Releasing the Lockdown: An Emerging Role for the Ubiquitin-Proteasome System in the Breakdown of Transient Protein Inclusions
Abstract
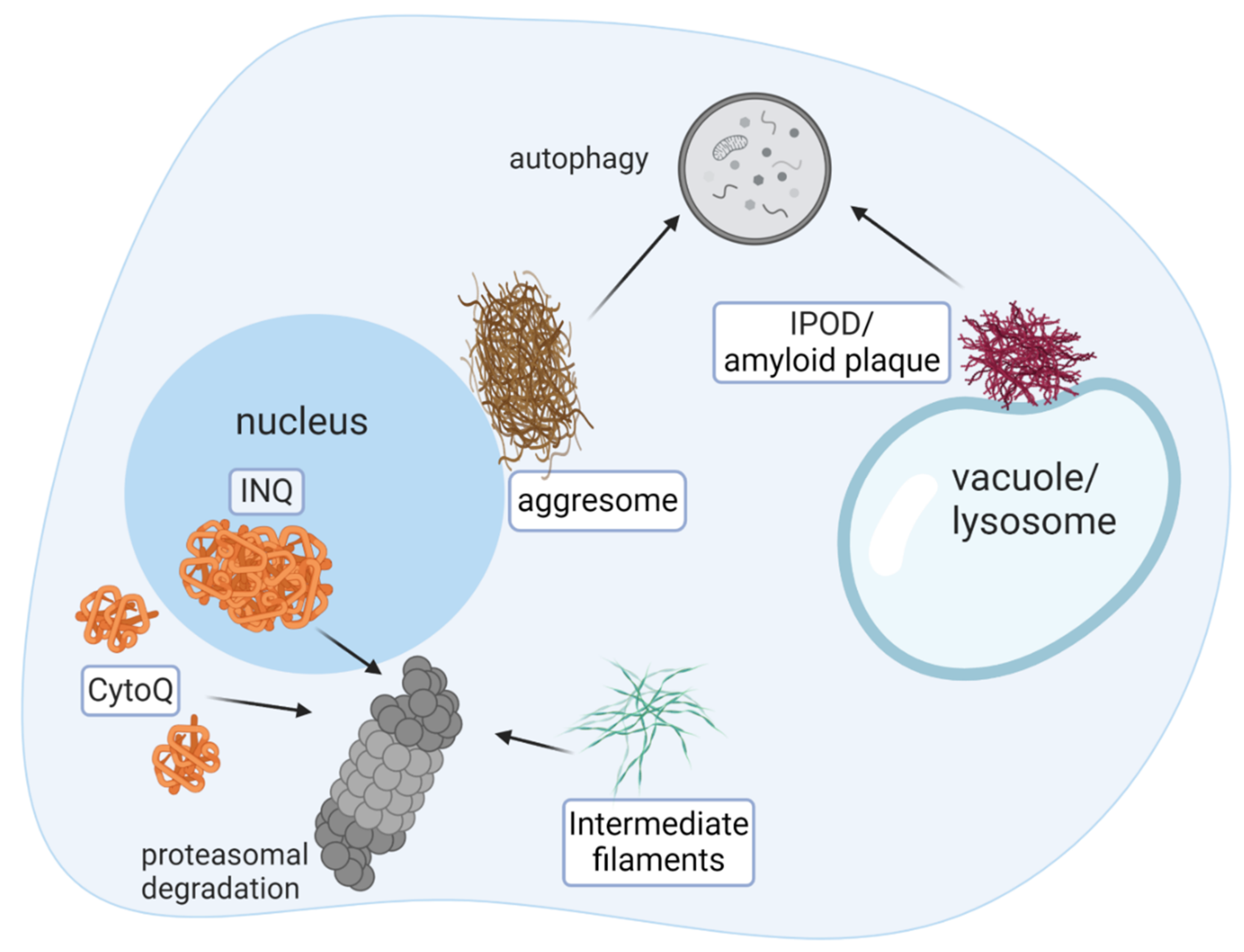
1. Introduction
2. Misfolded Proteins Are Extracted from Cell Inclusions and Sorted by Molecular Chaperones
3. A Role of the Ubiquitin-Proteasome System in Protein Quality-Control Triage Decisions
3.1. Proteasome Inhibition Induces Transient-QC Accumulation
3.2. Proteasomes Are Associated with Transient-QC Deposits
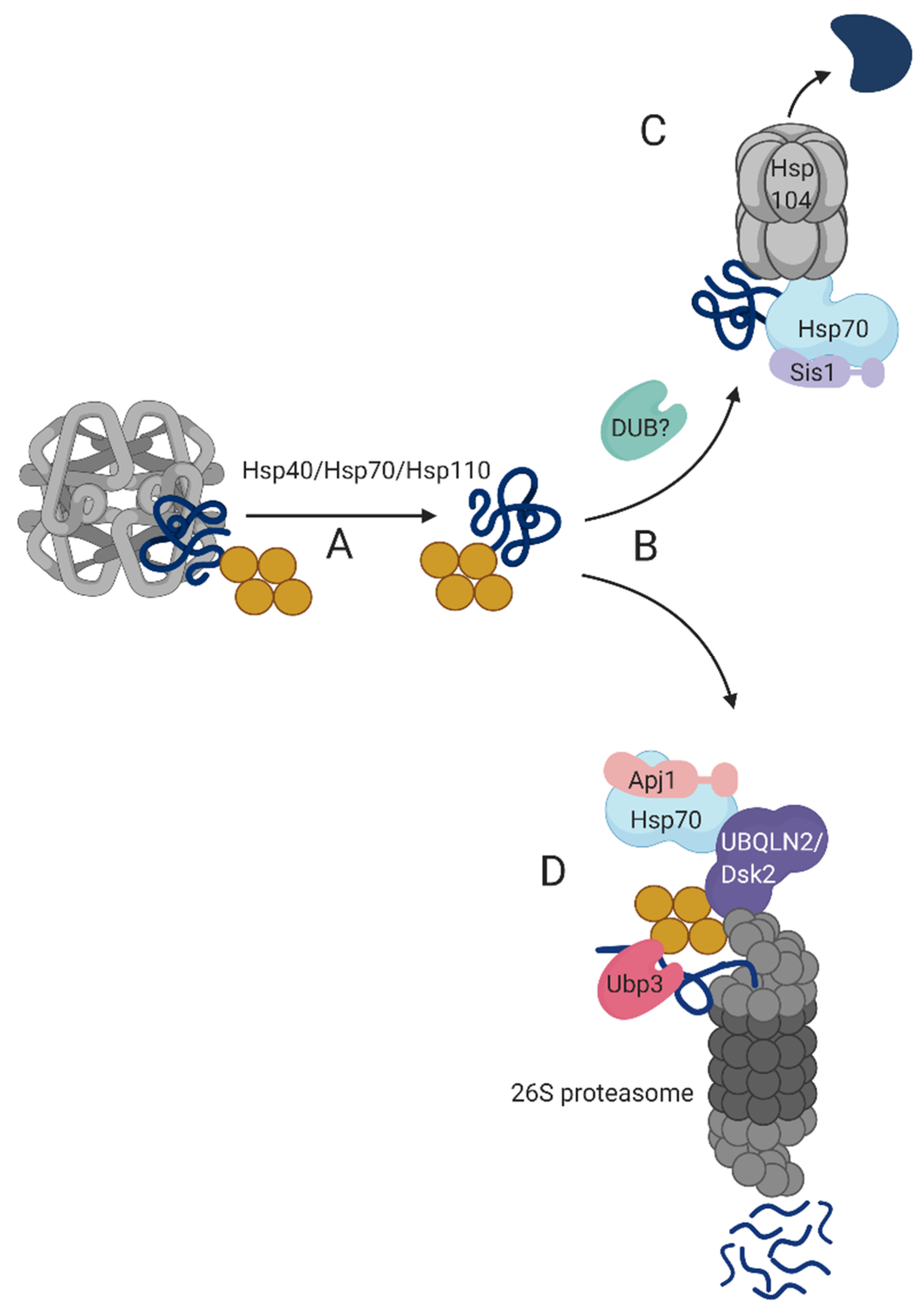
3.3. UBQLN2 Bridges between Transient-QC Bodies and the 26S Proteasome
3.4. Deubiquitylation Activity is Required for Transient-QC Clearance
4. A Putative Model for the Mechanism of Triage Decisions during Disaggregation
5. Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Alzheimer, A. A uber einen eigenartigen schweren erkrankungsprozess der hirninde. Neurol. Cent. 1906, 25, 1134. [Google Scholar]
- Hippius, H.; Neundörfer, G. The discovery of Alzheimer’s disease. Dialogues Clin. Neurosci. 2003, 5, 101–108. [Google Scholar] [PubMed]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef] [PubMed]
- Kopito, R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000, 10, 524–530. [Google Scholar] [CrossRef]
- Hao, R.; Nanduri, P.; Rao, Y.; Panichelli, R.S.; Ito, A.; Yoshida, M.; Yao, T.-P. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell 2013, 51, 819–828. [Google Scholar] [CrossRef]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef]
- Miller, S.B.M.; Ho, C.-T.; Winkler, J.; Khokhrina, M.; Neuner, A.; Mohamed, M.Y.H.; Guilbride, D.L.; Richter, K.; Lisby, M.; Schiebel, E.; et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015, 34, 778–797. [Google Scholar] [CrossRef]
- Escusa-Toret, S.; Vonk, W.I.M.; Frydman, J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 2013, 15, 1231–1243. [Google Scholar] [CrossRef]
- Sontag, E.M.; Samant, R.S.; Frydman, J. Mechanisms and functions of spatial protein quality control. Annu. Rev. Biochem. 2017, 86, 97–122. [Google Scholar] [CrossRef]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef]
