A Noninvasive Comparison Study between Human Gliomas with IDH1 and IDH2 Mutations by MR Spectroscopy
Abstract
1. Introduction
2. Results
2.1. Study Cohort
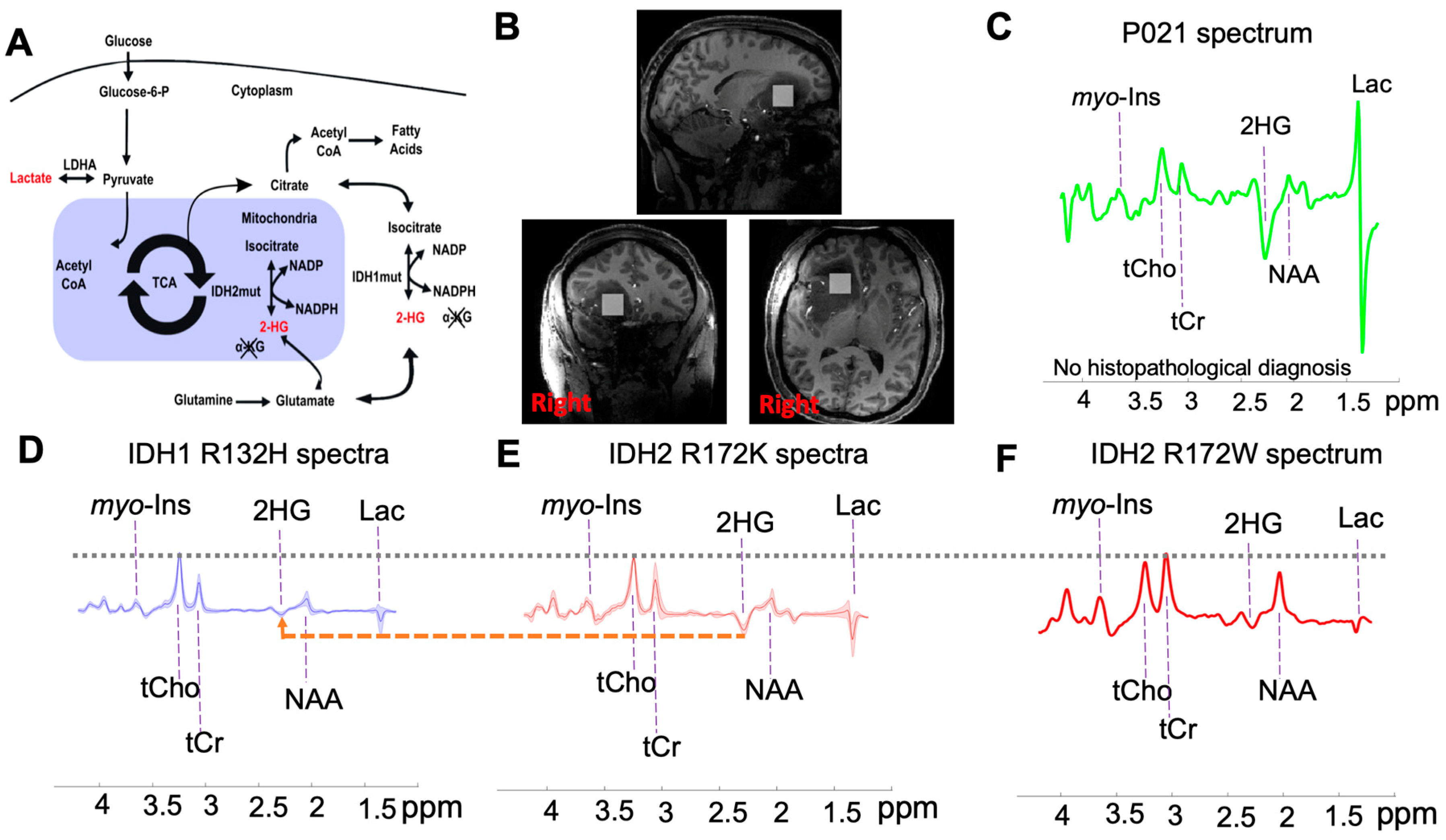
2.2. Subtyping IDH Mutant Gliomas
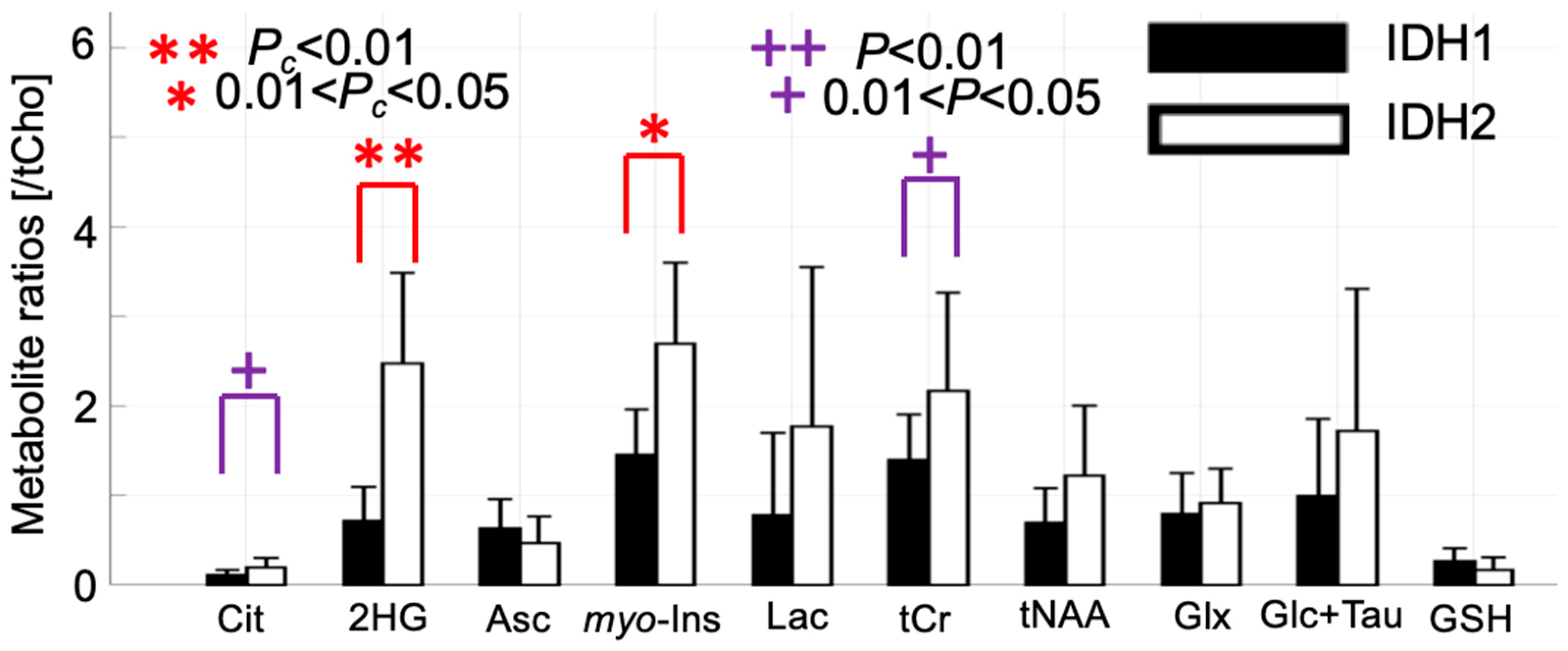
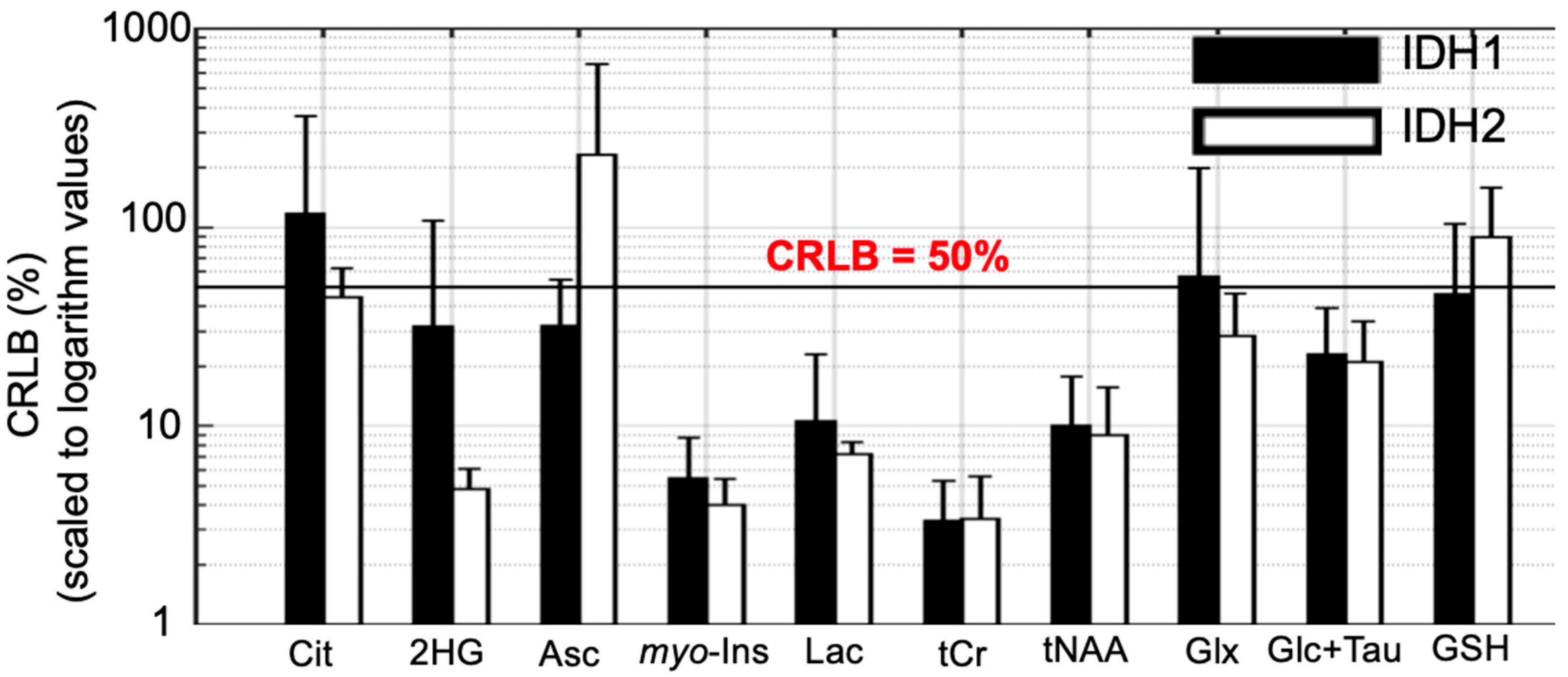
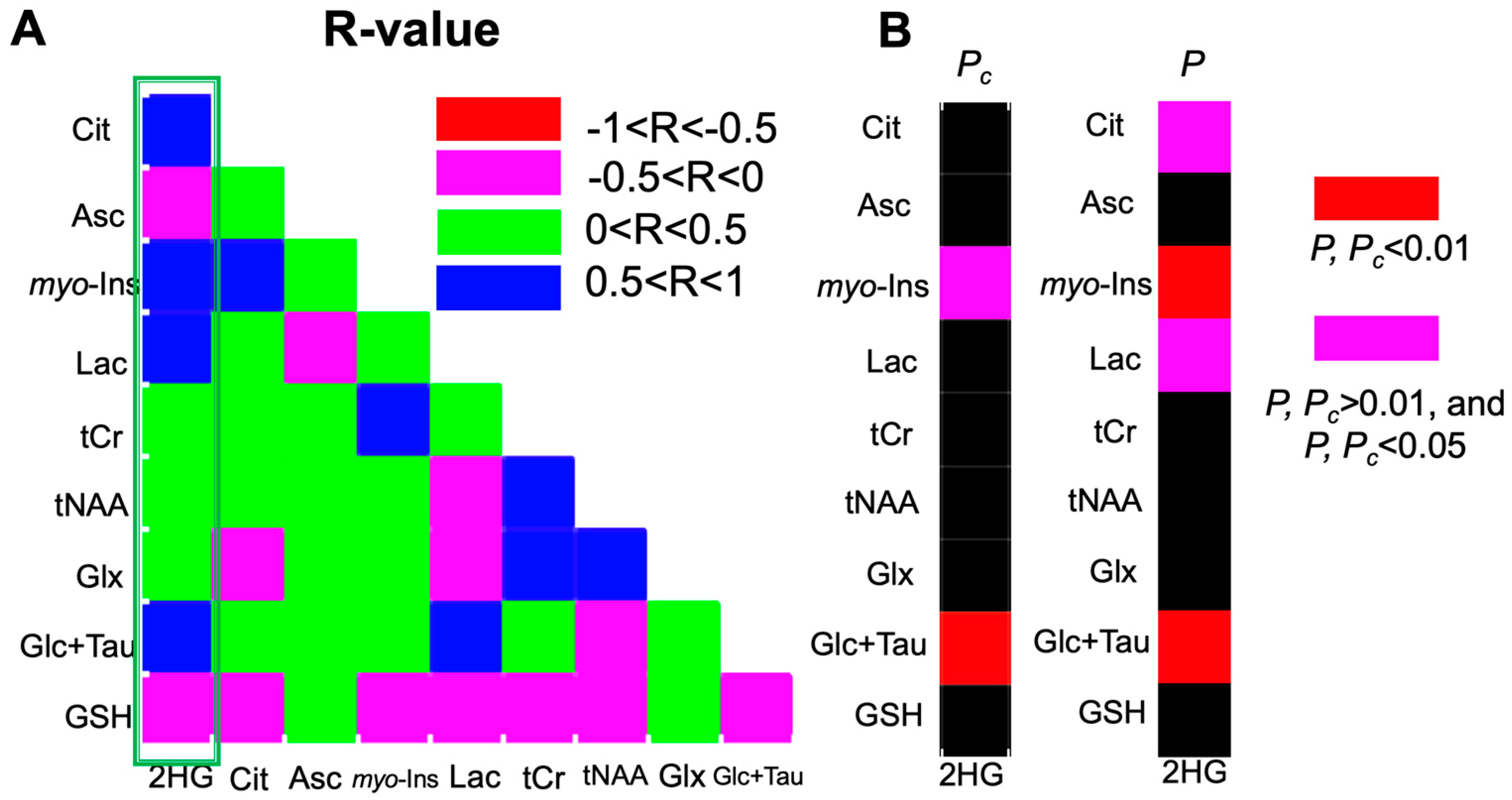
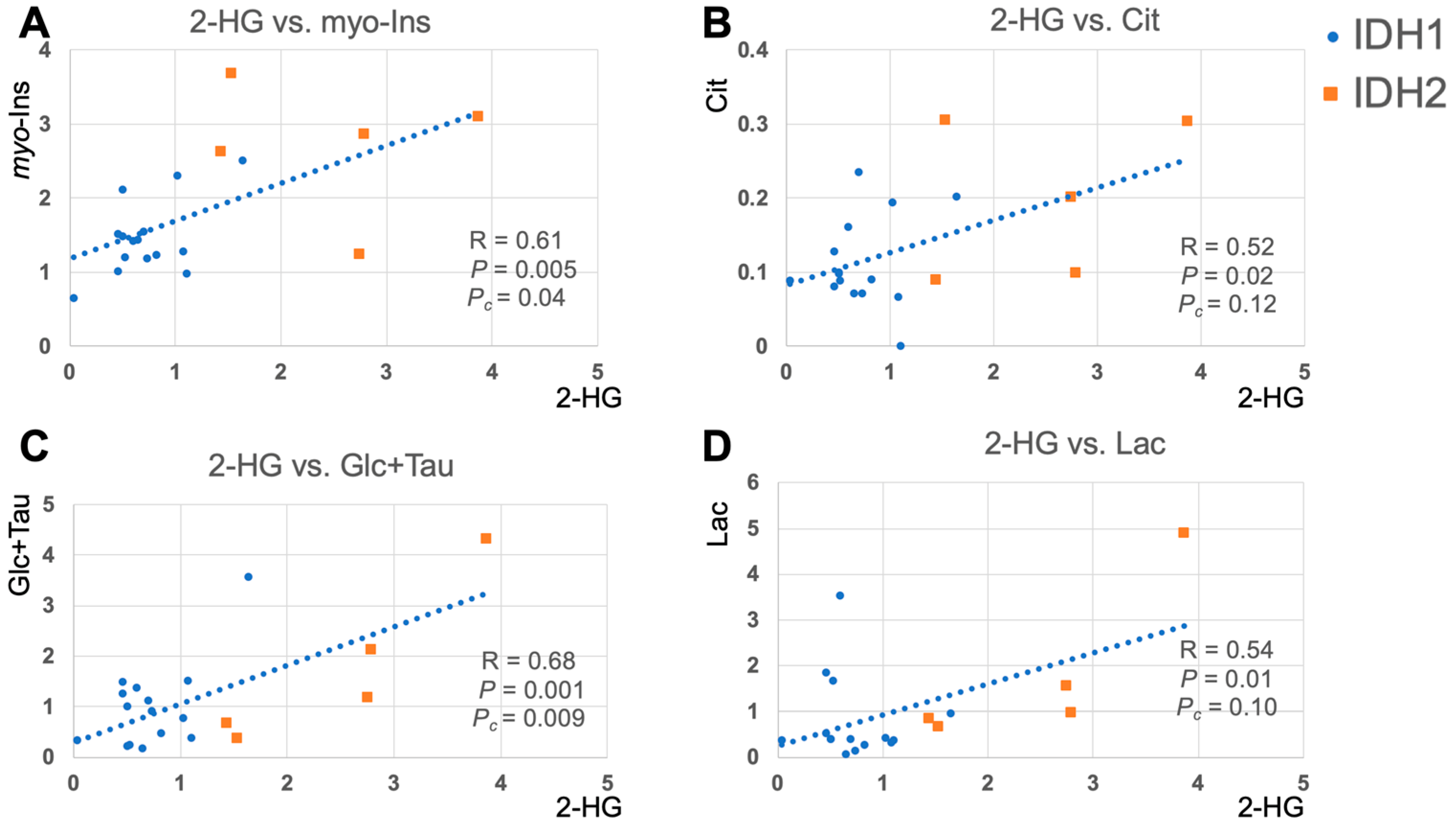
2.3. 2-HG and Associated Metabolites Concentrations
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Data Acquisition
4.3. Simulations
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta. Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Krell, D.; Mulholland, P.; Frampton, A.E.; Krell, J.; Stebbing, J.; Bardella, C. IDH mutations in tumorigenesis and their potential role as novel therapeutic targets. Future. Oncol. 2013, 9, 1923–1935. [Google Scholar] [CrossRef] [PubMed]
- De Carli, E.; Wang, X.; Puget, S. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 2248–2249. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ye, D.; Guan, K.L.; Xiong, Y. IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin. Cancer Res. 2012, 18, 5562–5571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Moore, L.M.; Li, X.; Yung, W.K.; Zhang, W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro. Oncol. 2013, 15, 1114–1126. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.; Wu, Y.Z.; Radmacher, M.D.; Mrozek, K.; Margeson, D.; Holland, K.B.; Whitman, S.P.; Becker, H.; Schwind, S.; et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J. Clin. Oncol. 