Ubiquitin-Like Modifiers: Emerging Regulators of Protozoan Parasites
Abstract
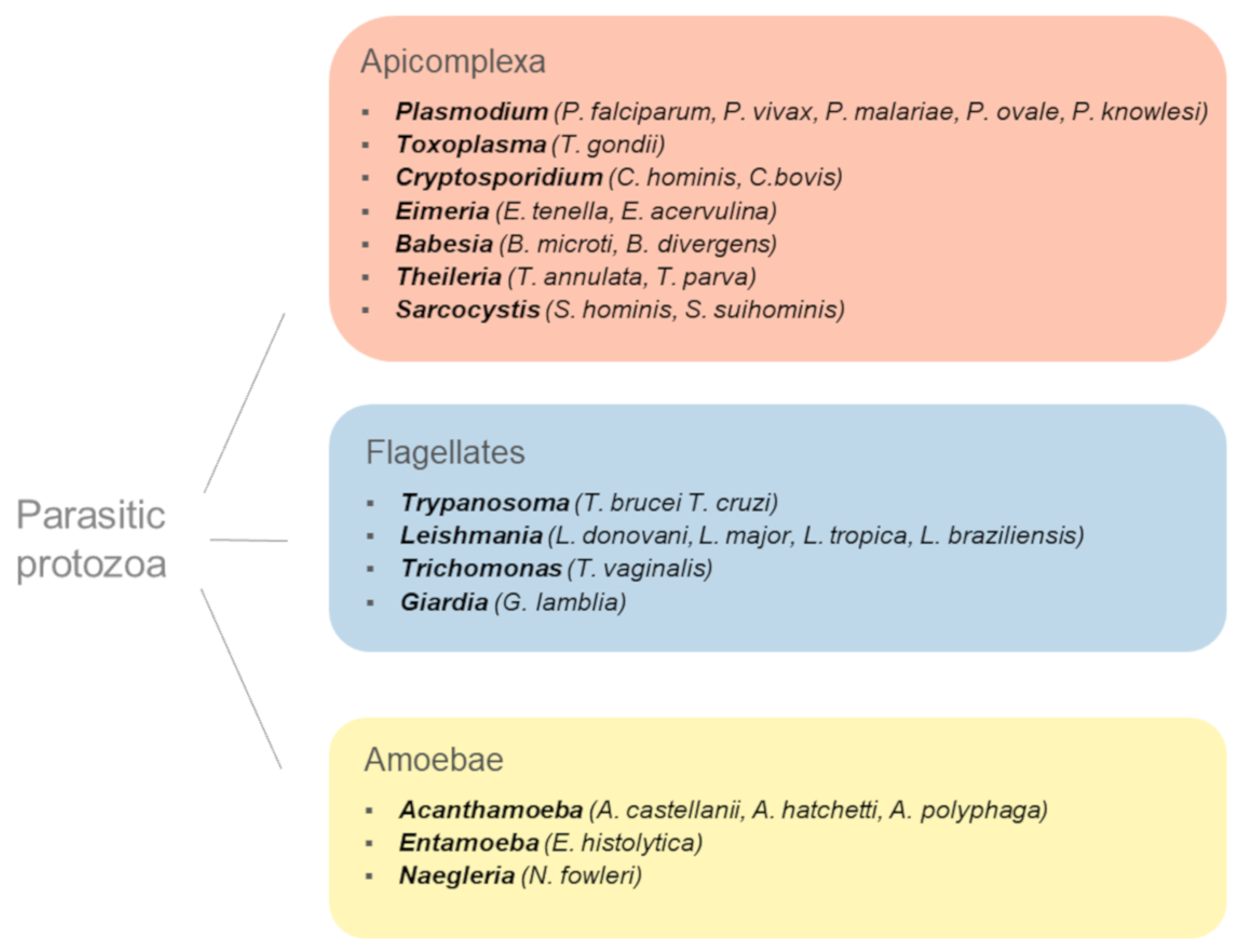
1. Introduction: When Parasitic Protozoa Met Ubls
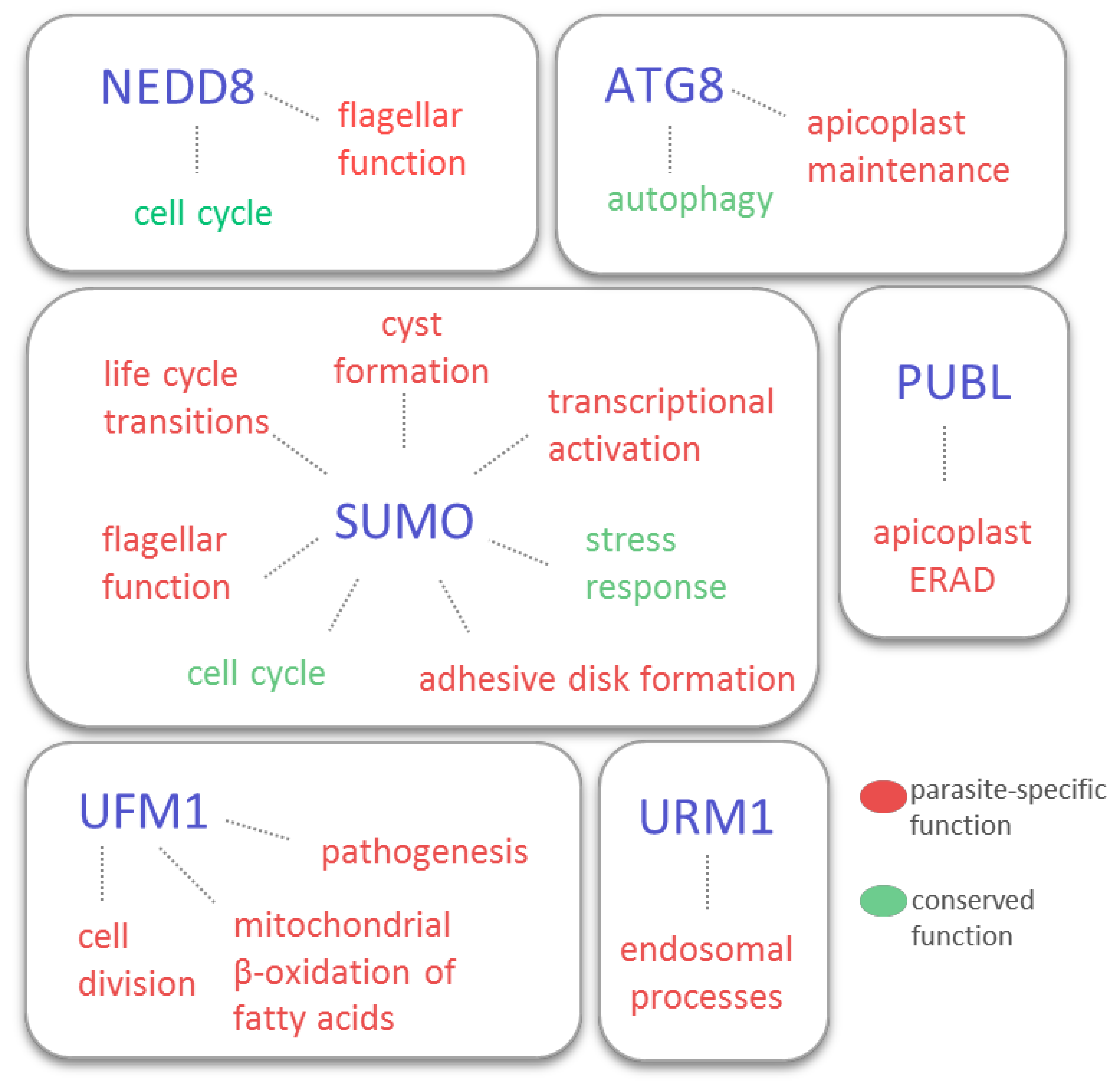
2. SUMO: Wrestling with Stress and More
3. NEDD8: Still a Mystery
4. ATG8 and ATG12: Moonlighting Autophagy Machinery
5. URM1: The Old-Timer
6. UFM1: Expect the Unexpected
7. PUBL: The New Kid on the Block?
8. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Cox, F.E.G. Systematics of the parasitic Protozoa. Trends Parasitol. 2002, 18, 108. [Google Scholar] [CrossRef]
- Cowman, A.F.; Healer, J.; Marapana, D.; Marsh, K. Malaria: Biology and Disease. Cell 2016, 167, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Georgiadou, S.P.; Makaritsis, K.P.; Dalekos, G.N. Leishmaniasis revisited: Current aspects on epidemiology, diagnosis and treatment. J. Transl. Intern. Med. 2015, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Brun, R.; Blum, J.; Chappuis, F.; Burri, C. Human African trypanosomiasis. Lancet 2010, 375, 148–159. [Google Scholar] [CrossRef]
- Tenter, A.M.; Heckeroth, A.R.; Weiss, L.M. Toxoplasma gondii: From animals to humans. Int. J. Parasitol. 2000, 30, 1217–1258. [Google Scholar] [CrossRef]
- Fletcher, S.M.; Stark, D.; Harkness, J.; Ellis, J.T. Enteric Protozoa in the Developed World: A Public Health Perspective. Clin. Microbiol. Rev. 2012, 25, 420–449. [Google Scholar] [CrossRef]
- Van Gerwen, O.; Muzny, C.A. Recent advances in the epidemiology, diagnosis, and management of Trichomonas vaginalis infection. F1000Research 2019, 8, 1666. [Google Scholar] [CrossRef]
- Maciver, S.K.; Piñero, J.E.; Lorenzo-Morales, J. Is Naegleria fowleri an Emerging Parasite? Trends Parasitol. 2020, 36, 19–28. [Google Scholar] [CrossRef]
- Capela, R.; Moreira, R.; Lopes, F. An Overview of Drug Resistance in Protozoal Diseases. Int. J. Mol. Sci. 2019, 20, 5748. [Google Scholar] [CrossRef]
- De Koning, H.P. Drug resistance in protozoan parasites. Emerg. Top. Life Sci. 2017, 1, 627–632. [Google Scholar]
- Schwede, A.; Krämer, S.; Carrington, M. How do trypanosomes change gene expression in response to the environment? Protoplasma 2011, 249, 223–238. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clayton, C.; Shapira, M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007, 156, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Foth, B.J.; Zhang, N.; Chaal, B.K.; Sze, S.K.; Preiser, P.R.; Bozdech, Z. Quantitative Time-course Profiling of Parasite and Host Cell Proteins in the Human Malaria Parasite Plasmodium falciparum. Mol. Cell. Proteom. 2011, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Yakubu, R.R.; Weiss, L.M.; De Monerri, N.C.S. Post-translational modifications as key regulators of apicomplexan biology: Insights from proteome-wide studies. Mol. Microbiol. 2017, 107, 1–23. [Google Scholar] [CrossRef]
- Chung, D.-W.D.; Ponts, N.; Cervantes, S.; Le Roch, K.G. Post-translational modifications in Plasmodium: More than you think! Mol. Biochem. Parasitol. 2009, 168, 123–134. [Google Scholar] [CrossRef]
- Zhang, N.; Jiang, N.; Zhang, K.; Zheng, L.; Zhang, D.; Sang, X.; Feng, Y.; Chen, R.; Yang, N.; Wang, X.; et al. Landscapes of Protein Posttranslational Modifications of African Trypanosoma Parasites. iScience 2020, 23, 101074. [Google Scholar] [CrossRef]
- Van Der Veen, A.G.; Ploegh, H.L. Ubiquitin-Like Proteins. Annu. Rev. Biochem. 2012, 81, 323–357. [Google Scholar] [CrossRef]
- Ponder, E.L.; Bogyo, M. Ubiquitin-Like Modifiers and Their Deconjugating Enzymes in Medically Important Parasitic Protozoa. Eukaryot. Cell 2007, 6, 1943–1952. [Google Scholar] [CrossRef]
- Ponts, N.; Yang, J.; Chung, D.-W.D.; Prudhomme, J.; Girke, T.; Horrocks, P.; Le Roch, K.G. Deciphering the Ubiquitin-Mediated Pathway in Apicomplexan Parasites: A Potential Strategy to Interfere with Parasite Virulence. PLoS ONE 2008, 3, e2386. [Google Scholar] [CrossRef]