- Fändrich, M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 2007, 64, 2066–2078. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Neurodegeneration. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 2015, 349, 1255555. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U. Protein misfolding diseases protein misfolding diseases. Annu. Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Duggan, M.; Torkzaban, B.; Ahooyi, T.M.; Khalili, K.; Gordon, J. Age-related neurodegenerative diseases. J. Cell. Physiol. 2020, 235, 3131–3141. [Google Scholar] [CrossRef]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein sorting factors coordinate the spatio-temporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef]
- Wooten, M.W.; Geetha, T.; Babu, J.R.; Seibenhener, M.L.; Peng, J.; Cox, N.; Diaz-Meco, M.-T.; Moscat, J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J. Biol. Chem. 2008, 283, 6783–6789. [Google Scholar] [CrossRef]
- Lu, K.; Psakhye, I.; Jentsch, S. A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy 2014, 10, 2381–2382. [Google Scholar] [CrossRef]
- Oling, D.; Eisele, F.; Kvint, K.; Nyström, T. Opposing roles of Ubp3-dependent deubiquitination regulate replicative life span and heat resistance. EMBO J. 2014, 33, 747–761. [Google Scholar] [CrossRef]
- Wang, N.; Ma, Q.; Peng, P.; Yu, Y.; Xu, S.; Wang, G.; Ying, Z.; Wang, H. Autophagy and ubiquitin-proteasome system coordinate to regulate the protein quality control of neurodegenerative disease-associated DCTN1. Neurotox. Res. 2020, 37, 48–57. [Google Scholar] [CrossRef]
- Sha, Z.; Schnell, H.M.; Ruoff, K.; Goldberg, A. Rapid induction of p62 and GABARAPL1 upon proteasome inhibition promotes survival before autophagy activation. J. Cell Biol. 2018, 217, 1757–1776. [Google Scholar] [CrossRef]
- Limanaqi, F.; Biagioni, F.; Gambardella, S.; Familiari, P.; Frati, A.; Fornai, F. Promiscuous roles of autophagy and proteasome in neurodegenerative proteinopathies. Int. J. Mol. Sci. 2020, 21, 3028. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, G.; Andréasson, C. Hsp70-Hsp110 chaperones deliver ubiquitin-dependent and -independent substrates to the 26S proteasome for proteolysis in yeast. J. Cell Sci. 2018, 131, jcs210948. [Google Scholar] [CrossRef] [PubMed]