2010, 28, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Boissel, N.; Nibourel, O.; Renneville, A.; Huchette, P.; Dombret, H.; Preudhomme, C. Differential prognosis impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood 2011, 117, 3696–3697. [Google Scholar] [CrossRef]
- Green, C.L.; Evans, C.M.; Zhao, L.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011, 118, 409–412. [Google Scholar] [CrossRef]
- Kim, M.M.; Parolia, A.; Dunphy, M.P.; Venneti, S. Non-invasive metabolic imaging of brain tumours in the era of precision medicine. Nat. Rev. Clin. Oncol. 2016, 13, 725–739. [Google Scholar] [CrossRef]
- Camelo-Piragua, S.; Jansen, M.; Ganguly, A.; Kim, J.C.; Cosper, A.K.; Dias-Santagata, D.; Nutt, C.L.; Iafrate, A.J.; Louis, D.N. A sensitive and specific diagnostic panel to distinguish diffuse astrocytoma from astrocytosis: Chromosome 7 gain with mutant isocitrate dehydrogenase 1 and p53. J. Neuropathol. Exp. Neurol. 2011, 70, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, R.J. Impending Impact of Molecular Pathology on Classifying Adult Diffuse Gliomas. Cancer Control 2015, 22, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Ganji, S.K.; DeBerardinis, R.J.; Hatanpaa, K.J.; Rakheja, D.; Kovacs, Z.; Yang, X.L.; Mashimo, T.; Raisanen, J.M.; Marin-Valencia, I.; et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 2012, 18, 624–629. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Kim, G.S.; Gerstner, E.; Batchelor, T.; Tzika, A.A.; Fantin, V.R.; Vander Heiden, M.G.; Sorensen, A.G. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med. 2012, 4, 116ra4. [Google Scholar] [CrossRef]
- Pope, W.B.; Prins, R.M.; Albert Thomas, M.; Nagarajan, R.; Yen, K.E.; Bittinger, M.A.; Salamon, N.; Chou, A.P.; Yong, W.H.; Soto, H.; et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J. Neurooncol. 2012, 107, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Berrington, A.; Voets, N.L.; Plaha, P.; Larkin, S.J.; McCullagh, J.; Stacey, R.; Yildirim, M.; Schofield, C.J.; Jezzard, P.; Cadoux-Hudson, T.; et al. Improved localisation for 2-hydroxyglutarate detection at 3T using long-TE semi-LASER. Tomography 2016, 2, 94–105. [Google Scholar]
- Andronesi, O.C.; Loebel, F.; Bogner, W.; Marjanska, M.; Vander Heiden, M.G.; Iafrate, A.J.; Dietrich, J.; Batchelor, T.T.; Gerstner, E.R.; Kaelin, W.G.; et al. Treatment Response Assessment in IDH-Mutant Glioma Patients by Noninvasive 3D Functional Spectroscopic Mapping of 2-Hydroxyglutarate. Clin. Cancer Res. 2016, 22, 1632–1641. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Arrillaga-Romany, I.C.; Ly, K.I.; Bogner, W.; Ratai, E.M.; Reitz, K.; Iafrate, A.J.; Dietrich, J.; Gerstner, E.R.; Chi, A.S.; et al. Pharmacodynamics of mutant-IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2-hydroxyglutarate. Nat. Commun. 2018, 9, 1474. [Google Scholar] [CrossRef]