- Castellanos, I.C.; Calvo, E.P.; Wasserman, M. A new gene inventory of the ubiquitin and ubiquitin-like conjugation pathways in Giardia intestinalis. Mem. Inst. Oswaldo Cruz 2020, 115, e190242. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.; Sharma, G.; Gupta, P.; Tiwari, S. In silico analysis of ubiquitin/ubiquitin-like modifiers and their conjugating enzymes in Entamoeba species. Parasitol. Res. 2012, 111, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Buerger, S.; Groettrup, M. The ubiquitin-like modifier FAT10 in antigen processing and antimicrobial defense. Mol. Immunol. 2015, 68, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Guerra, S.; Sánchez-Madrid, F.; Bustos-Morán, E.; Blas-Rus, N.; Martin-Cófreces, N.B. ISGylation – A key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 2017, 130, 2961–2969. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Balaji, S.; Iyer, L.M.; Aravind, L. Small but versatile: The extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct 2007, 2, 18. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2017, 118, 889–918. [Google Scholar] [CrossRef]
- Ronau, J.A.; Beckmann, J.F.; Hochstrasser, M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Res. 2016, 26, 441–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, W.-G.; Xu, X. Ubiquitin-like modifications in the DNA damage response. Mutat. Res. Mol. Mech. Mutagen. 2017, 56–75. [Google Scholar] [CrossRef]
- Witze, E.S.; Old, W.M.; A Resing, K.; Ahn, N.G. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 2007, 4, 798–806. [Google Scholar] [CrossRef]
- Sylvestersen, K.B.; Young, C.; Nielsen, M.L. Advances in characterizing ubiquitylation sites by mass spectrometry. Curr. Opin. Chem. Biol. 2013, 17, 49–58. [Google Scholar] [CrossRef]
- De Monerri, N.C.S.; Yakubu, R.R.; Chen, A.L.; Bradley, P.J.; Nieves, E.; Weiss, L.M.; Kim, K. The Ubiquitin Proteome of Toxoplasma gondii Reveals Roles for Protein Ubiquitination in Cell-Cycle Transitions. Cell Host Microbe 2015, 18, 621–633. [Google Scholar]
- Green, J.L.; Wu, Y.; Encheva, V.; Lasonder, E.; Prommaban, A.; Kunzelmann, S.; Christodoulou, E.; Grainger, M.; Truongvan, N.; Bothe, S.; et al. Ubiquitin activation is essential for schizont maturation in Plasmodium falciparum blood-stage development. PLoS Pathog. 2020, 16, e1008640. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Barysch, S.V.; Karaca, S.; Dittner, C.; Hsiao, H.-H.; Diaz, M.B.; Herzig, S.; Urlaub, H.; Melchior, F. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 2013, 20, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Keuss, M.J.; Hjerpe, R.; Hsia, O.; Gourlay, R.; Burchmore, R.; Trost, M.; Kurz, T. Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death. EMBO J. 2019, 38, e100024. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, M.; Xi, J.; Luo, F.; Li, A. PTM-ssMP: A Web Server for Predicting Different Types of Post-translational Modification Sites Using Novel Site-specific Modification Profile. Int. J. Biol. Sci. 2018, 14, 946–956. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, Y.; Zheng, Y.; Jiang, S.; Liu, W.; Mu, W.; Liu, Z.; Zhao, Y.; Xue, Y.; Ren, J. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014, 42, W325–W330. [Google Scholar] [CrossRef]
- Sharma, A.; Lysenko, A.; López, Y.; Dehzangi, A.; Sharma, R.; Reddy, H.; Sattar, A.; Tsunoda, T. HseSUMO: Sumoylation site prediction using half-sphere exposures of amino acids residues. BMC Genom. 2019, 19, 982. [Google Scholar] [CrossRef]
- Dehzangi, A.; López, Y.; Taherzadeh, G.; Sharma, A.; Tsunoda, T. SumSec: Accurate Prediction of Sumoylation Sites Using Predicted Secondary Structure. Molecules 2018, 23, 3260. [Google Scholar] [CrossRef]