- Shiber, A.; Breuer, W.; Brandeis, M.; Ravid, T. Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol. Biol. Cell 2013, 24, 2076–2087. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Wickner, S.; Maurizi, M.R. Protein quality control: Triage by chaperones and proteases. Genes Dev. 1997, 11, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Bukau, B.; Kampinga, H.H. Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 2018, 69, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Lum, R.; Tkach, J.M.; Vierling, E.; Glover, J.R. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 2004, 279, 29139–29146. [Google Scholar] [CrossRef]
- Kaimal, J.M.; Kandasamy, G.; Gasser, F.; Andréasson, C. Coordinated Hsp110 and Hsp104 activities power protein disaggregation in saccharomyces cerevisiae. Mol. Cell. Biol. 2017, 37, e00027-17. [Google Scholar] [CrossRef]
- Gao, X.; Carroni, M.; Nussbaum-Krammer, C.; Mogk, A.; Nillegoda, N.B.; Szlachcic, A.; Guilbride, D.L.; Saibil, H.R.; Mayer, M.P.; Bukau, B. Human Hsp70 disaggregase reverses parkinson’s-linked α-synuclein amyloid fibrils. Mol. Cell 2015, 59, 781–793. [Google Scholar] [CrossRef]
- Nillegoda, N.B.; Kirstein, J.; Szlachcic, A.; Berynskyy, M.; Stank, A.; Stengel, F.; Arnsburg, K.; Gao, X.; Scior, A.; Aebersold, R.; et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 2015, 524, 247–251. [Google Scholar] [CrossRef]
- Shorter, J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 2011, 6, e26319. [Google Scholar] [CrossRef]
- Hoppe, T.; Branzei, D. Stefan Jentsch (1955–2016)-Maestro of the ubiquitin family. EMBO J. 2017, 36, 1–2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- den Brave, F.; Cairo, L.V.; Jagadeesan, C.; Ruger-Herreros, C.; Mogk, A.; Bukau, B.; Jentsch, S. Chaperone-mediated protein disaggregation triggers proteolytic clearance of intra-nuclear protein inclusions. Cell Rep. 2020, 31, 107680. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.T.; Grousl, T.; Shatz, O.; Jawed, A.; Ruger-Herreros, C.; Semmelink, M.; Zahn, R.; Richter, K.; Bukau, B.; Mogk, A. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.E.; Nolte, A.M.; Kamiya, E.; Hines, J.K. Three J-proteins impact Hsp104-mediated variant-specific prion elimination: A new critical role for a low. Curr. Genet. 2020, 66, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Serlidaki, D.; van Waarde, M.A.W.H.; Rohland, L.; Wentink, A.S.; Dekker, S.L.; Kamphuis, M.J.; Boertien, J.M.; Brunsting, J.F.; Nillegoda, N.B.; Bukau, B.; et al. Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J. Biol. Chem. 2020, 295, 7301–7316. [Google Scholar] [CrossRef]