- Emir, U.E.; Larkin, S.J.; de Pennington, N.; Voets, N.; Plaha, P.; Stacey, R.; Al-Qahtani, K.; McCullagh, J.; Schofield, C.J.; Clare, S.; et al. Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations. Cancer Res. 2016, 76, 43–49. [Google Scholar] [CrossRef]
- Parker, S.J.; Metallo, C.M. Metabolic consequences of oncogenic IDH mutations. Pharmacol. Ther. 2015, 152, 54–62. [Google Scholar] [CrossRef]
- Olivares, O.; Dabritz, J.H.M.; King, A.; Gottlieb, E.; Halsey, C. Research into cancer metabolomics: Towards a clinical metamorphosis. Semin. Cell Dev. Biol. 2015, 43, 52–64. [Google Scholar] [CrossRef]
- Amatangelo, M.D.; Quek, L.; Shih, A.; Stein, E.M.; Roshal, M.; David, M.D.; Marteyn, B.; Farnoud, N.R.; de Botton, S.; Bernard, O.A.; et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 2017, 130, 732–741. [Google Scholar] [CrossRef]
- Julia-Sape, M.; Candiota, A.P.; Arus, C. Cancer metabolism in a snapshot: MRS(I). NMR Biomed. 2019, e4054. [Google Scholar] [CrossRef]
- Ward, P.S.; Lu, C.; Cross, J.R.; Abdel-Wahab, O.; Levine, R.L.; Schwartz, G.K.; Thompson, C.B. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J. Biol. Chem. 2013, 288, 3804–3815. [Google Scholar] [CrossRef]
- Jin, G.; Reitman, Z.J.; Spasojevic, I.; Batinic-Haberle, I.; Yang, J.; Schmidt-Kittler, O.; Bigner, D.D.; Yan, H. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS ONE 2011, 6, e16812. [Google Scholar] [CrossRef]