- Chang, C.-C.; Tung, C.-H.; Chen, C.-W.; Tu, C.-H.; Chu, Y.-W. SUMOgo: Prediction of sumoylation sites on lysines by motif screening models and the effects of various post-translational modifications. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Swapna, L.S.; Parkinson, J. Genomics of apicomplexan parasites. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 254–273. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Wu, Z.; Zhang, L.; Ji, P.; Cai, Y.; Luo, S.; Wang, H.; Li, H. Genome mining offers a new starting point for parasitology research. Parasitol. Res. 2015, 114, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Swearingen, K.E.; Lindner, S.E. Plasmodium Parasites Viewed through Proteomics. Trends Parasitol. 2018, 34, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Hemelaar, J.; Galardy, P.J.; Borodovsky, A.; Kessler, B.M.; Ploegh, H.L.; Ovaa, H. Chemistry-Based Functional Proteomics: Mechanism-Based Activity-Profiling Tools for Ubiquitin and Ubiquitin-like Specific Proteases. J. Proteome Res. 2004, 3, 268–276. [Google Scholar] [CrossRef]
- Mulder, M.P.C.; Witting, K.; Berlin, I.; Pruneda, J.N.; Wu, K.-P.; Chang, J.-G.; Merkx, R.; Bialas, J.; Groettrup, M.; Vertegaal, A.C.; et al. A cascading activity-based probe sequentially targets E1–E2–E3 ubiquitin enzymes. Nat. Methods 2016, 12, 523–530. [Google Scholar] [CrossRef]
- Ekkebus, R.; Van Kasteren, S.I.; Kulathu, Y.; Scholten, A.; Berlin, I.; Geurink, P.P.; De Jong, A.; Goerdayal, S.; Neefjes, J.; Heck, A.J.; et al. On Terminal Alkynes That Can React with Active-Site Cysteine Nucleophiles in Proteases. J. Am. Chem. Soc. 2013, 135, 2867–2870. [Google Scholar] [CrossRef]
- Brownell, J.E.; Sintchak, M.D.; Gavin, J.M.; Liao, H.; Bruzzese, F.J.; Bump, N.J.; Soucy, T.A.; Milhollen, M.A.; Yang, X.; Burkhardt, A.L.; et al. Substrate-Assisted Inhibition of Ubiquitin-like Protein-Activating Enzymes: The NEDD8 E1 Inhibitor MLN4924 Forms a NEDD8-AMP Mimetic In Situ. Mol. Cell 2010, 37, 102–111. [Google Scholar] [CrossRef]
- Ernst, A.; Avvakumov, G.; Tong, J.; Fan, Y.; Zhao, Y.; Alberts, P.; Persaud, A.; Walker, J.R.; Neculai, A.-M.; Neculai, D.; et al. A Strategy for Modulation of Enzymes in the Ubiquitin System. Science 2013, 339, 590–595. [Google Scholar] [CrossRef]
- Lv, Z.; Yuan, L.; Atkison, J.H.; Williams, K.M.; Vega, R.; Sessions, E.H.; Divlianska, D.B.; Davies, C.; Chen, Y.; Olsen, S.K. Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme. Nat. Commun. 2018, 9, 5145. [Google Scholar] [CrossRef]
- Wu, H.Q.; Baker, D.; Ovaa, H. Small molecules that target the ubiquitin system. Biochem. Soc. Trans. 2020, 48, 479–497. [Google Scholar] [CrossRef]
- Karpiyevich, M.; Adjalley, S.; Mol, M.; Ascher, D.B.; Mason, B.; Noort, G.J.V.D.H.V.; Laman, H.; Ovaa, H.; Lee, M.C.S.; Artavanis-Tsakonas, K. Nedd8 hydrolysis by UCH proteases in Plasmodium parasites. PLoS Pathog. 2019, 15, e1008086. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, K.; Misaghi, S.; Comeaux, C.A.; Catic, A.; Spooner, E.; Duraisingh, M.T.; Ploegh, H.L. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol. Microbiol. 2006, 61, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Frickel, E.-M.; Quesada, V.; Muething, L.; Gubbels, M.-J.; Spooner, E.; Ploegh, H.L.; Artavanis-Tsakonas, K. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell. Microbiol. 2007, 9, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, K.; Weihofen, W.A.; Antos, J.M.; Coleman, B.I.; Comeaux, C.A.; Duraisingh, M.T.; Gaudet, R.; Ploegh, H.L. Characterization and Structural Studies of the Plasmodium falciparum Ubiquitin and Nedd8 Hydrolase UCHL3. J. Biol. Chem. 2009, 285, 6857–6866. [Google Scholar] [CrossRef] [PubMed]
- Ponder, E.L.; Albrow, V.E.; Leader, B.A.; Békés, M.; Mikolajczyk, J.; Fonović, U.P.; Shen, A.; Drag, M.; Xiao, J.; Deu, E.; et al. Functional Characterization of a SUMO Deconjugating Protease of Plasmodium falciparum Using Newly Identified Small Molecule Inhibitors. Chem. Biol. 2011, 18, 711–721. [Google Scholar] [CrossRef]
- Grzybek, M.; Golonko, A.; Górska, A.; Szczepaniak, K.; Strachecka, A.; Lass, A.; Lisowski, P. The CRISPR/Cas9 system sheds new lights on the biology of protozoan parasites. Appl. Microbiol. Biotechnol. 2018, 102, 4629–4640. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, C.; Otto, T.D.; Oberstaller, J.; Liao, X.; Adapa, S.R.; Udenze, K.; Bronner, I.F.; Casandra, D.; Mayho, M.; et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 2018, 360, eaap7847. [Google Scholar] [CrossRef]