- Preston, G.M.; Guerriero, C.J.; Metzger, M.B.; Michaelis, S.; Brodsky, J.L. Substrate insolubility dictates Hsp104-dependent endoplasmic-reticulum-associated degradation. Mol. Cell 2018, 70, 242–253. [Google Scholar] [CrossRef]
- García-Mata, R.; Bebök, Z.; Sorscher, E.J.; Sztul, E.S. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1999, 146, 1239–1254. [Google Scholar] [CrossRef]
- Hjerpe, R.; Bett, J.S.; Keuss, M.J.; Solovyova, A.; McWilliams, T.G.; Johnson, C.; Sahu, I.; Varghese, J.; Wood, N.; Wightman, M.; et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 2016, 166, 935–949. [Google Scholar] [CrossRef]
- Andersson, V.; Hanzén, S.; Liu, B.; Molin, M.; Nyström, T. Enhancing protein disaggregation restores proteasome activity in aged cells. Aging 2013, 5, 802–812. [Google Scholar] [CrossRef]
- Guo, Q.; Lehmer, C.; Martínez-Sánchez, A.; Rudack, T.; Beck, F.; Hartmann, H.; Pérez-Berlanga, M.; Frottin, F.; Hipp, M.S.; Hartl, F.U.; et al. In situ structure of neuronal C9orf72 Poly-GA aggregates reveals proteasome recruitment. Cell 2018, 172, 696–705. [Google Scholar] [CrossRef]
- Bäuerlein, F.J.B.; Saha, I.; Mishra, A.; Kalemanov, M.; Martínez-Sánchez, A.; Klein, R.; Dudanova, I.; Hipp, M.S.; Hartl, F.U.; Baumeister, W.; et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell 2017, 171, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097. [Google Scholar] [CrossRef] [PubMed]
- Cliffe, R.; Sang, J.C.; Kundel, F.; Finley, D.; Klenerman, D.; Ye, Y. Filamentous aggregates are fragmented by the proteasome holoenzyme. Cell Rep. 2019, 26, 2140–2149. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Kim, M.; Marshall, R.S.; Vierstra, R.D.; Vierling, E. HSP101 interacts with the proteasome and promotes the clearance of ubiquitylated protein aggregates. Plant Physiol. 2019, 180, 1829–1847. [Google Scholar] [CrossRef]
- Martín-Aparicio, E.; Yamamoto, A.; Hernández, F.; Hen, R.; Avila, J.; Lucas, J.J. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington’s disease. J. Neurosci. 2001, 21, 8772–8781. [Google Scholar] [CrossRef]
- Carpenter, K.; Bell, R.B.; Yunus, J.; Amon, A.; Berchowitz, L.E. Phosphorylation-mediated clearance of amyloid-like assemblies in meiosis. Dev. Cell 2018, 45, 392–405.e6. [Google Scholar] [CrossRef]
