An Isotopic Ratio Outlier Analysis Approach for Global Metabolomics of Biosynthetically Talented Actinomycetes
Abstract
1. Introduction
2. Results
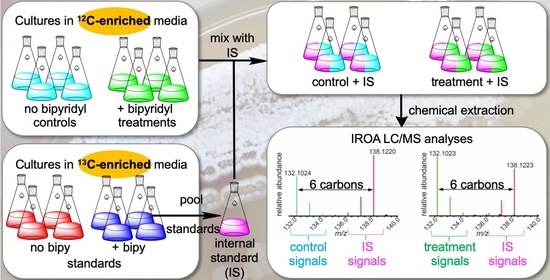
2.1. IROA Approach Enabling Detection of Metabolites Whose Production Is Initiated or Terminated in Response to Experimental Treatment
2.2. N. dassonvillei Metabolome Revealed by Untargeted IROA UHPLC/MS
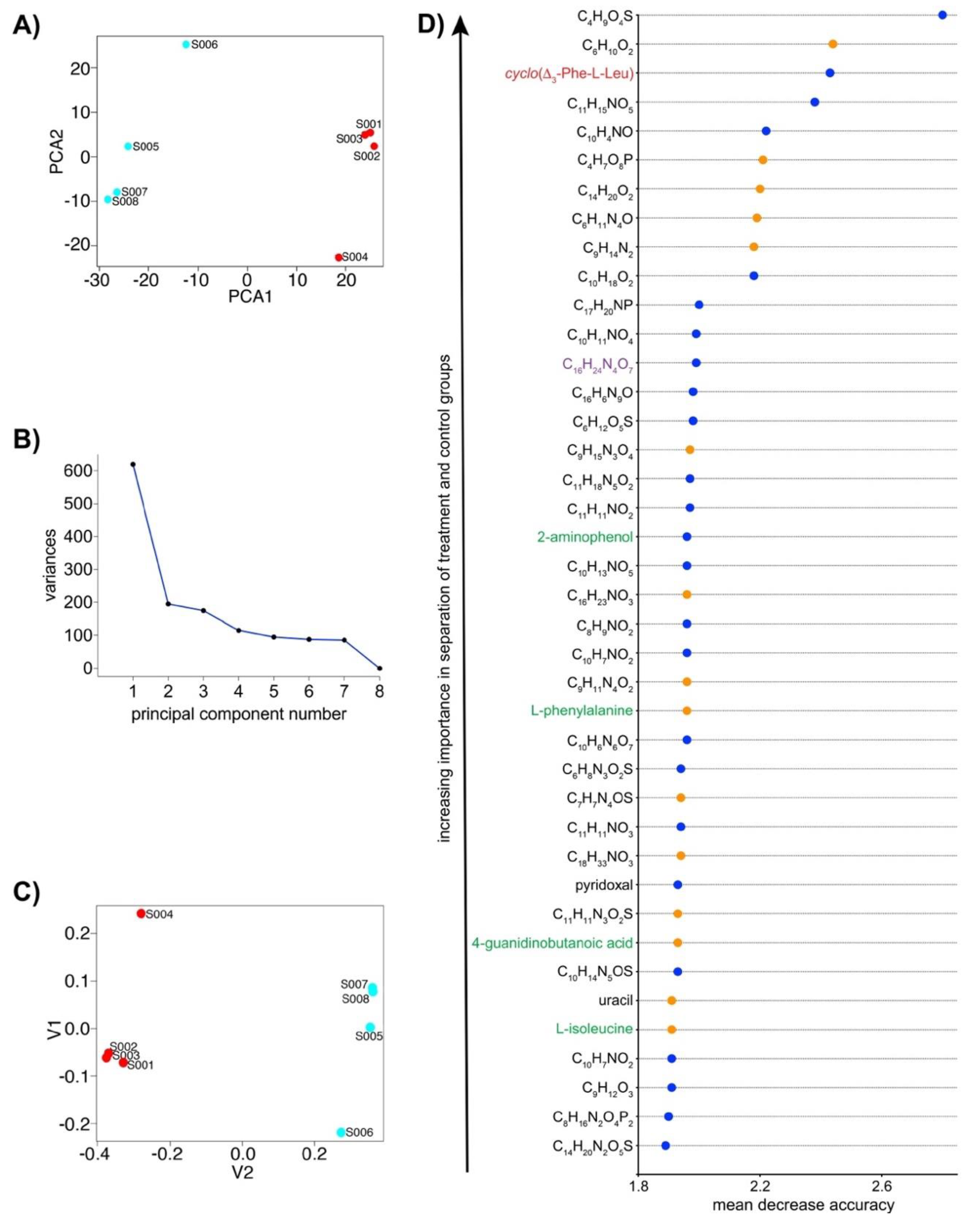
2.3. IROA LC/MS Differentiates Treatment and Control N. dassonvillei Metabolomes and Suggests the Production of Novel Metabolites
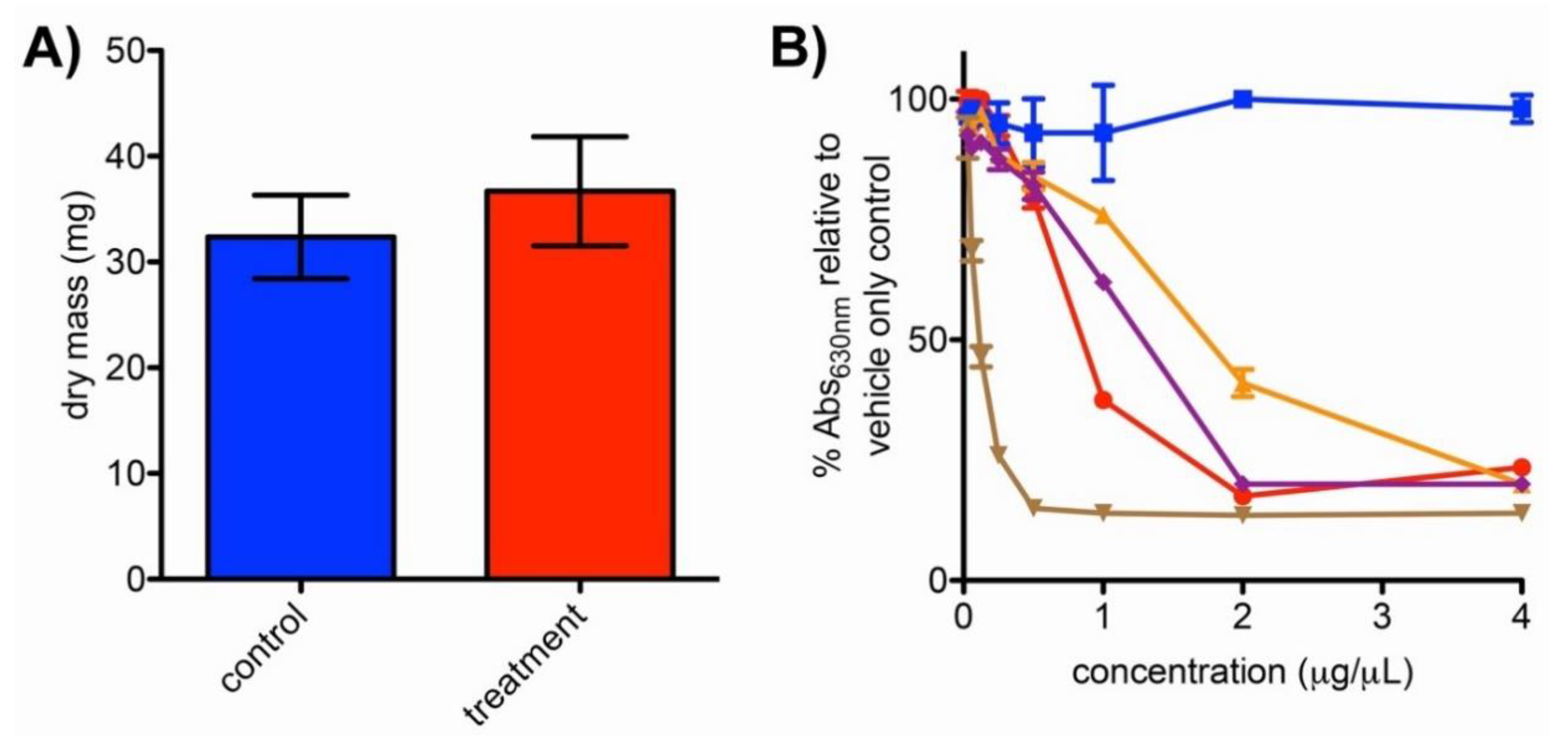
2.4. Impacts of Bipyridyl Treatment on N. dassonvillei Biomass and Siderophore Production
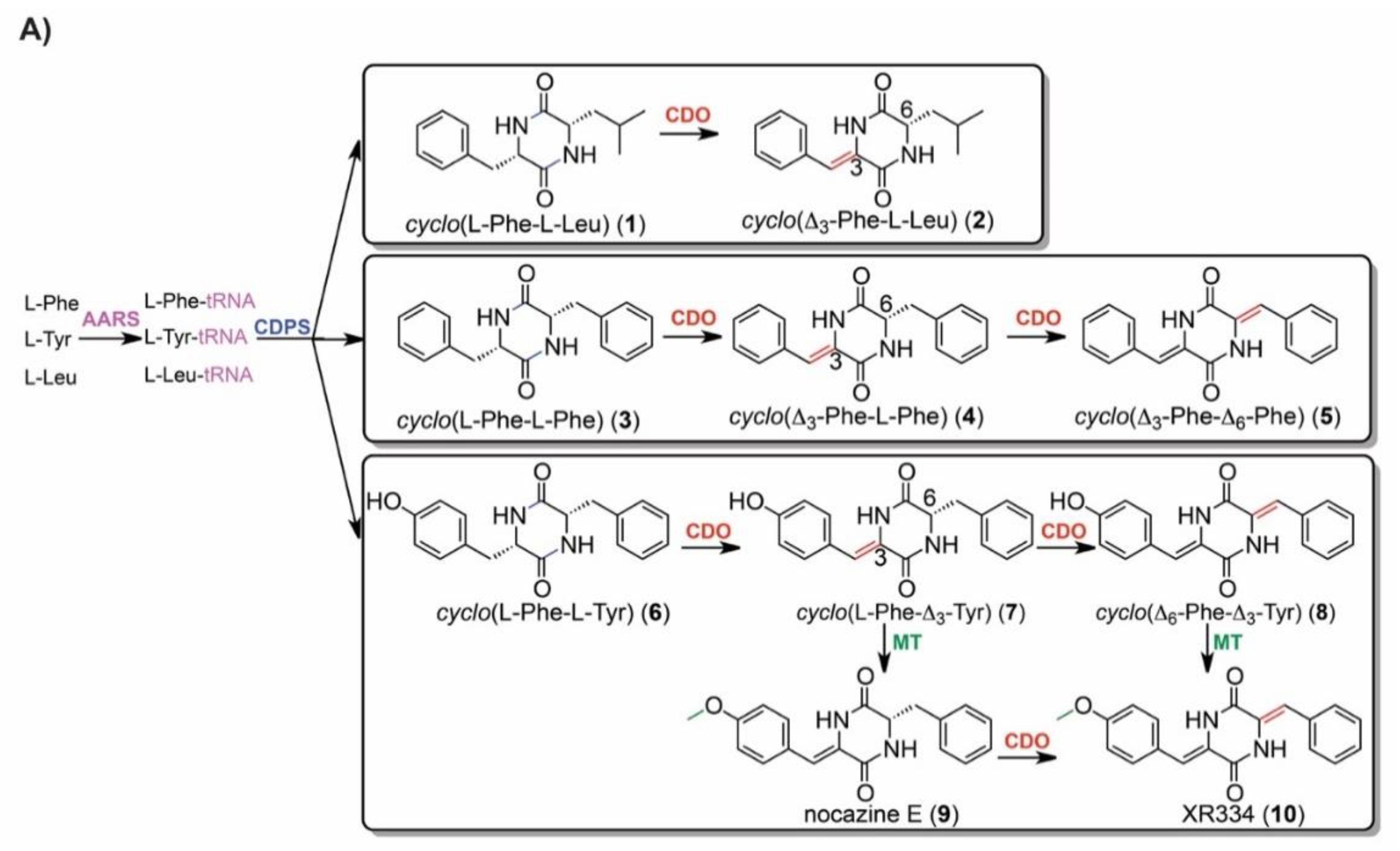
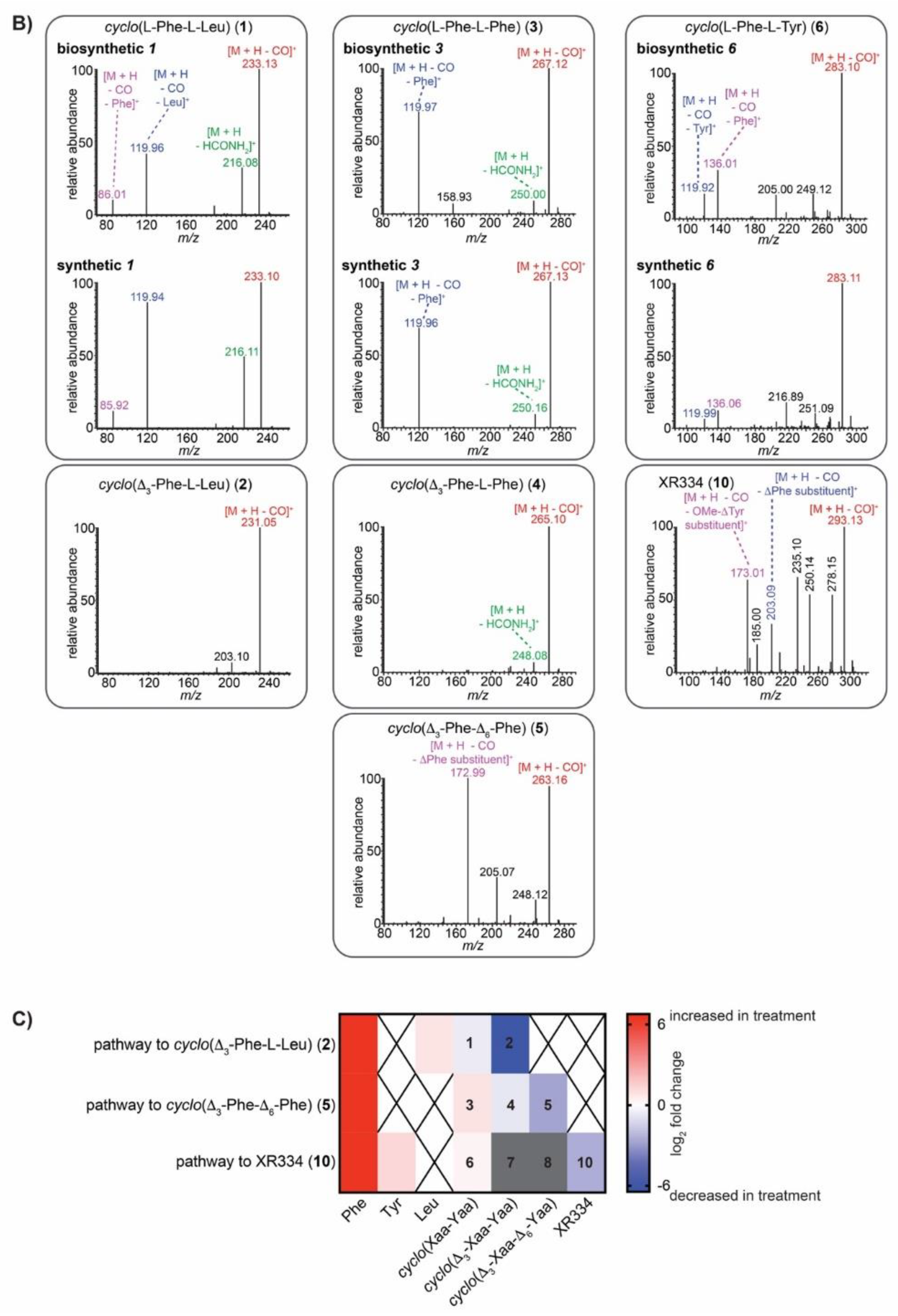
2.5. IROA Enables Rapid Assessment of the Perturbation of Targeted Metabolic Pathways
3. Discussion
4. Materials and Methods
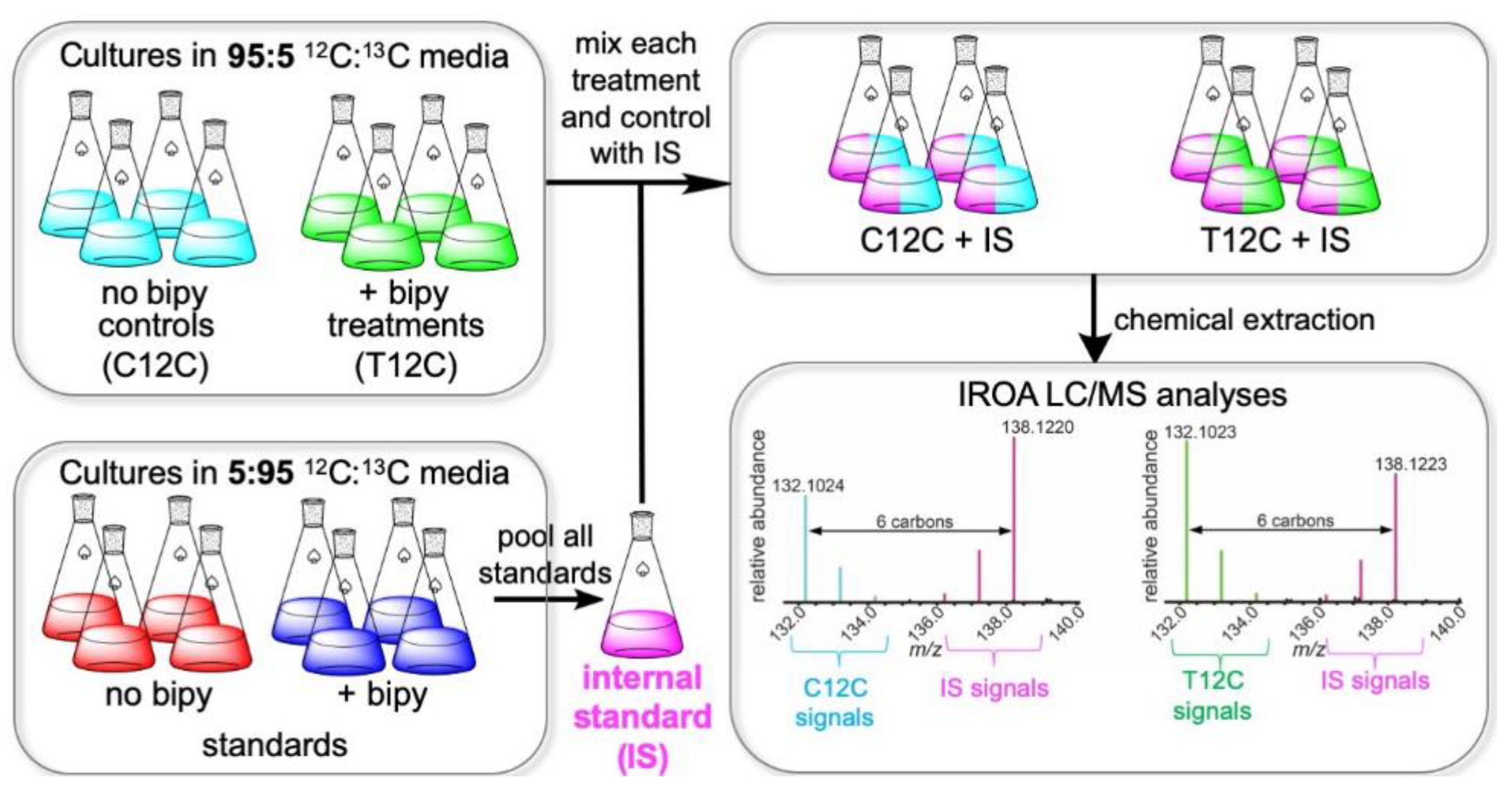
4.1. Nocardiopsis dassonvillei Cultures and Fermentation for IROA Experiments
4.2. Chemical Extraction and Preparation of Samples for UHPLC/MS Metabolomics
4.3. UHPLC/MS Analysis
4.4. Data Processing and Analyses
4.5. Evaluation of Biomass and Siderophore Production