- Reitman, Z.J.; Jin, G.; Karoly, E.D.; Spasojevic, I.; Yang, J.; Kinzler, K.W.; He, Y.; Bigner, D.D.; Vogelstein, B.; Yan, H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA 2011, 108, 3270–3275. [Google Scholar] [CrossRef]
- Nagashima, H.; Sasayama, T.; Tanaka, K.; Kyotani, K.; Sato, N.; Maeyama, M.; Kohta, M.; Sakata, J.; Yamamoto, Y.; Hosoda, K.; et al. Myo-inositol concentration in MR spectroscopy for differentiating high grade glioma from primary central nervous system lymphoma. J. Neurooncol. 2018, 136, 317–326. [Google Scholar] [CrossRef]
- Wang, C.; McKeithan, T.W.; Gong, Q.; Zhang, W.; Bouska, A.; Rosenwald, A.; Gascoyne, R.D.; Wu, X.; Wang, J.; Muhammad, Z.; et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood 2015, 126, 1741–1752. [Google Scholar] [CrossRef]
- Wang, H.Y.; Tang, K.; Liang, T.Y.; Zhang, W.Z.; Li, J.Y.; Wang, W.; Hu, H.M.; Li, M.Y.; Wang, H.Q.; He, X.Z.; et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J. Exp. Clin. Cancer Res. 2016, 35, 86. [Google Scholar] [CrossRef]
- Jalbert, L.E.; Elkhaled, A.; Phillips, J.J.; Neill, E.; Williams, A.; Crane, J.C.; Olson, M.P.; Molinaro, A.M.; Berger, M.S.; Kurhanewicz, J.; et al. Metabolic Profiling of IDH Mutation and Malignant Progression in Infiltrating Glioma. Sci. Rep. 2017, 7, 44792. [Google Scholar] [CrossRef]