- Walters, K.J.; Kleijnen, M.F.; Goh, A.M.; Wagner, G.; Howley, P.M. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 2002, 41, 1767–1777. [Google Scholar] [CrossRef]
- Kleijnen, M.F.; Alarcon, R.M.; Howley, P.M. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol. Biol. Cell 2003, 14, 3868–3875. [Google Scholar] [CrossRef]
- Kleijnen, M.F.; Shih, A.H.; Zhou, P.; Kumar, S.; Soccio, R.E.; Kedersha, N.L.; Gill, G.; Howley, P.M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 2000, 6, 409–419. [Google Scholar] [CrossRef]
- Funakoshi, M.; Sasaki, T.; Nishimoto, T.; Kobayashi, H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 2002, 99, 745–750. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Ohtake, F.; Arai, N.; Kaiho, A.; Yasuda, S.; Tanaka, K.; Saeki, Y. In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 Axis contributes to k48-linked chain specificity of the proteasome. Mol. Cell 2017, 66, 488–502. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Goldberg, A.L. UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc. Natl. Acad. Sci. USA 2018, 115, E11642–E11650. [Google Scholar] [CrossRef] [PubMed]
- Munari, F.; Barracchia, C.G.; Franchin, C.; Parolini, F.; Capaldi, S.; Romeo, A.; Bubacco, L.; Assfalg, M.; Arrigoni, G.; D’Onofrio, M. Semisynthetic and enzyme-mediated conjugate preparations illuminate the ubiquitination-dependent aggregation of tau protein. angew. Chem. Int. Ed. Engl. 2020, 59, 6607–6611. [Google Scholar] [CrossRef]
- Morimoto, D.; Walinda, E.; Fukada, H.; Sou, Y.-S.; Kageyama, S.; Hoshino, M.; Fujii, T.; Tsuchiya, H.; Saeki, Y.; Arita, K.; et al. The unexpected role of polyubiquitin chains in the formation of fibrillar aggregates. Nat. Commun. 2015, 6, 6116. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, Y.; Hagai, T.; Levy, Y. Nonspecific yet decisive: Ubiquitination can affect the native-state dynamics of the modified protein. Protein Sci. 2015, 24, 1580–1592. [Google Scholar] [CrossRef]
- Baker, R.T.; Tobias, J.W.; Varshavsky, A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J. Biol. Chem. 1992, 267, 23364–23375. [Google Scholar]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef]