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Berdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Hale, C.R.; Nilewski, C.; Ioannidou, H.A. Constructing molecular complexity and diversity: Total synthesis of natural products of biological and medicinal importance. Chem. Soc. Rev. 2012, 41, 5185–5238. [Google Scholar] [CrossRef] [PubMed]
- Behie, S.W.; Bonet, B.; Zacharia, V.M.; McClung, D.J.; Traxler, M.F. Molecules to Ecosystems: Actinomycete Natural Products in situ. Front. Microbiol. 2017, 7, 2149. [Google Scholar] [CrossRef]
- Carlson, E.E. Natural Products as Chemical Probes. ACS Chem. Biol. 2010, 5, 639–653. [Google Scholar] [CrossRef]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef]
- Nguyen, Q.T.; Merlo, M.E.; Medema, M.H.; Jankevics, A.; Breitling, R.; Takano, E. Metabolomics methods for the synthetic biology of secondary metabolism. FEBS Lett. 2012, 586, 2177–2183. [Google Scholar] [CrossRef]
- Van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Bilyk, O.; Luzhetskyy, A. Metabolic engineering of natural product biosynthesis in actinobacteria. Curr. Opin. Biotechnol. 2016, 42, 98–107. [Google Scholar] [CrossRef]
- Gaudencio, S.P.; Pereira, F. Dereplication: Racing to speed up the natural products discovery process. Nat. Prod. Rep. 2015, 32, 779–810. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Grkovic, T.; Balasubramanian, S.; Kamel, M.S.; Quinn, R.J.; Hentschel, U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015, 33, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a Cryptic Antifungal Compound from Streptomyces albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef] [PubMed]
- Adnani, N.; Chevrette, M.G.; Adibhatla, S.N.; Zhang, F.; Yu, Q.; Braun, D.R.; Nelson, J.; Simpkins, S.W.; McDonald, B.R.; Myers, C.L.; et al. Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Derewacz, D.K.; Covington, B.C.; McLean, J.A.; Bachmann, B.O. Mapping Microbial Response Metabolomes for Induced Natural Product Discovery. ACS Chem Biol 2015, 10, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Adnani, N.; Vazquez-Rivera, E.; Adibhatla, S.N.; Ellis, G.A.; Braun, D.R.; Bugni, T.S. Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach. Mar. Drugs 2015, 13, 6082–6098. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Deng, Z.; Liu, T. Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab. Eng. 2018, 50, 74–84. [Google Scholar] [CrossRef]
- Wu, C.; Choi, Y.H.; van Wezel, G.P. Metabolic profiling as a tool for prioritizing antimicrobial compounds. J. Ind. Microbiol. Biotechnol. 2015, 43, 299–312. [Google Scholar] [CrossRef]
- Aretz, I.; Meierhofer, D. Advantages and Pitfalls of Mass Spectrometry Based Metabolome Profiling in Systems Biology. Int. J. Mol. Sci. 2016, 17, 632. [Google Scholar] [CrossRef]