- Bisdas, S.; Chadzynski, G.L.; Braun, C.; Schittenhelm, J.; Skardelly, M.; Hagberg, G.E.; Ethofer, T.; Pohmann, R.; Shajan, G.; Engelmann, J.; et al. MR spectroscopy for in vivo assessment of the oncometabolite 2-hydroxyglutarate and its effects on cellular metabolism in human brain gliomas at 9.4T. J. Magn. Reson. Imaging 2016, 44, 823–833. [Google Scholar] [CrossRef]
- Gruetter, R.; Tkac, I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn. Reson. Med. 2000, 43, 319–323. [Google Scholar] [CrossRef]
- Teeuwisse, W.M.; Brink, W.M.; Haines, K.N.; Webb, A.G. Simulations of high permittivity materials for 7 T neuroimaging and evaluation of a new barium titanate-based dielectric. Magn. Reson. Med. 2012, 67, 912–918. [Google Scholar] [CrossRef]
- van de Bank, B.L.; Emir, U.E.; Boer, V.O.; van Asten, J.J.; Maas, M.C.; Wijnen, J.P.; Kan, H.E.; Oz, G.; Klomp, D.W.; Scheenen, T.W. Multi-center reproducibility of neurochemical profiles in the human brain at 7 T. NMR Biomed. 2015, 28, 306–316. [Google Scholar] [CrossRef]
- Kreis, R. The trouble with quality filtering based on relative Cramer-Rao lower bounds. Magn. Reson. Med. 2016, 75, 15–18. [Google Scholar] [CrossRef]
| Subject ID | Age, Gender | Prior Oncological Treatment | Diagnosis | IHC | DNA Sequencing | 2HG/tCho | 2HG CRLB |
|---|---|---|---|---|---|---|---|
| P001 | 29, M | None | Astrocytoma (WHO grade 2) | +ve | N/A | 0.60 | 20 |
| P002 | 30, M | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 0.70 | 8 |
| P003 | 35, F | Radiotherapy | Anaplastic oligoastrocytoma (WHO grade 3) | +ve | N/A | 0.46 | 30 |