- Muñoz, C.; Francisco, J.S.; Gutiérrez, B.; González, J. Role of the Ubiquitin-Proteasome Systems in the Biology and Virulence of Protozoan Parasites. BioMed Res. Int. 2015, 2015, 1–13. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef]
- Flotho, A.; Melchior, F. Sumoylation: A Regulatory Protein Modification in Health and Disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Tempé, D.; Piechaczyk, M.; Bossis, G. SUMO under stress. Biochem. Soc. Trans. 2008, 36, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Reiter, K.; Mukhopadhyay, D.; Zhang, H.; Boucher, L.E.; Kumar, N.; Bosch, J.; Matunis, M.J. Identification of Biochemically Distinct Properties of the Small Ubiquitin-related Modifier (SUMO) Conjugation Pathway inPlasmodium falciparum. J. Biol. Chem. 2013, 288, 27724–27736. [Google Scholar] [CrossRef] [PubMed]
- Reiter, K.H.; Ramachandran, A.; Xia, X.; Boucher, L.E.; Bosch, J.; Matunis, M.J. Characterization and Structural Insights into Selective E1-E2 Interactions in the Human and Plasmodium falciparum SUMO Conjugation Systems. J. Biol. Chem. 2015, 291, 3860–3870. [Google Scholar] [CrossRef] [PubMed]
- Rujimongkon, K.; Mungthin, M.; Tummatorn, J.; Ampawong, S.; Adisakwattana, P.; Boonyuen, U.; Reamtong, O. Proteomic analysis of Plasmodium falciparum response to isocryptolepine derivative. PLoS ONE 2019, 14, e0220871. [Google Scholar] [CrossRef] [PubMed]
- Issar, N.; Roux, E.; Mattei, D.; Scherf, A. Identification of a novel post-translational modification in Plasmodium falciparum: Protein sumoylation in different cellular compartments. Cell. Microbiol. 2008, 10, 1999–2011. [Google Scholar] [CrossRef]
- Mundwiler-Pachlatko, E.; Beck, H.-P. Maurer’s clefts, the enigma of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2013, 110, 19987–19994. [Google Scholar] [CrossRef]
- Sindhe, K.M.V.; Wu, W.; Legac, J.; Zhang, Y.-K.; Easom, E.E.; Cooper, R.A.; Plattner, J.J.; Freund, Y.R.; DeRisi, J.L.; Rosenthal, P.J. Plasmodium falciparum Resistance to a Lead Benzoxaborole Due to Blocked Compound Activation and Altered Ubiquitination or Sumoylation. mBio 2020, 11. [Google Scholar] [CrossRef]
- Oakley, M.S.; Verma, N.; Zheng, H.; Anantharaman, V.; Takeda, K.; Gao, Y.; Myers, T.G.; Pham, P.T.; Mahajan, B.; Kumar, N.; et al. Molecular Markers of Radiation Induced Attenuation in Intrahepatic Plasmodium falciparum Parasites. PLoS ONE 2016, 11, e0166814. [Google Scholar] [CrossRef]
- Cerqueira, G.C.; Cheeseman, I.H.; Schaffner, S.F.; Nair, S.; McDew-White, M.; Phyo, A.P.; Ashley, E.A.; Melnikov, A.; Rogov, P.; Birren, B.W.; et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017, 18, 78. [Google Scholar] [CrossRef]
- Crater, A.K.; Roscoe, S.; Fahim, A.; Ananvoranich, S. Toxoplasma ubiquitin-like protease 1, a key enzyme in sumoylation and desumoylation pathways, is under the control of non-coding RNAs. Int. J. Parasitol. 2018, 48, 867–880. [Google Scholar] [CrossRef]
- Braun, L.; Cannella, D.; Pinheiro, A.M.; Kieffer, S.; Belrhali, H.; Garin, J.; Hakimi, M.-A. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int. J. Parasitol. 2009, 39, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhai, B.-T.; He, J.-J.; Elsheikha, H.M.; Li, J.-X.; Zhu, X.-Q.; Yang, X.Y. Transcriptional changes in Toxoplasma gondii in response to treatment with monensin. Parasites Vectors 2020, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Charvat, R.A.; Arrizabalaga, G. Oxidative stress generated during monensin treatment contributes to altered Toxoplasma gondii mitochondrial function. Sci. Rep. 2016, 6, 22997. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Wang, T.; Fan, K.; Tu, X. The small ubiquitin-like modifier (SUMO) is essential in cell cycle regulation in Trypanosoma brucei. Exp. Cell Res. 2010, 316, 704–715. [Google Scholar] [CrossRef]
- Klein, C.A.; Droll, R.; Clayton, C. SUMOylation in Trypanosoma brucei. PeerJ 2013, 1, 180. [Google Scholar] [CrossRef]
- Ye, K.; Zhang, X.; Ni, J.; Liao, S.; Tu, X. Identification of enzymes involved in SUMOylation in Trypanosoma brucei. Sci. Rep. 2015, 5, srep10097. [Google Scholar] [CrossRef]