- Covington, B.C.; McLean, J.A.; Bachmann, B.O. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 2017, 34, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Krug, D.; Müller, R. Secondary metabolomics: The impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Prod. Rep. 2014, 31, 768–783. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Nuzillard, J.-M.; van der Hooft, J.J.J.; Renault, J.-H.; Bertrand, S. Accelerating Metabolite Identification in Natural Product Research: Toward an Ideal Combination of Liquid Chromatography—High- Resolution Tandem Mass Spectrometry and NMR Profiling, in Silico Databases, and Chemometrics. Anal. Chem. 2018, 91, 704–742. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies Interactions Stimulate Diversification of the Streptomyces coelicolor Secreted Metabolome. MBio 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Xu, Y.; Kersten, R.D.; Liu, W.-T.; Meehan, M.J.; Moore, B.S.; Bandeira, N.; Dorrestein, P.C. Connecting Chemotypes and Phenotypes of Cultured Marine Microbial Assemblages by Imaging Mass Spectrometry. Angew. Chem. Int. Ed. Engl. 2011, 50, 5839–5842. [Google Scholar] [CrossRef]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012, 84, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Chanana, S.; Thomas, C.; Braun, D.; Hou, Y.; Wyche, T.; Bugni, T. Natural Product Discovery Using Planes of Principal Component Analysis in R (PoPCAR). Metabolites 2017, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Wu, C.H.; Moree, W.J.; Lamsa, A.; Medema, M.H.; Zhao, X.; Gavilan, R.G.; Aparicio, M.; Atencio, L.; Jackson, C.; et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA 2013, 110, E2611–E2620. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- De Jong, F.A.; Beecher, C. Addressing the current bottlenecks of metabolomics: Isotopic Ratio Outlier Analysis™, an isotopic-labeling technique for accurate biochemical profiling. Bioanalysis 2012, 4, 2303–2314. [Google Scholar] [CrossRef]
- Clendinen, C.S.; Stupp, G.S.; Wang, B.; Garrett, T.J.; Edison, A.S. 13C Metabolomics: NMR and IROA for Unknown Identification. Curr. Metab. 2016, 4, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Stupp, G.S.; Clendinen, C.S.; Ajredini, R.; Szewc, M.A.; Garrett, T.J.; Menger, R.F.; Yost, R.A.; Beecher, C.; Edison, A.S. Isotopic Ratio Outlier Analysis Global Metabolomics of Caenorhabditis elegans. Anal. Chem. 2013, 85, 11858–11865. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Moir, R.; Willis, I.; Beecher, C.; Tsai, Y.