C.A. Structural diversity of mitochondria: Functional implications. Ann. N. Y. Acad. Sci. 2008, 1147, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Langer, T. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef]
- Pan, Y.; Cao, M.; Liu, J.; Yang, Q.; Miao, X.; Go, V.L.W.; Lee, P.W.N.; Xiao, G.G. Metabolic regulation in mitochondria and drug resistance. Adv. Exp. Med. Biol. 2017, 1038, 149–171. [Google Scholar]
- Nadege, B.; Patrick, L.; Rodrigue, R. Mitochondria: From bioenergetics to the metabolic regulation of carcinogenesis. Front. Biosci. 2009, 14, 4015–4034. [Google Scholar] [CrossRef]
- Okamoto, K.; Shaw, J.M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005, 39, 503–536. [Google Scholar] [CrossRef]
- Zungu, M.; Schisler, J.C.; Essop, M.F.; McCudden, C.; Patterson, C.; Willis, M.S. Regulation of AMPK by the ubiquitin proteasome system. Am. J. Pathol. 2011, 178, 4–11. [Google Scholar] [CrossRef]
- Ronnebaum, S.M.; Patterson, C.; Schisler, J.C. Minireview: Hey U(PS): Metabolic and proteolytic homeostasis linked via AMPK and the ubiquitin proteasome system. Mol. Endocrinol. 2014, 28, 1602–1615. [Google Scholar] [CrossRef]
- Sun, R.C.; Denko, N.C. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014, 19, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Yang, L.; Choi, H.H.; Baddour, J.; Achreja, A.; Liu, Y.; Li, Y.; Li, J.; Wan, G.; Huang, C.; et al. Amplification of USP13 drives ovarian cancer metabolism. Nat. Commun. 2016, 7, 13525. [Google Scholar] [CrossRef] [PubMed]
- Lavie, J.; De Belvalet, H.; Sonon, S.; Ion, A.M.; Dumon, E.; Melser, S.; Lacombe, D.; Dupuy, J.W.; Lalou, C.; Bénard, G. Ubiquitin-Dependent Degradation of Mitochondrial Proteins Regulates Energy Metabolism. Cell Rep. 2018, 23, 2852–2863. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Zhang, V.; El-Hattab, A.; Ficicioglu, C.; Shinawi, M.; Lines, M.; Schulze, A.; McNutt, M.; Gotway, G.; Tian, X.; et al. FBXL4 defects are common in patients with congenital lactic acidemia and encephalomyopathic mitochondrial DNA depletion syndrome. Clin Genet. 2017, 91, 634–639. [Google Scholar] [CrossRef]
- Gai, X.; Ghezzi, D.; Johnson, M.A.; Biagosch, C.A.; Shamseldin, H.E.; Haack, T.B.; Reyes, A.; Tsukikawa, M.; Sheldon, C.A.; Srinivasan, S.; et al. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 2013, 93, 482–495. [Google Scholar] [CrossRef]
- Alsina, D.; Lytovchenko, O.; Schab, A.; Atanassov, I.; Schober, F.A.; Jiang, M.; Koolmeister, C.; Wedell, A.; Taylor, R.W.; Wredenberg, A.; et al. FBXL 4 deficiency increases mitochondrial removal by autophagy. EMBO Mol. Med. 2020, 12, e11659. [Google Scholar] [CrossRef]
- Vincow, E.S.; Merrihew, G.; Thomas, R.E.; Shulman, N.J.; Beyer, R.P.; MacCoss, M.J.; Pallanck, L.J. The PINK1–Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 6400–6405. [Google Scholar] [CrossRef]
- Bogenhagen, D.F.; Haley, J.D. Pulse-chase SILAC–based analyses reveal selective oversynthesis and rapid turnover of mitochondrial protein components of respiratory complexes. J. Biol. Chem. 2020, 295, 2544–2554. [Google Scholar] [CrossRef]
- Livnat-Levanon, N.; Kevei, E.; Kleifeld, O.; Krutauz, D.; Segref, A.; Rinaldi, T.; Erpapazoglou, Z.; Cohen, M.; Reis, N.; Hoppe, T.; et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014, 7, 1371–1380. [Google Scholar] [CrossRef]
- Youle, R.J. Mitochondria-Striking a balance between host and endosymbiont. Science 2019, 365. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulkshane, P.; Ram, J.; Glickman, M.H. Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability. Biomolecules 2020, 10, 1559. https://doi.org/10.3390/biom10111559
Sulkshane P, Ram J, Glickman MH. Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability. Biomolecules. 2020; 10(11):1559. https://doi.org/10.3390/biom10111559
Chicago/Turabian StyleSulkshane, Prasad, Jonathan Ram, and Michael H Glickman. 2020. "Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability" Biomolecules 10, no. 11: 1559. https://doi.org/10.3390/biom10111559
APA StyleSulkshane, P., Ram, J., & Glickman, M. H. (2020). Ubiquitination of Intramitochondrial Proteins: Implications for Metabolic Adaptability. Biomolecules, 10(11), 1559. https://doi.org/10.3390/biom10111559