- Bayona, J.C.; Nakayasu, E.S.; Laverrière, M.; Aguilar, C.; Sobreira, T.J.P.; Choi, H.; Nesvizhskii, A.I.; Almeida, I.C.; Cazzulo, J.J.; Alvarez, V.E. SUMOylation Pathway inTrypanosoma cruzi: Functional Characterization and Proteomic Analysis of Target Proteins. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Iribarren, P.A.; Berazategui, M.A.; Cazzulo, J.J.; Alvarez, V.E. Biosynthesis of SUMOylated Proteins in Bacteria Using the Trypanosoma brucei Enzymatic System. PLoS ONE 2015, 10, e0134950. [Google Scholar] [CrossRef] [PubMed]
- Annoura, T.; Makiuchi, T.; Sariego, I.; Aoki, T.; Nara, T. SUMOylation of Paraflagellar Rod Protein, PFR1, and Its Stage-Specific Localization in Trypanosoma cruzi. PLoS ONE 2012, 7, e37183. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Xu, C.; Zhang, J.; Zhang, X.; Tu, X. Solution structure of SUMO fromTrypanosoma bruceiand its interaction with Ubc9. Proteins: Struct. Funct. Bioinform. 2009, 76, 266–269. [Google Scholar] [CrossRef]
- Iribarren, P.A.; Di Marzio, L.A.; Berazategui, M.A.; De Gaudenzi, J.G.; Alvarez, V.E. SUMO polymeric chains are involved in nuclear foci formation and chromatin organization in Trypanosoma brucei procyclic forms. PLoS ONE 2018, 13, e0193528. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, J.; Zhang, W.; Liu, H.; Fang, J.; Xie, H. An improved workflow for identifying ubiquitin/ubiquitin-like protein conjugation sites from tandem mass spectra. Proteomics 2013, 13, 2579–2584. [Google Scholar] [CrossRef] [PubMed]
- Iribarren, P.A.; Berazategui, M.A.; Bayona, J.C.; Almeida, I.C.; Cazzulo, J.J.; Alvarez, V.E. Different proteomic strategies to identify genuine SUMO targets and their modification sites in Trypanosoma brucei procyclic forms. Cell. Microbiol. 2015, 17, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Yu, Z.; Liu, Y.; Wang, T.; Wei, Y.; Li, Z. The Aurora B kinase in Trypanosoma brucei undergoes post-translational modifications and is targeted to various subcellular locations through binding to TbCPC1. Mol. Microbiol. 2013, 91, 256–274. [Google Scholar] [CrossRef] [PubMed]
- Obado, S.O.; Bot, C.; Echeverry, M.C.; Bayona, J.C.; Alvarez, V.E.; Taylor, M.; Kelly, J.M. Centromere-associated topoisomerase activity in bloodstream form Trypanosoma brucei. Nucleic Acids Res. 2011, 39, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Malureanu, L.; Jeganathan, K.B.; Kao, E.; Sustmann, C.; Tahk, S.; Shuai, K.; Grosschedl, R.; Van Deursen, J.M. Resolution of Sister Centromeres Requires RanBP2-Mediated SUMOylation of Topoisomerase IIα. Cell 2008, 133, 103–115. [Google Scholar] [CrossRef]
- Takahashi, Y.; Yong-Gonzalez, V.; Kikuchi, Y.; Strunnikov, A. SIZ1/SIZ2 Control of Chromosome Transmission Fidelity Is Mediated by the Sumoylation of Topoisomerase II. Genetics 2006, 172, 783–794. [Google Scholar] [CrossRef]
- López-Farfán, D.; Bart, J.-M.; Rojas-Barros, D.I.; Navarro, M. SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in Trypanosoma brucei. PLoS Pathog. 2014, 10, e1004545. [Google Scholar] [CrossRef]
- Horn, D. Antigenic variation in African trypanosomes. Mol. Biochem. Parasitol. 2014, 195, 123–129. [Google Scholar] [CrossRef]
- Saura, A.; A Iribarren, P.; Rojas-Barros, D.; Bart, J.M.; López-Farfán, D.; Andrés-León, E.; Vidal-Cobo, I.; Boehm, C.; E Alvarez, V.; Field, M.C.; et al. SUMOylated SNF2PH promotes variant surface glycoprotein expression in bloodstream trypanosomes. EMBO Rep. 2019, 20, 48029. [Google Scholar] [CrossRef]
- Portman, N.; Gull, K. The paraflagellar rod of kinetoplastid parasites: From structure to components and function. Int. J. Parasitol. 2010, 40, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Vranych, C.V.; Merino, M.C.; Zamponi, N.; Touz, M.C.; Rópolo, A.S. SUMOylation in Giardia lamblia: A Conserved Post-Translational Modification in One of the Earliest Divergent Eukaryotes. Biomolecules 2012, 2, 312–330. [Google Scholar] [CrossRef] [PubMed]