| P004 | 34, M | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 1.64 | 7 |
| P005 | 47, M | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 0.46 | 20 |
| P006 | 57, F | None | Astrocytoma (WHO grade 2) | +ve | N/A | 0.73 | 9 |
| P007 | 51, M | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 0.51 | 9 |
| P008 | 48, M | None | Anaplastic oligodendroglioma (WHO grade 3) | +ve | N/A | 0.52 | 21 |
| P009 | 53, M | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 0.82 | 6 |
| P010 | 45, M | None | Oligodendroglioma (WHO grade 2) | +ve | N/A | 0.65 | 12 |
| P011 | 44, F | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 0.50 | 13 |
| P012 | 20, M | None | Astrocytoma (WHO grade 2) | +ve | N/A | 0.04 | 306 |
| P013 | 36, M | None | Anaplastic astrocytoma (WHO grade 3) | +ve | N/A | 1.10 | 4 |
| P014 | 51, M | None | Oligodendroglioma (WHO grade 2) | +ve | N/A | 1.08 | 5 |
| P015 | 36, M | None | Astrocytoma (WHO grade 2) | +ve | N/A | 1.02 | 7 |
| P016 | 27, F | None | Oligodendroglioma (WHO grade 2) | -ve | IDH2 R172K | 2.79 | 4 |
| P017 | 29, F | None | Oligodendroglioma (WHO grade 2) | -ve | IDH2 R172K | 2.76 | 3 |
| P018 | 32, F | Chemotherapy | Oligodendroglioma (WHO grade 2) | -ve | IDH2 R172K | 3.87 | 5 |
| P019 | 19, F | None | Oligodendroglioma (WHO grade 2) | -ve | IDH2 R172W | 1.54 | 6 |
| P020 | 26, M | None | Anaplastic astrocytoma (WHO grade 3) | -ve | IDH2 R172K | 1.44 | 6 |