-H.; Garrett, T.J.; Yost, R.A.; Kurland, I.J. Isotopic Ratio Outlier Analysis of the S. cerevisiae Metabolome Using Accurate Mass Gas Chromatography/Time-of-Flight Mass Spectrometry: A New Method for Discovery. Anal. Chem. 2016, 88, 2747–2754. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Moir, R.; Willis, I.; Seethapathy, S.; Biniakewitz, R.; Kurland, I. Enhanced Isotopic Ratio Outlier Analysis (IROA) Peak Detection and Identification with Ultra-High Resolution GC-Orbitrap/MS: Potential Application for Investigation of Model Organism Metabolomes. Metabolites 2018, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Vinayavekhin, N.; Saghatelian, A. Regulation of Alkyl-dihydrothiazole-carboxylates (ATCs) by Iron and the Pyochelin Gene Cluster in Pseudomonas aeruginosa. ACS Chem. Biol. 2009, 4, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Lapidus, A.; Nolan, M.; Lucas, S.; Del Rio, T.G.; Tice, H.; Cheng, J.-F.; Tapia, R.; Han, C.; Goodwin, L.; et al. Complete genome sequence of Nocardiopsis dassonvillei type strain (IMRU 509T). Stand. Genom. Sci. 2010, 3, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Li, H.W.; Zhi, X.Y.; Yao, J.C.; Zhou, Y.; Tang, S.K.; Klenk, H.P.; Zhao, J.; Li, W.J. Comparative genomic analysis of the genus Nocardiopsis provides new insights into its genetic mechanisms of environmental adaptability. PLoS ONE 2013, 8, e61528. [Google Scholar] [CrossRef] [PubMed]