- Di Genova, B.M.; Da Silva, R.C.; Da Cunha, J.P.C.; Gargantini, P.R.; Mortara, R.A.; Tonelli, R.R. Protein SUMOylation is Involved in Cell-cycle Progression and Cell Morphology in Giardia lamblia. J. Eukaryot. Microbiol. 2016, 64, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Vranych, C.V.; Rivero, M.R.; Merino, M.C.; Mayol, G.F.; Zamponi, N.; Maletto, B.A.; Pistoresi-Palencia, M.C.; Touz, M.C.; Rópolo, A.S. SUMOylation and deimination of proteins: Two epigenetic modifications involved in Giardia encystation. Biochim. Biophys. Acta 2014, 1843, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Touz, M.C.; Ropolo, A.S.; Rivero, M.R.; Vranych, C.V.; Conrad, J.T.; Svärd, S.G.; Nash, T.E. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 2008, 121, 2930–2938. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Wang, W.; Xiong, Y. Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction. Subcell. Biochem. 2017, 83, 323–347. [Google Scholar] [PubMed]
- Brown, J.S.; Jackson, S.P. Ubiquitylation, neddylation and the DNA damage response. Open Biol. 2015, 5, 150018. [Google Scholar] [CrossRef]
- Enchev, R.I.; Schulman, B.A.; Peter, M. Protein neddylation: Beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 2015, 16, 30–44. [Google Scholar] [CrossRef]
- Hjerpe, R.; Thomas, Y.; Kurz, T. NEDD8 Overexpression Results in Neddylation of Ubiquitin Substrates by the Ubiquitin Pathway. J. Mol. Biol. 2012, 421, 27–29. [Google Scholar] [CrossRef]
- Rabut, G.; Peter, M. Function and regulation of protein neddylation. EMBO Rep. 2008, 9, 969–976. [Google Scholar] [CrossRef]
- Liao, S.; Hu, H.; Wang, T.; Tu, X.; Li, Z. The Protein Neddylation Pathway in Trypanosoma brucei: Functional characterization and substrate identification. J. Biol. Chem. 2016, 292, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Barth, E.; Hübler, R.; Baniahmad, A.; Marz, M. The Evolution of COP9 Signalosome in Unicellular and Multicellular Organisms. Genome Biol. Evol. 2016, 8, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Leaton, L.A.; Farr, L.; Barfield, A.; Moonah, S. Interaction between parasite-encoded JAB1/CSN5 and macrophage migration inhibitory factor proteins attenuates its proinflammatory function. Sci. Rep. 2018, 8, 10241. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rigden, D.J.; Michels, P.A.M.; Ginger, M. Autophagy in protists: Examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy 2009, 5, 784–794. [Google Scholar] [CrossRef]
- Duszenko, M.; Ginger, M.; Brennand, A.; Gualdrón-López, M.; Colombo, M.I.; Coombs, G.H.; Coppens, I.; Jayabalasingham, B.; Langsley, G.; De Castro, S.L.; et al. Autophagy in protists. Autophagy 2011, 7, 127–158. [Google Scholar] [CrossRef]
- Williams, R.A.; Woods, K.L.; Juliano, L.; Mottram, J.C.; Coombs, G.H. Characterisation of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy 2009, 5, 159–172. [Google Scholar] [CrossRef]
- Picazarri, K.; Nakada-Tsukui, K.; Nozaki, T. Autophagy during Proliferation and Encystation in the Protozoan Parasite Entamoeba invadens. Infect. Immun. 2007, 76, 278–288. [Google Scholar] [CrossRef]
- Moon, E.-K.; Hong, Y.; Chung, D.-I.; Kong, H.-H. Identification of Atg8 Isoform in Encysting Acanthamoeba. Korean J. Parasitol. 2013, 51, 497–502. [Google Scholar] [CrossRef]
- Cárdenas-Zúñiga, R.; Sánchez-Monroy, V.; Bermúdez-Cruz, R.M.; Rodríguez, M.A.; Serrano-Luna, J.; Shibayama, M. Ubiquitin-like Atg8 protein is expressed during autophagy and the encystation process in Naegleria gruberi. Parasitol. Res. 2016, 116, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, M.S.; Miranda-Ozuna, J.F.T.; Salazar-Villatoro, L.; Vázquez-Calzada, C.; Ávila-González, L.; González-Robles, A.; Ortega-López, J.; Arroyo, R. Biogenesis of Autophagosome in Trichomonas vaginalis during Macroautophagy Induced by Rapamycin-treatment and Iron or Glucose Starvation Conditions. J. Eukaryot. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-Y.; Chen, R.-M.; Lin, H.-C.; Cheng, W.-H.; Lin, H.-A.; Lin, W.-N.; Huang, P.-J.; Chiu, C.-H.; Tang, P. Potential role of autophagy in proteolysis in Trichomonas vaginalis. J. Microbiol. Immunol. Infect. 2019, 52, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, V.E.; Kosec, G.; Sant’Anna, C.; Turk, V.; Cazzulo, J.J.; Eturk, B. Autophagy Is Involved in Nutritional Stress Response and Differentiation in Trypanosoma cruzi. J. Biol. Chem. 2008, 283, 3454–3464. [Google Scholar] [CrossRef]