| P021 | 29, F | None | N/A | N/A | N/A | 12.82 | 3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Voets, N.L.; Larkin, S.J.; de Pennington, N.; Plaha, P.; Stacey, R.; McCullagh, J.S.O.; Schofield, C.J.; Clare, S.; Jezzard, P.; et al. A Noninvasive Comparison Study between Human Gliomas with IDH1 and IDH2 Mutations by MR Spectroscopy. Metabolites 2019, 9, 35. https://doi.org/10.3390/metabo9020035
Shen X, Voets NL, Larkin SJ, de Pennington N, Plaha P, Stacey R, McCullagh JSO, Schofield CJ, Clare S, Jezzard P, et al. A Noninvasive Comparison Study between Human Gliomas with IDH1 and IDH2 Mutations by MR Spectroscopy. Metabolites. 2019; 9(2):35. https://doi.org/10.3390/metabo9020035
Chicago/Turabian StyleShen, Xin, Natalie L. Voets, Sarah J. Larkin, Nick de Pennington, Puneet Plaha, Richard Stacey, James S. O. McCullagh, Christopher J. Schofield, Stuart Clare, Peter Jezzard, and et al. 2019. "A Noninvasive Comparison Study between Human Gliomas with IDH1 and IDH2 Mutations by MR Spectroscopy" Metabolites 9, no. 2: 35. https://doi.org/10.3390/metabo9020035
APA StyleShen, X., Voets, N. L., Larkin, S. J., de Pennington, N., Plaha, P., Stacey, R., McCullagh, J. S. O., Schofield, C. J., Clare, S., Jezzard, P., Cadoux-Hudson, T., Ansorge, O., & Emir, U. E. (2019). A Noninvasive Comparison Study between Human Gliomas with IDH1 and IDH2 Mutations by MR Spectroscopy. Metabolites, 9(2), 35. https://doi.org/10.3390/metabo9020035