- Lee, M.J.; Lee, B.H.; Hanna, J.; King, R.W.; Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteom. 2011, 10, R110.003871. [Google Scholar] [CrossRef]
- Mao, P.; Smerdon, M.J. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J. Biol. Chem. 2010, 285, 37542–37550. [Google Scholar] [CrossRef]
- Kriegenburg, F.; Jakopec, V.; Poulsen, E.G.; Nielsen, S.V.; Roguev, A.; Krogan, N.; Gordon, C.; Fleig, U.; Hartmann-Petersen, R. A chaperone-assisted degradation pathway targets kinetochore proteins to ensure genome stability. PLoS Genet. 2014, 10, e1004140. [Google Scholar] [CrossRef]
- De Poot, S.A.H.; Tian, G.; Finley, D. Meddling with Fate: The proteasomal deubiquitinating enzymes. J. Mol. Biol. 2017, 429, 3525–3545. [Google Scholar] [CrossRef] [PubMed]
- Baxter, B.K.; Craig, E.A. Isolation of UBP3, encoding a de-ubiquitinating enzyme, as a multicopy suppressor of a heat-shock mutant strain of S. cerevisiae. Curr. Genet. 1998, 33, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Bukau, B. Role of sHsps in organizing cytosolic protein aggregation and disaggregation. Cell Stress Chaperones 2017, 22, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Yen, H.-C.S.; Xu, Q.; Chou, D.M.; Zhao, Z.; Elledge, S.J. Global protein stability profiling in mammalian cells. Science 2008, 322, 918–923. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reiss, Y.; Gur, E.; Ravid, T. Releasing the Lockdown: An Emerging Role for the Ubiquitin-Proteasome System in the Breakdown of Transient Protein Inclusions. Biomolecules 2020, 10, 1168. https://doi.org/10.3390/biom10081168
Reiss Y, Gur E, Ravid T. Releasing the Lockdown: An Emerging Role for the Ubiquitin-Proteasome System in the Breakdown of Transient Protein Inclusions. Biomolecules. 2020; 10(8):1168. https://doi.org/10.3390/biom10081168
Chicago/Turabian StyleReiss, Yuval, Elisheva Gur, and Tommer Ravid. 2020. "Releasing the Lockdown: An Emerging Role for the Ubiquitin-Proteasome System in the Breakdown of Transient Protein Inclusions" Biomolecules 10, no. 8: 1168. https://doi.org/10.3390/biom10081168
APA StyleReiss, Y., Gur, E., & Ravid, T. (2020). Releasing the Lockdown: An Emerging Role for the Ubiquitin-Proteasome System in the Breakdown of Transient Protein Inclusions. Biomolecules, 10(8), 1168. https://doi.org/10.3390/biom10081168