- Cull, B.; Godinho, J.L.P.; Rodrigues, J.C.F.; Frank, B.; Schurigt, U.; Williams, R.A.M.; Coombs, G.H.; Mottram, J.C. Glycosome turnover inLeishmania majoris mediated by autophagy. Autophagy 2014, 10, 2143–2157. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Berry, L.; Sullivan, W.J.; Besteiro, S. Autophagy participates in the unfolded protein response in Toxoplasma gondii. FEMS Microbiol. Lett. 2017, 364, 364. [Google Scholar] [CrossRef]
- Arisue, N.; Hashimoto, T. Phylogeny and evolution of apicoplasts and apicomplexan parasites. Parasitol. Int. 2015, 64, 254–259. [Google Scholar] [CrossRef]
- McFadden, G.I.; Yeh, E. The apicoplast: Now you see it, now you don’t. Int. J. Parasitol. 2017, 47, 137–144. [Google Scholar] [CrossRef]
- Kitamura, K.; Kishi-Itakura, C.; Tsuboi, T.; Sato, S.; Kita, K.; Ohta, N.; Mizushima, N. Autophagy-Related Atg8 Localizes to the Apicoplast of the Human Malaria Parasite Plasmodium falciparum. PLoS ONE 2012, 7, e42977. [Google Scholar] [CrossRef]
- Tomlins, A.M.; Ben Rached, F.; Williams, R.A.; Proto, W.R.; Coppens, I.; Ruch, U.; Gilberger, T.W.; Coombs, G.H.; Mottram, J.C.; Muller, S.; et al. Plasmodium falciparum ATG8 implicated in both autophagy and apicoplast formation. Autophagy 2013, 9, 1540–1552. [Google Scholar] [CrossRef]
- Cervantes, S.; Bunnik, E.M.; Saraf, A.; Conner, C.M.; Escalante, A.; E Sardiu, M.; Ponts, N.; Prudhomme, J.; Florens, L.; Le Roch, K.G. The multifunctional autophagy pathway in the human malaria parasite, Plasmodium falciparum. Autophagy 2013, 10, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Lévêque, M.F.; Berry, L.; Cipriano, M.J.; Nguyen, H.M.; Striepen, B.; Besteiro, S. Autophagy-Related Protein ATG8 Has a Noncanonical Function for Apicoplast Inheritance in Toxoplasma gondii. mBio 2015, 6, e01446-15. [Google Scholar]
- Kong-Hap, M.A.; Mouammine, A.; Daher, W.; Berry, L.; Lebrun, M.; Dubremetz, J.-F.; Besteiro, S. Regulation of ATG8 membrane association by ATG4 in the parasitic protistToxoplasma gondii. Autophagy 2013, 9, 1334–1348. [Google Scholar] [CrossRef] [PubMed]
- Walczak, M.; Ganesan, S.M.; Niles, J.C.; Yeh, E. ATG8 Is Essential Specifically for an Autophagy-Independent Function in Apicoplast Biogenesis in Blood-Stage Malaria Parasites. mBio 2018, 9, e02021-17. [Google Scholar] [CrossRef]
- Walker, D.M.; Mahfooz, N.; Kemme, K.A.; Patel, V.C.; Spangler, M.; Drew, M.E. Plasmodium falciparum Erythrocytic Stage Parasites Require the Putative Autophagy Protein PfAtg7 for Normal Growth. PLoS ONE 2013, 8, e67047. [Google Scholar] [CrossRef][Green Version]
- Hain, A.U.; Weltzer, R.R.; Hammond, H.; Jayabalasingham, B.; Dinglasan, R.R.; Graham, D.R.M.; Colquhoun, D.R.; Coppens, I.; Bosch, J. Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J. Struct. Biol. 2012, 180, 551–562. [Google Scholar] [CrossRef]
- Hain, A.U.; Bartee, D.; Sanders, N.G.; Miller, A.S.; Sullivan, D.J.; Levitskaya, J.; Meyers, C.L.F.; Bosch, J. Identification of an Atg8-Atg3 Protein–Protein Interaction Inhibitor from the Medicines for Malaria Venture Malaria Box Active in Blood and Liver Stage Plasmodium falciparum Parasites. J. Med. Chem. 2014, 57, 4521–4531. [Google Scholar] [CrossRef]
- Hain, A.U.P.; Miller, A.S.; Levitskaya, J.; Bosch, J. Virtual Screening and Experimental Validation Identify Novel Inhibitors of thePlasmodium falciparumAtg8-Atg3 Protein-Protein Interaction. ChemMedChem 2016, 11, 900–910. [Google Scholar] [CrossRef]
- Villa, S.; Legnani, L.; Colombo, D.; Gelain, A.; Lammi, C.; Bongiorno, D.; Ilboudo, D.P.; McGee, K.E.; Bosch, J.; Grazioso, G. Structure-based drug design, synthesis and biological assays of P. falciparum Atg3–Atg8 protein–protein interaction inhibitors. J. Comput. Mol. Des. 2018, 32, 473–486. [Google Scholar] [CrossRef]