- Giessen, T.W.; von Tesmar, A.M.; Marahiel, M.A. Insights into the generation of structural diversity in a tRNA-dependent pathway for highly modified bioactive cyclic dipeptides. Chem. Biol. 2013, 20, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Kam Ho, T. The Random Subspace Method for Constructing Decision Forests. IEEE Trans. Pattern Anal. 1998, 20, 832–844. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1986, 160, 47–56. [Google Scholar] [CrossRef]
- Guo, Y.-C.; Cao, S.-X.; Zong, X.-K.; Liao, X.-C.; Zhao, Y.-F. ESI-MSn study on the fragmentation of protonated cyclic-dipeptides. Spectrosc. Int. J. 2009, 23, 131–139. [Google Scholar] [CrossRef]
- Klementz, D.; Doring, K.; Lucas, X.; Telukunta, K.K.; Erxleben, A.; Deubel, D.; Erber, A.; Santillana, I.; Thomas, O.S.; Bechthold, A.; et al. StreptomeDB 2.0—An extended resource of natural products produced by streptomycetes. Nucleic Acids Res. 2016, 44, D509–D514. [Google Scholar] [CrossRef] [PubMed]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Barona-Gomez, F.; Wong, U.; Giannakopulos, A.E.; Derrick, P.J.; Challis, G.L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 2004, 126, 16282–16283. [Google Scholar] [CrossRef] [PubMed]
| ESI Mode | |||
|---|---|---|---|
| Positive | Negative | Total | |
| IROA peak pairs detected | 1243 | 89 | 1332 |
| Identified metabolites | 85 | 22 | 107 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carey, J.; Nguyen, T.; Korchak, J.; Beecher, C.; de Jong, F.; Lane, A.L. An Isotopic Ratio Outlier Analysis Approach for Global Metabolomics of Biosynthetically Talented Actinomycetes. Metabolites 2019, 9, 181. https://doi.org/10.3390/metabo9090181
Carey J, Nguyen T, Korchak J, Beecher C, de Jong F, Lane AL. An Isotopic Ratio Outlier Analysis Approach for Global Metabolomics of Biosynthetically Talented Actinomycetes. Metabolites. 2019; 9(9):181. https://doi.org/10.3390/metabo9090181
Chicago/Turabian StyleCarey, Jordan, Thanh Nguyen, Jennifer Korchak, Christopher Beecher, Felice de Jong, and Amy L. Lane. 2019. "An Isotopic Ratio Outlier Analysis Approach for Global Metabolomics of Biosynthetically Talented Actinomycetes" Metabolites 9, no. 9: 181. https://doi.org/10.3390/metabo9090181
APA StyleCarey, J., Nguyen, T., Korchak, J., Beecher, C., de Jong, F., & Lane, A. L. (2019). An Isotopic Ratio Outlier Analysis Approach for Global Metabolomics of Biosynthetically Talented Actinomycetes. Metabolites, 9(9), 181. https://doi.org/10.3390/metabo9090181