- Pang, Y.; Yamamoto, H.; Sakamoto, H.; Oku, M.; Mutungi, J.K.; Sahani, M.H.; Kurikawa, Y.; Kita, K.; Noda, N.N.; Sakai, Y.; et al. Evolution from covalent conjugation to non-covalent interaction in the ubiquitin-like ATG12 system. Nat. Struct. Mol. Biol. 2019, 26, 289–296. [Google Scholar] [CrossRef]
- Wang, F.; Liu, M.; Qiu, R.; Ji, C.N. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell 2011, 2, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Pabis, M.; Termathe, M.; E Ravichandran, K.; Kienast, S.D.; Krutyhołowa, R.; Sokołowski, M.; Jankowska, U.; Grudnik, P.; A Leidel, S.; Glatt, S. Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation. EMBO J. 2020, e105087. [Google Scholar]
- Zhang, W.; Zhang, J.; Xu, C.; Wang, T.; Zhang, X.; Tu, X. Solution structure of Urm1 fromTrypanosoma brucei. Proteins 2009, 75, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Sharma, P.; Selvapandiyan, A.; Salotra, P. Leishmania donovani-specific Ub-related modifier-1: An early endosome-associated ubiquitin-like conjugation inLeishmania donovani. Mol. Microbiol. 2015, 99, 597–610. [Google Scholar] [CrossRef]
- Gerakis, Y.; Quintero, M.; Li, H.; Hetz, C. The UFMylation System in Proteostasis and Beyond. Trends Cell Biol. 2019, 29, 974–986. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, X. UFMylation: A Unique & Fashionable Modification for Life. Genom. Proteom. Bioinform. 2016, 14, 140–146. [Google Scholar]
- Gannavaram, S.; Sharma, P.; Duncan, R.; Salotra, P.; Nakhasi, H.L. Mitochondrial Associated Ubiquitin Fold Modifier-1 Mediated Protein Conjugation in Leishmania donovani. PLoS ONE 2011, 6, e16156. [Google Scholar] [CrossRef]
- Gannavaram, S.; Davey, S.; Lakhal-Naouar, I.; Duncan, R.; Nakhasi, H.L. Deletion of Ubiquitin Fold Modifier Protein Ufm1 Processing Peptidase Ufsp in L. donovani Abolishes Ufm1 Processing and Alters Pathogenesis. PLoS Negl. Trop. Dis. 2014, 8, e2707. [Google Scholar] [CrossRef]
- Gannavaram, S.; Connelly, P.S.; Daniels, M.P.; Duncan, R.; Salotra, P.; Nakhasi, H.L. Deletion of mitochondrial associated ubiquitin fold modifier protein Ufm1 in Leishmania donovani results in loss of β-oxidation of fatty acids and blocks cell division in the amastigote stage. Mol. Microbiol. 2012, 86, 187–198. [Google Scholar] [CrossRef]
- Fellows, J.D.; Cipriano, M.J.; Agrawal, S.; Striepen, B. A Plastid Protein That Evolved from Ubiquitin and Is Required for Apicoplast Protein Import in Toxoplasma gondii. mBio 2017, 8, e00950-17. [Google Scholar] [CrossRef]
- Agrawal, S.; Van Dooren, G.G.; Beatty, W.L.; Striepen, B. Genetic Evidence that an Endosymbiont-derived Endoplasmic Reticulum-associated Protein Degradation (ERAD) System Functions in Import of Apicoplast Proteins. J. Biol. Chem. 2009, 284, 33683–33691. [Google Scholar] [CrossRef] [PubMed]
| SUMO | NEDD8 | ATG8 | ATG12 | URM1 | UFM1 | ISG15 | FAT10 | |
|---|---|---|---|---|---|---|---|---|
| Plasmodium | + | + | + | + | + | − | − | − |
| Toxoplasma | + | + | + | + | + | + | − | − |
| Trypanosoma | + | + | + | + | + | + | − | − |
| Leishmania | + | + | + | + | + | + | − | − |
| Giardia | + | + | − | − | + | + | − | − |
| Entamoeba | + | + | + | − | + | − | − | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karpiyevich, M.; Artavanis-Tsakonas, K. Ubiquitin-Like Modifiers: Emerging Regulators of Protozoan Parasites. Biomolecules 2020, 10, 1403. https://doi.org/10.3390/biom10101403
Karpiyevich M, Artavanis-Tsakonas K. Ubiquitin-Like Modifiers: Emerging Regulators of Protozoan Parasites. Biomolecules. 2020; 10(10):1403. https://doi.org/10.3390/biom10101403
Chicago/Turabian StyleKarpiyevich, Maryia, and Katerina Artavanis-Tsakonas. 2020. "Ubiquitin-Like Modifiers: Emerging Regulators of Protozoan Parasites" Biomolecules 10, no. 10: 1403. https://doi.org/10.3390/biom10101403
APA StyleKarpiyevich, M., & Artavanis-Tsakonas, K. (2020). Ubiquitin-Like Modifiers: Emerging Regulators of Protozoan Parasites. Biomolecules, 10(10), 1403. https://doi.org/10.3390/biom10101403




